Abstract
Autophagy, a conserved self-eating process for the bulk degradation of cytoplasmic materials, involves double-membrane autophagosomes formed when an isolation membrane emerges and their direct fusion with lysosomes for degradation. For the early biogenesis of autophagosomes and their later degradation in lysosomes, membrane fusion is necessary, although different sets of genes and autophagy-related proteins involved in distinct fusion steps have been reported. To clarify the molecular mechanism of membrane fusion in autophagy, to not only expand current knowledge of autophagy, but also benefit human health, this review discusses key findings that elucidate the unique membrane dynamics of autophagy.
Keywords: autophagy, membrane fusion, SNARE
1. Introduction
The term autophagy, from the Greek auto, meaning “self,” and phagy, meaning “eat” [1], refers to a conserved self-eating process initiated by the emergence of a cup-shaped, single membrane-bound, pre-autophagosomal structure that elongates and expands during the process. Gradually, spherical, double membrane-bound structures called autophagosomes form amid cytoplasmic surroundings and mature by docking and fusing with endosomes or lysosomes, if not both [2]. The outer membrane of a completed autophagosome fuses with the vacuole, and the inner membrane vesicles and cargo are released into the vacuolar lumen for degradation [3].
Autophagy is an evolutionarily conserved process in eukaryotes that regulates numerous physiological and pathological processes [4]. Research has shown that autophagy not only serves as a cellular mechanism of waste disposal, but is also an adaptive response that provides nutrients and energy upon exposure to various stressors [5]. In that light, the phenomenon itself explains to some extent why cells would digest their own components [6]. However, autophagic dysfunction has also been implicated in cellular quality control, responses to stress, development, lifespan, and a range of infectious and other diseases in humans, including cancer, neurodegenerative diseases, and diabetes [7–9].
Autophagosome membranes have been proposed to originate from several sources, including the endoplasmic reticulum (ER), Golgi apparatus, mitochondria, and the plasma membrane [10, 11]. The fusion process of membranes is required for the closure of isolation membranes into autophagosomes (i.e., early biogenesis of autophagosomes) and the maturation of autophagosomes via fusion with endosomes or lysosomes to form autolysosomes (i.e., late degradation in lysosomes) [12]. Although many proteins involved in membrane fusion have been identified, key regulatory mechanisms of autophagic membrane tethering and fusion remain ambiguous.
2. Membrane fusion in autophagosome biogenesis
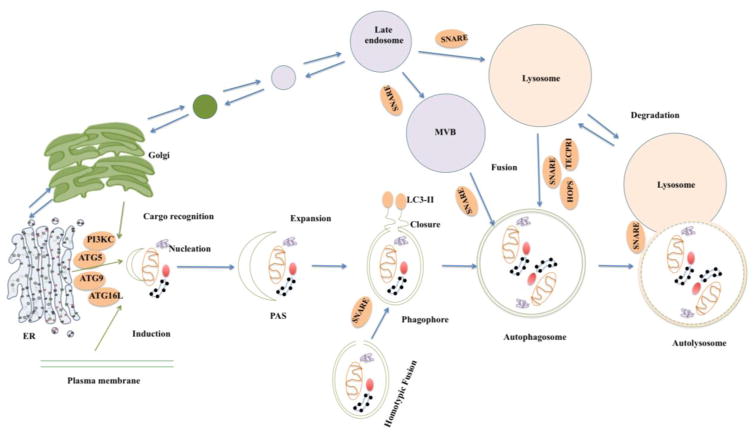
Autophagy is an intracellular degradation process characterized by unique membrane trafficking [13]. Autophagosome biogenesis is usually divided into three continuous steps—phagophore formation, phagophore elongation, and isolation membrane sealing—which result in the formation of a double membrane autophagosome [14]. It is widely accepted that autophagosome formation occurs with dynamic rearrangements of the cellular membrane, initiated by the emergence of an isolation membrane from phagophore assembly sites (PAS) or the pre-autophagosomal structure [15]. This unique membrane structure, called a phagophore, is formed de novo by nucleation on a preexisting membrane [16]. The phagophore engulfs part of the cytoplasm, extends, and ultimately forms a double-membrane autophagosome [17], which delivers its contents to the vacuole or lysosome for degradation [18].
The recruitment of autophagy-associated proteins and membranes results in an elongated double-membrane sheet, which curves itself into the cup-like phagophore. This process is essential for autophagosome biogenesis and acts as a precursor to autophagosome formation [19]. Since the discovery of yeast Atg-related proteins (Atg), autophagosome formation has been dissected at the molecular level [20]. Studies on yeast have found that phagophore formation needs 20–30-nm vesicles containing the integral membrane protein Atg9 [21]. In yeast, Atg9-positive vesicles are generated through Golgi-related secretory and endosomal pathways [22, 23], In human cells, by some contrast, Atg9 exists within the trans-Golgi network in human cells, where it recycles early and late endosomes [24].
In the early phase of autophagosome initiation, precursor vesicles fuse to form the phagophore, a process shown to be mediated by soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) proteins [25, 26]. Membrane-localized SNARE proteins zip themselves into an α-helical bundle that can bridge the two membrane sites tightly together and provide the tether force to fuse two membrane vesicles [27]. Given the presence of a conserved glutamate (Q) or arginine (R) residue in the SNARE motif, SNARE proteins can be classified into three Q-SNARE proteins – Qa, Qb, and Qc – and one R-SNARE protein [28]. SNARE proteins involved in early autophagosome formation include all members of the appropriate classes and, in principle, should be sufficient to drive fusion [29].
