Abstract
Nicotinic acetylcholine receptors are ligand gated ion channels that mediate fast chemical neurotransmission at the neuromuscular junction and play diverse signaling roles in the central nervous system. The nicotinic receptor has been a model system for cell surface receptors, and specifically for ligand-gated ion channels, for well over a century1,2. In addition to the receptors’ prominent roles in the development of the fields of pharmacology and neurobiology, nicotinic receptors are important therapeutic targets for neuromuscular disease, addiction, epilepsy, and for neuromuscular blocking agents used during surgery2–4. The overall architecture of the receptor was described in landmark studies of the nicotinic receptor isolated from the electric organ of Torpedo marmorata5. Structures of a soluble ligand binding domain have provided atomic-scale insights into receptor-ligand interactions6, while high-resolution structures of other members of the pentameric receptor superfamily provide touchstones for an emerging allosteric gating mechanism7. All available high-resolution structures are of homopentameric receptors. However, the vast majority of pentameric receptors (called Cys-loop receptors in eukaryotes) present physiologically are heteromeric. Here we present the X-ray crystallographic structure of the human α4β2 nicotinic receptor, the most abundant nicotinic subtype in the brain. This structure provides insights into the architectural principles governing ligand recognition, heteromer assembly, ion permeation and desensitization in this prototypical receptor class.
The α4