At the initial stage of autophagosome biogenesis process, yeast Atg9-containing vesicles and tubules derived from the secretory pathway are translocated to vacuoles to form the PAS. Atg9-containing vesicles help to recruit Atg proteins to the PAS in the absence of Atg14 [21], the autophagy-specific subunit of phosphatidylinositol (PI) 3-kinase recruited by the N-terminal HORMA domain of Atg13 [30]. Research in human cells has shown that when SNARE-mediated fusion is blocked, the recruitment of Atg proteins can nevertheless proceed to the stage involving Atg16 [31]. This dynamic implies that fusion in the early stage of autophagosome generation facilitates the recruitment of Atg proteins in the absence of checkpoints for recruited proteins. In human cells, Atg16L1 vesicles are similar in size and could correspond to Atg9 vesicles in yeast, although that hypothesis remains unconfirmed [32]. A study addressing HeLa cells reported an accumulation of small Atg16L-positive vesicles when VAMP7, syntaxin-7, syntaxin-8, and Vti1b were depleted, thereby underscoring the importance of SNAREs in the fusion of small vesicles required for autophagosome biogenesis [31].
Little is known about mechanisms involved in the closure of the phagophore membrane. Since the phagophore is a double-membrane structure, its closure constitutes a fusion of a narrow opening that differs from fusion processes in other cells. The Rab protein family (i.e., Ras-related proteins) has been identified in autophagy, in which Rab proteins travel between the active GTP-bound state and an inactive GDP-bound state, as well as recruit effector proteins to mediate trafficking between different compartments [33]. Rab1 is known to regulate vesicular transport between the ER and Golgi apparatus, and a recent study identified its importance in omegasome formation [34]. In yeast, the transport protein particle (TRAPP) III complex acts as an autophagy-specific guanine nucleotide exchange factor for the yeast homolog of Rab1 known as YPT1 [35]. Atg17 recruits TRAPP III to PAS in order to activate YPT1 and stimulate the recruitment of Atg1 to the PAS [36]. TRAPP III and YPT1/Rab1 might facilitate the tethering of Atg9 vesicles. However, further studies are needed to fully understand membrane fusion events in the autophagosome closure process.
3. Membrane fusion in autophagosome maturation
Upon the formation of double-membrane vacuoles containing cytoplasmic material, autophagosomes start their stepwise maturation process, including fusion with both endosomes and lysosomes. Autophagosome formation and fusion with lysosomes lasts 3–5 min and occurs only after autophagosomes are fully sealed; if the fusion occurs before that event, then the cytosolic cargo will be released or left attached to the cytosolic side of the lysosomal membrane, where it cannot be degraded [37]. Therefore, those fusion processes must be regulated precisely. Fusion factors can recognize only fully matured autophagosomes, which occurs via an unknown mechanism, though a signal may involve the Atg8 family of proteins. This family of ubiquitin-like proteins is involved in sealing the phagophore to form the autophagosome [38]. Research with yeast has suggested that the loss of Atg8 from the outer autophagosomal membrane signals fusion competency by allowing the removal of the autophagy-initiating machinery [38, 39]. In mammalian cells, however, it remains unclear whether Atg8 removal as a fusion signal is necessary. For one, in mammalian cells, mature autophagosomes lack phagophore-associated proteins such as Atg16 and the ULK1 complex, which implies their removal before fusion with the lysosome. Nevertheless, the mechanism for removing phagopore-associated proteins is currently unknown, as is whether such removal is a prerequisite for fusion.
Immunogold labeling studies [40, 41] and subcellular fractionation studies [42] both indicate that endosomes and lysosomes fuse with autophagosomes during the multistep maturation process, which entails several fusion events with vesicles originating from the endolysosomal compartment. Generally speaking, autophagosome maturation starts by autophagosome fusion with early endosomes, followed by fusion with multivesicular endosomes, which ultimately forms amphisomes. The final step of fusion involves dense lysosomes and allows lysosome enzymes to degrade the cargo of autophagosomes.
Membrane tethers provide specificity and facilitate the docking and fusion process by acting as a bridge between opposite membranes or by stimulating SNARE protein complex formation. In the tethering process, Rab proteins function to recruit and activate tethering factors [43]. The homotypic fusion and protein sorting (HOPS) complex consists of six subunits--VPS11, VPS16, VPS18, VPS33A, VPS39, and VPS41--all of which can coprecipitate with syntaxin-17 (STX17), knock down VPS33A-, VPS16-, or VPS39-blocked autophagic flux, and cause the accumulation of STX17- and LC3-positive autophagosomes in mammalian cells. Furthermore, the knockdown of VPS33A affects the endocytic pathway [44]. Those findings suggest that the HOPS complex promotes autophagosome-lysosome fusion via its tethering functions and interaction with STX17.
In yeast, the HOPS complex has also been shown to bind to and revise the vacuolar SNARE proteins Vam3p, Vam7p, and Vti1p in order to enhance the efficiency of fusion [43, 45]. Another tether factor is tectonin β-propeller repeat-containing protein 1 (TECPR1), a component of the autophagy network identified by its interaction with Atg5 [46] and also implicated in selective autophagy against bacterial pathogens [47]. Our earlier study showed that TECPR1 binds to the Atg12-Atg5 conjugate and phosphatidylinositol 3-phosphate (PtdIns[3]P) to promote autophagosome-lysosome fusion. TECPR1 and Atg16 form mutually exclusive complexes with the Atg12-Atg5 conjugate, and TECPR1 binds PtdIns(3)P upon association with the Atg12-Atg5 conjugate. In all, TECPR1 localizes to mature autophagosomes and lysosomes, and its loss leads to autophagosome accumulation [48]. In effect, TECPR1 could function as a tethering factor in lysosome and autophagosome fusion, though it remains unknown whether TECPR1 tethers SNARE or Rab proteins.
4. SNARE proteins in autophagic fusion
Membrane fusion activity is involved in most steps of autophagy, in which degradation faces its final barrier in the fusion of autophagosomes and lysosomes. Previous findings strongly suggest that proteins of the SNARE family play a major role in the fusion of transport vesicles with acceptor membranes [26, 49]. Despite considerable sequence divergence, their mechanism of action is conserved; different SNARE proteins assemble into complexes (i.e., QabcR-complex) in a combinatorial fashion with helps from several accessory proteins, which in effect drives membrane fusion [14, 50]. Nearly all SNARE proteins are tail-anchored proteins, which have a single transmembrane domain at the extreme C-terminus and N-terminal fragments that are exposed to cytosol [25]. Some SNARE proteins appear in diverse cellular locations and collaborate via the formation of SNARE protein complexes, which for a parallel four-helix bundle during membrane fusion [25]. Since SNARE proteins have been recognized as key components of protein complexes that drive membrane fusion involved in most steps of autophagy, the machinery is also adaptable for diverse fusion reactions, including those in cell growth, membrane repair, synaptic transmission, and autophagy [51].
Without a doubt, the autophagic fusion process is precisely regulated by SNARE and other proteins (Table 1). Fusion between late endosomes and lysosomes, the last barrier for autophagic degradation, uses syntaxin-7 (Qa), Vti1b (Qb), and syntaxin-8 (Qc) on the late endosomes and vesicle-associated membrane protein 7 (VAMP7) (R) on the lysosomes [32, 52]. It has been suggested that Vam3 (Qa), Vam7 (Qc), Ykt6 (R), and Vti1 (Qb) are involved in autophagosome-vacuole fusion in yeast and that VAMP7 (R), VAMP8 (R), and Vti1 (Qb) in such fusion in mammals [53–55]. It was long assumed that phagophore elongation depended on de novo lipid addition and that SNAREs did not play any role in autophagosome biogenesis. Recent studies, however, have suggested that SNARE proteins have roles in autophagy other than simply mediating autophagosome-lysosome fusion [23, 26, 31, 56, 57]. In what follows, we discuss recent work that reveals how SNARE proteins influence autophagosome biogenesis and autophagosome-lysosome fusion (Fig. 1).
Table 1.
Proteins involved in autophagic membrane fusion.
| Protein name | Location | Function | Reference |
|---|---|---|---|
| Sec22 | ER, Golgi | Regulates Atg9 tubular network formation and autophagosome-vacuole fusion in yeast | Cell 2011 [26] |
| VAMP3 | Vacuole R/v-SNARE |
Involved in the fusion of Atg9- and Atg16L1-positive membranes | Cell 2013 [88] |
| VAMP7 | Vacuole R/v-SNARE |
Required for late endosome and lysosome fusion Atg16L1 precursor homotypic fusion | BBA 2009 [55] Cell 2011 [31] |
| VAMP3-VAMP7 | Vacuole R/v-SNARE |
Mammalian MVB-autophagosome fusion | BBA 2009 [55] MCB 1998 [89] |
| Vti1b | Q/t-SNARE | Regulates Atg16L1 homotypic fusion involved in Atg16L1 precursor formation and late endosome-lysosome fusion | Cell 2011 [31] Nature Rev. Mol. Cell Biol. 2006 [53] |
| Vtilb-VAMP8 | R/v-SNARE | Mammalian autophagosome lysosome fusion | MBC 2010 [54] |
| VAMP8 | R/v-SNARE | Mammalian autophagosome lysosome fusion | MBC 2010 [54] |
| STX17 | Autophagosome-localized Q/t-SNARE |
Mammalian autophagosome lysosome fusion | Cell 2012 [61] |
| SNAP29 | Q/t-SNARE | Mammalian autophagosome lysosome fusion | Nat Cell Biol, 2014 [65] Nature 2015 [12] |
| Atg8/LC3 | Atg8-LC3-PE conjugation, present on both inner and outer membranes of autophagosome | Ubiquitin-like protein that is required for autophagosome formation, tethering, and hemifusion | EMBO J 2000 [82] Cell 2007 [81] |
| Atg12 | present on the outer side of the isolation membrane | Ubiquitin-like protein conjugates with Atg5; Atg12-Atg5-Atg16L1 complex promotes Atg8-PE formation and tethers membranes, which is essential for proper elongation of the | EMBO J 2012 [87] |
| Atg14(L) | Class III PI3K complex | Promotes membrane tethering and fusion of autophagosomes to endolysosomes | Nature 2015 [12] |
Figure 1.
SNARE-mediated membrane fusions in autophagy process
In yeast, autophagosomes appear to be generated at the PAS localized near the vacuole [58]. These structures need to somehow undergo fusion and reorganization in order to generate the isolation membrane and, in turn, the autophagosome. At the PAS, several proteins required for autophagosome formation localize in a hierarchical manner [59]. Studies with yeast have found that Atg9 localizes to incompletely characterized compartments composed of vesicles and tubules [21], as well as the exocytic Q/t-SNAREs Sso1-2 and Sec9 are required for organizing Atg9 into tubulovesicular clusters as loss-of-function mutations in the exocytic Q/t-SNAREs result in the absence of tubules in Atg9 membrane clusters [26]. These SNARE proteins may function together with endosomal Q/t-SNARE Tlg2 and R/v-SNARE Sec22, which suggests a role of SNARE proteins in an early fusion event. Interestingly, the absence of these SNARE proteins abolish the formation of that tubular network and cause the formation of small Atg9 vesicles, which do not appear to undergo homotypic fusions, which further suggests that the SNARE proteins are involved in mediating the homotypic fusion of Atg9 vesicles [23].
Autophagic activity also requires the homotypic fusion of Atg16L precursors in order to form mature autophagosomes [60]. When autophagy is stimulated by starvation or rapamycin treatment, the homotypic fusion of Atg16L1 precursors increases, thereby indicating that homotypic fusion is essential for subsequent autophagosome formation [31, 56]. This step depends on SNARE protein VAMP7 together with partner SNARE proteins syntaxin-7, syntaxin-8, and Vti1b. However, it is likely that SNARE proteins VAMP7, syntaxin-7, syntaxin-8, and Vti1b in mammalian cells regulate a different process required for autophagosome formation than that in yeast SNARE proteins [31, 55, 57]. The inhibition of SNARE protein-dependent fusion decreases the size of autophagic precursors and thereby inhibits their subsequent maturation into autophagosomes. Thus, the fusion of small, apparently single-membrane phagophore precursor vesicles that forms tubulovesicular structures could be a mechanism that contributes to the genesis of double-membraned phagophores or autophagosomes. Blocking VAMP7 endocytosis by knocking down Hrb, which is its adaptor at the plasma membrane, prompts the same phenotype as VAMP7 knockdown [31]. Taken together, SNARE proteins could regulate autophagosome formation in both yeast and mammalian cells.
Autophagosomes and late endosomes are the two major organelles that can fuse with lysosomes specialized for degradation. The most obvious vesicle fusion events in autophagy occur between autophagosomes and lysosomes [32]. Recent genetic evidence suggests that a set of SNARE proteins, including STX17, synaptosomal-associated protein 29 (SNAP29), and vesicle-associated membrane protein 8 (VAMP8), are essential for autophagosomes’ fusion with lysosomes [61, 62]. In one way or another, membrane fusion during autophagy appears to proceed via a SNARE protein-dependent mechanism. The Q-SNARE protein STX17 translocates to the outer membrane of the completed autophagosome, and the packed hairpin-like structure causes the exposure of hydrophobic residues [62]. In contrast to other SNARE proteins, STX17 has two TMDs, whose unique hairpin-like structure is important for localizing STX17 to the autophagosome. STX17 interacts with SNAP29 and VAMP8, thereby mediating fusion between autophagosomes and lysosomes [61]. Required for autophagosome-lysosome fusion, VAMP8 is thus speculated to be a partner SNARE protein on the lysosomal membrane.
Increased O-GlcNAcylation has been proposed to inhibit autophagy activity, whereas the inhibition of O-GlcNAcylation increases autophagic flux and reduces cytotoxicity caused by the mutant huntingtin exon 1 protein [63, 64]. O-linked β-N-acetylglucosamine (O-GlcNAc) transferase (OGT) mediates the O-GlcNAcylation of SNARE protein SNAP29 and regulates autophagy in a nutrient-dependent manner. In mammalian cells, OGT knockdown, or the mutation of O-GlcNAc sites in SNAP29, promotes the formation of a SNAP29-containing a SNARE protein complex, increases fusion between autophagosomes and both endosomes and lysosomes, and promotes autophagic flux [65]. It has been reported that O-GlcNAc-modified SNAP-29 reduces the binding affinity with partner SNARE proteins and thus attenuates the assembly of the SNAP-29-containing SNARE protein complex. O-GlcNAc-modification of SNAP-29 could create steric hindrance that affects SNARE protein assembly and function, thus preventing the untimely or ectopic formation of the SNARE protein complex. Remarkably, the depletion of ogt-1 has a similar effect on autophagy in Caenorhabditis elegans, while the expression of an O-GlcNAc-defective SNAP29 mutant facilitates the autophagic degradation of protein aggregates [66].
5. SNARE-associated proteins in autophagic fusion
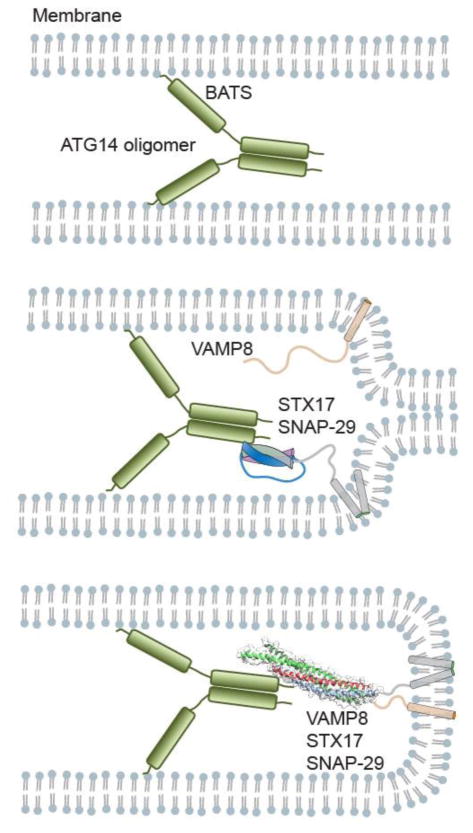
As discussed above, ER-derived SNARE proteins STX17, SNAP29, and endolysosome-localized VAMP8 converge on complete autophagosomes and mediate autophagosome-lysosome fusion [54, 61, 62]. However, it remains unclear whether those SNARE proteins are fusion competent and how their fusogenic activity is specifically regulated during autophagy. It has long been speculated that class III phosphatidylinositol 3-kinase (PI3KC3) nucleates the initiating autophagosome membrane and is required for autophagy and endocytosis via the assembly of distinct protein complexes [67]. ATG14/Barkor/ATG14L, an essential autophagy specific regulator of the class III phosphatidylinositol 3-kinase (PtdIns3K) complex, is concentrated on the curved autophagic membrane enriched in PtdIns(3)P via the Barkor/ATG14(L) autophagosome targeting sequence (BATS) domain in a stress-inducible manner [68]. That finding indicates that Barkor/ATG14(L) functions as a membrane curvature sensor and targeting factor for PI3KC3 to autophagosomes recruited by STX17 [68].
In addition to its localization to phagophores [69–73], ATG14 also localizes to mature autophagosomes and controls the fusogenic activity of the autophagic SNARE protein complex, both spatially and temporally [11, 12, 68, 69]. We have previously shown that ATG14 binds directly to the STX17-SNAP29 binary complex on autophagosomes and promotes STX17-SNAP29-VAMP8-mediated autophagosome fusion with lysosomes. ATG14 homo-oligomerization is required for SNARE protein binding and fusion between autophagosomes and lysosomes, yet is dispensable for PtdIns3K stimulation and autophagosome biogenesis [12]. These data suggest ATG14’s pivotal role in autophagy-specific autophagosome–endolysosome fusion activity. Crystal structure analysis reveals that the autophagic SNARE protein complex forms a parallel four α-helix bundle characteristic of all fusion-competent SNARE protein complexes. The fusogenic activity of autophagic SNARE proteins has also been tested by protein-reconstituted proteoliposome ensemble lipid- and content-mixing assays and membrane morphologies examined by cryo-electron microscopy [74]. We found that ATG14 enhances the hemifusion and complete fusion of proteoliposomes reconstituted with autophagic SNARE proteins. The SNARE protein complex STX17-SNAP29-VAMP8 acts as a positive fusogen, and ATG14 is a critical switch to control the complex-mediated autophagic membrane fusion [74].
ATG14 participates directly in membrane tethering, as a single-vesicle assay has shown, the activity of which is due not only to membrane curvature sensing and requires the membrane-binding BATS domain. As such, the ATG14 fusion-enhancing effect is specific to autophagic SNARE proteins. Interestingly, a STX17 binding-deficient mutant of ATG14 with a coil-coiled domain (CCD) deletion retains its membrane-tethering function, but loses its fusion-enhancing activity. Therefore, the interaction of ATG14 and STX17 necessarily contributes to fusion enhancement [11]. Size exclusion chromatography coupled with multi-angle static light scattering (SEC-MALS) experiments have also indicated that recombinant ATG14 exists in monomeric, dimeric, and tetrameric states and that the amino terminus of ATG14 was required for homo-oligomerization. In particular, the dimeric form of ATG14 is more active in membrane tethering, whereas ATG14 homo-oligomerization-deficient mutants completely abolish its interaction with the autophagic SNARE protein STX17 [12]. Accordingly, ATG14 homo-oligomerization plays a critical role in the interaction with autophagic SNARE proteins. Autophagosome fusion with a lysosome examined in a combination of biochemical, cell biology, and genetic approaches showed results that strongly support the conclusion that ATG14 homo-oligomerization is indispensable in autophagosome-lysosome fusion.
Yet, it remains unclear how ATG14 promotes membrane fusion mediated by autophagic SNARE proteins. ATG14 is homo-oligomerized through cysteine repeats and localizes to complete autophagosomes, after which it binds to the STX17 SNARE protein motif through its CCD, thereby allowing it to function as a clasp that stabilizes the α-helical structure formed by STX17 and SNAP29. As a result, ATG14 promotes and stabilizes the STX17-SNAP29 binary t-SNARE protein complex assembly, which it primes for interaction with endolysosome-localized VAMP8 in order to promote membrane fusion between autophagosomes and endolysosomes [74]. Following fusion, ATG14 dissociates from the cis-SNARE protein complex and is retrieved from autolysosomes. In that light, it has been suggested that ATG14 could be recruited again to complete autophagosomes once fusion commences. The retrieval of ATG14 and possibly STX17 from fused autolysosomes would thereby greatly enhance fusion efficiency.
6. Other autophagy-related proteins in autophagic fusion
During the past few years, the analysis of ATG gene products has progressed rapidly, and many of their mammalian homologs have been identified and characterized. Although the mechanisms underlying autophagosome formation are only poorly understood, more than 35 ATG genes involved in core autophagy machinery were originally identified in yeast genetic screens [20], primarily because little is known about their biochemical activities. In fact, 18 of those ATG genes have been proven necessary for autophagosome formation [14, 20]. Phagophore maturation and, in turn, autophagosome formation require membrane expansion and fusion, which are regulated by two ubiquitylation-like reactions in the Atg12–Atg5 protein and Atg8 lipid conjugation systems [5]. One ubiquitin-like protein, Atg12, is conjugated to protein Atg5, with which it stimulates the linkage of Atg8 to the lipid phosphatidylethanolamine (PE) present in specific intracellular membranes to generate Atg8-PE [75]. The conjugation of Atg8 and PE is reversible, and Atg8 can be rendered competent for conjugation to PE by a proteolytic processing event, catalyzed by Atg4, which functions as a deconjugation enzyme [39]. The Atg8 proteins appear to play multiple roles in autophagy process. They have been shown to recruit cytoplasmic cargo to the isolation membrane and then contribute to the completion of autophagosome formation by localizing on the isolation membranes and autophagosomes [76, 77]. Atg8 could also directly support membrane fusion events that drive the expansion and closure of autophagosomes. All its function is associated with the membrane-binding state [78].
Ohsumi and colleagues used an in vitro system to reconstitute Atg8-PE conjugation in the presence of liposomes. They showed that liposomes containing Atg8-PE aggregate in an Atg8-PE dose-dependent manner [79]. Those Atg8 oligomers, upon PE conjugation, have been proposed to mediate membrane tethering of liposomes to which the Atg8 was anchored. Membrane tethering mediated by Atg8-PE thereafter leads to membrane fusion and thereby allows the completion of autophagosome formation by promoting vesicular carrier fusion [80]. In addition to the crucial role of Atg8-PE in autophagy, Atg4-mediated delipidation of Atg8, which is required to facilitate autophagosome maturation and fusion with the vacuole [38]. In mammalian cells, functional inhibition of the Atg8 conjugation system results in isolation membranes that apparently fail to achieve closure, suggesting that Atg8-PE is essential for a rather late step of autophagosome formation [77]. These results strongly support the hypothesis that Atg8/LC3 mediates membrane tethering and hemifusion, and thereby facilitates the expansion of the phagophore [81]. LC3, one of Atg8’s homologs in mammalian cells, has been used in various studies as a reliable marker for the induction and progression of autophagy [82]. Similar to yeast Atg8, LC3 has been reported to promote tethering and membrane fusion. This activity is mediated by positively charged and hydrophobic amino acids in the N-terminal α-helices [83]. SNARE proteins facilitate most of the fusion events in eukaryotic cells [84], however, unlike LC3, the SNARE machinery contains transmembrane domains, which are critical for their fusion activity. This work has illuminated an unanticipated cellular function of an ubiquitin-like protein and described a membrane tethering and fusion reaction unmediated by SNAREs. However, their in vitro fusion reactions involved liposomes with an extremely high concentration (55 %) of phosphatidylethanolamine (PE) molecules, which is known to promote docking and hemifusion [85]. Recently, Klionsky and colleagues showed Atg8 does not act as a fusogen with physiological PE concentrations [26].
The membrane-remodeling activity of Atg8 proteins is associated with their capacity to promote the tethering and hemifusion of liposomes. The recruitment of Atg8 to the autophagosome follows a second ubiquitin-like system that mediates the conjugation of ubiquitin-like Atg12 and Atg5 [86]. The Atg12-Atg5 conjugate associates non-covalently with Atg16, which can form homo-oligomers through a CCD, which allows Atg16 to cross-link multiple Atg12-Atg5 conjugates into a single, large protein complex [86]. Acting as an E3-ubiquitin ligase, the Atg5-Atg12-Atg16 complex might facilitate the conjugation of Atg8 to PE during classical ubiquitin conjugation reactions. All components of the complex are required for efficient promotion of Atg8 conjugation with PE in a fully reconstituted system. In addition to its Atg8-PE promoting activity, the Atg5-Atg12-Atg16 complex efficiently tethers membranes in an Atg16-dependent manner in vitro; membrane binding is mediated by Atg5, inhibited by Atg12, and activated by Atg16. The membrane binding site in Atg5 is essential for both autophagy and the cytoplasm-to-vacuole transport pathway at a step preceding Atg8-PE conjugation and vesicle closure [87]. Thus, the Atg5-Atg12-Atg16 complex might contribute to the initial tethering of vesicular precursors during autophagosome formation.
7. Outlook
As an evolutionally conserved intracellular degradation pathway, autophagy acts as a double-edged sword to some degree. On one hand, it can function primarily as a cytoprotective mechanism; while on the other hand, its activation can detrimentally allow cancer cells to become resistant to chemotherapy. Accordingly, a greater understanding of the regulatory pathways that control autophagy is urgently needed.
Autophagy is characterized by the fusion of autophagosomes with endosomes and lysosomes. In addition to the late autophagosome-lysosome fusion for degradation, membrane fusion activity is also required for the early elongation and closure of isolation membranes into completed autophagosomes. Autophagic fusion is probably closely regulated by SNARE proteins and other factors, including ATG14, SNARE protein STX17, and ubiquitin-like protein Atg8/LC3. In support of that conclusion, studies have also shown that loss-of-function mutations of those membrane fusion-associated factors lead to a significant inhibition of autophagy. Nevertheless, the exact molecular mechanism of autophagic membrane fusion remains far from clear and thus a major topic of investigation. Since membrane fusion activity could act as a switch to spatially and temporally regulate the autophagic flux in human diseases due to its dysregulation, dissecting the fusion machinery is essential to understanding the exact roles of autophagy in specific disease contexts. Furthermore, it will facilitate the development of therapeutics that can specifically modulate autophagy.
Figure 2.
ATG14 promotes STX17-SNAP29-VAMP8-mediated autophagosome fusion with a lysosome.
Acknowledgments
This work was supported by the 973-Program Funding (2015CB856300, 2015CB553602), Welch Foundation (I-1864), the Cancer Prevention & Research Institute of Texas (RP140320), and National Institutes of Health R01 (GM116908).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levine B, Klionsky DJ. Development by Self-Digestion: Molecular Mechanisms and Biological Functions of Autophagy. Developmental Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Klionsky DJ, Emr SD. Autophagy as a Regulated Pathway of Cellular Degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol. 2010;12(9):814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 5.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–52. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maiuri MC, et al. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8(9):741–52. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 7.Beau I, Mehrpour M, Codogno P. Autophagosomes and human diseases. Int J Biochem Cell Biol. 2011;43(4):460–4. doi: 10.1016/j.biocel.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Mizushima N, et al. Autophagy fights disease through cellular self-digestion. Nature. 2008;451(7182):1069–75. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(7):651–62. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 10.Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol. 2010;12(9):831–5. doi: 10.1038/ncb0910-831. [DOI] [PubMed] [Google Scholar]
- 11.Hamasaki M, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495(7441):389–93. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 12.Diao J, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520(7548):563–6. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizushima N. Autophagy: process and function. Genes & Development. 2007;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 14.Lamb CA, Yoshimori T, Tooze SA. The autophagosome: origins unknown, biogenesis complex. Nat Rev Mol Cell Biol. 2013;14(12):759–74. doi: 10.1038/nrm3696. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki K, et al. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. The EMBO Journal. 2001;20(21):5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carlsson SR, Simonsen A. Membrane dynamics in autophagosome biogenesis. Journal of Cell Science. 2015;128(2):193–205. doi: 10.1242/jcs.141036. [DOI] [PubMed] [Google Scholar]
- 17.Hara T, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. The Journal of cell biology. 2008;181(3):497–510. doi: 10.1083/jcb.200712064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanley RE, Ragusa MJ, Hurley JH. The beginning of the end: how scaffolds nucleate autophagosome biogenesis. Trends in Cell Biology. 2014;24(1):73–81. doi: 10.1016/j.tcb.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamasaki M, Shibutani ST, Yoshimori T. Up-to-date membrane biogenesis in the autophagosome formation. Curr Opin Cell Biol. 2013;25(4):455–60. doi: 10.1016/j.ceb.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 20.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–32. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 21.Mari M, et al. An Atg9-containing compartment that functions in the early steps of. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21(22):3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto H, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198(2):219–33. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orsi A, et al. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Molecular biology of the cell. 2012;23(10):1860–1873. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sudhof TC, Rothman JE. Membrane Fusion: Grappling with SNARE and SM Proteins. Science. 2009;323(5913):474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair U, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146(2):290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Söllner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362(6418):318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 28.Fasshauer D, et al. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q-and R-SNAREs. Proceedings of the national academy of sciences. 1998;95(26):15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber T, et al. SNAREpins: Minimal Machinery for Membrane Fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 30.Jao CC, et al. A HORMA domain in Atg13 mediates PI 3-kinase recruitment in autophagy. Proceedings of the National Academy of Sciences. 2013;110(14):5486–5491. doi: 10.1073/pnas.1220306110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreau K, et al. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146(2):303–17. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreau K, Renna M, Rubinsztein DC. Connections between SNAREs and autophagy. Trends Biochem Sci. 2013;38(2):57–63. doi: 10.1016/j.tibs.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 34.Mochizuki Y, et al. Phosphatidylinositol 3-phosphatase myotubularin-related protein 6 (MTMR6) is regulated by small GTPase Rab1B in the early secretory and autophagic pathways. Journal of Biological Chemistry. 2013;288(2):1009–1021. doi: 10.1074/jbc.M112.395087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch-Day MA, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proceedings of the National Academy of Sciences. 2010;107(17):7811–7816. doi: 10.1073/pnas.1000063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang J, et al. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proceedings of the National Academy of Sciences. 2013;110(24):9800–9805. doi: 10.1073/pnas.1302337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ganley IG. Autophagosome maturation and lysosomal fusion. Essays In Biochemistry. 2013;55:65–78. doi: 10.1042/bse0550065. [DOI] [PubMed] [Google Scholar]
- 38.Nair U, et al. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy. 2012;8(5):780–93. doi: 10.4161/auto.19385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu ZQ, et al. Dual roles of Atg8-PE deconjugation by Atg4 in autophagy. Autophagy. 2012;8(6):883–92. doi: 10.4161/auto.19652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka Y, et al. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406(6798):902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- 41.Eskelinen EL, et al. Inhibition of Autophagy in Mitotic Animal Cells. Traffic. 2002;3(12):878–893. doi: 10.1034/j.1600-0854.2002.31204.x. [DOI] [PubMed] [Google Scholar]
- 42.Berg TO, et al. Isolation and Characterization of Rat Liver Amphisomes: EVIDENCE FOR FUSION OF AUTOPHAGOSOMES WITH BOTH EARLY AND LATE ENDOSOMES. Journal of Biological Chemistry. 1998;273(34):21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 43.Orr A, et al. Yeast vacuolar HOPS, regulated by its kinase, exploits affinities for acidic lipids and Rab: GTP for membrane binding and to catalyze tethering and fusion. Molecular biology of the cell. 2015;26(2):305–315. doi: 10.1091/mbc.E14-08-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang P, et al. The HOPS complex mediates autophagosome–lysosome fusion through interaction with syntaxin 17. Molecular biology of the cell. 2014;25(8):1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Starai VJ, Hickey CM, Wickner W. HOPS proofreads the trans-SNARE complex for yeast vacuole fusion. Mol Biol Cell. 2008;19(6):2500–8. doi: 10.1091/mbc.E08-01-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Behrends C, et al. Network organization of the human autophagy system. Nature. 2010;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogawa M, et al. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell host & microbe. 2011;9(5):376–389. doi: 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 48.Chen D, et al. A Mammalian Autophagosome Maturation Mechanism Mediated by TECPR1 and the Atg12-Atg5 Conjugate. Molecular Cell. 2012;45(5):629–641. doi: 10.1016/j.molcel.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brunger AT, Cipriano DJ, Diao J. Towards reconstitution of membrane fusion mediated by SNAREs and other synaptic proteins. Critical Reviews in Biochemistry & Molecular Biology. 2015:50. doi: 10.3109/10409238.2015.1023252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kloepper TH, Kienle CN, Fasshauer D. An elaborate classification of SNARE proteins sheds light on the conservation of the eukaryotic endomembrane system. Mol Biol Cell. 2007;18(9):3463–71. doi: 10.1091/mbc.E07-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chernomordik LV, Kozlov MM. Mechanics of membrane fusion. Nature Structural & Molecular Biology. 2008;15(7):675–683. doi: 10.1038/nsmb.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Furuta N, Amano A. SNARE mediates autophagosome–lysosome fusion. Journal of Oral Biosciences. 2012;54(2):83–85. [Google Scholar]
- 53.Jahn R, Scheller RH. SNAREs--engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7(9):631–43. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 54.Furuta N, et al. Combinational soluble N-ethylmaleimide-sensitive factor attachment protein receptor proteins VAMP8 and Vti1b mediate fusion of antimicrobial and canonical autophagosomes with lysosomes. Mol Biol Cell. 2010;21(6):1001–10. doi: 10.1091/mbc.E09-08-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fader CM, et al. TI-VAMP/VAMP7 and VAMP3/cellubrevin: two v-SNARE proteins involved in specific steps of the autophagy/multivesicular body pathways. Biochim Biophys Acta. 2009;1793(12):1901–16. doi: 10.1016/j.bbamcr.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 56.Ravikumar B, et al. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12(8):747–57. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pryor PR, et al. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 2004;5(6):590–5. doi: 10.1038/sj.embor.7400150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. NATURE CELL BIOLOGY V. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 59.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy. 2010;6(6):764–776. doi: 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stroupe C. Autophagy: cells SNARE selves. Curr Biol. 2011;21(18):R697–9. doi: 10.1016/j.cub.2011.08.017. [DOI] [PubMed] [Google Scholar]
- 61.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151(6):1256–69. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Takats S, et al. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013;201(4):531–9. doi: 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marsh SA, et al. Cardiac O-GlcNAcylation blunts autophagic signaling in the diabetic heart. Life Sci. 2013;92(11):648–56. doi: 10.1016/j.lfs.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kumar A, et al. Decreased O-linked GlcNAcylation protects from cytotoxicity mediated by huntingtin exon1 protein fragment. J Biol Chem. 2014;289(19):13543–53. doi: 10.1074/jbc.M114.553321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo B, et al. O-GlcNAc-modification of SNAP-29 regulates autophagosome maturation. Nat Cell Biol. 2014;16(12):1215–26. doi: 10.1038/ncb3066. [DOI] [PubMed] [Google Scholar]
- 66.Wang P, et al. O-GlcNAc cycling mutants modulate proteotoxicity in Caenorhabditis elegans models of human neurodegenerative diseases. Proc Natl Acad Sci U S A. 2012;109(43):17669–74. doi: 10.1073/pnas.1205748109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Noda T, et al. Regulation of membrane biogenesis in autophagy via PI3P dynamics. Semin Cell Dev Biol. 2010;21(7):671–6. doi: 10.1016/j.semcdb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 68.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) PNAS. 2011;108(19):7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Matsunaga K, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190(4):511–21. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Itakura E, et al. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19(12):5360–72. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11(4):468–76. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun Q, Fan W, Zhong Q. Regulation of Beclin 1 in autophagy. Autophagy. 2009;5(5):713–716. doi: 10.4161/auto.5.5.8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11(4):385–96. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 74.Liu R, Zhi X, Zhong Q. ATG14 controls SNARE-mediated autophagosome fusion with a lysosome. Autophagy. 2015;11(5):847–9. doi: 10.1080/15548627.2015.1037549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanada T, et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J Biol Chem. 2007;282(52):37298–302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- 76.Yoshimoto K. Processing of ATG8s, Ubiquitin-Like Proteins, and Their Deconjugation by ATG4s Are Essential for Plant Autophagy. The Plant Cell Online. 2004;16(11):2967–2983. doi: 10.1105/tpc.104.025395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Slobodkin MR, Elazar Z. The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy. Essays Biochem. 2013;55:51–64. doi: 10.1042/bse0550051. [DOI] [PubMed] [Google Scholar]
- 78.Kabeya Y, et al. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. Journal of Cell Science. 2004;117(13):2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 79.Subramani S, Farre JC. A ubiquitin-like protein involved in membrane fusion. Cell. 2007;130(1):18–20. doi: 10.1016/j.cell.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 80.Xie Z, Nair U, Klionsky DJ. Atg8 Controls Phagophore Expansion during Autophagosome Formation. Molecular Biology of the Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130(1):165–78. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 82.Kabeya Y, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19(21):5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Weidberg H, et al. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev Cell. 2011;20(4):444–54. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 84.Diao JJ, et al. Studying protein-reconstituted proteoliposome fusion with content indicators in vitro. Bioessays. 2013;35(7):658–665. doi: 10.1002/bies.201300010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lai Y, et al. Lipid molecules influence early stages of yeast SNARE-mediated membrane fusion. Phys Biol. 2015;12(2):025003. doi: 10.1088/1478-3975/12/2/025003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9(10):1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 87.Romanov J, et al. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31(22):4304–17. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Puri C, et al. Diverse autophagosome membrane sources coalesce in recycling endosomes. Cell. 2013;154(6):1285–99. doi: 10.1016/j.cell.2013.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sato TK, Darsow T, Emr SD. Vam7p, a SNAP-25-Like Molecule, and Vam3p, a Syntaxin Homolog, Function Together in Yeast Vacuolar Protein Traffickin. MOLECULAR AND CELLULAR BIOLOGY. 1998;18(9):5308–5319. doi: 10.1128/mcb.18.9.5308. [DOI] [PMC free article] [PubMed] [Google Scholar]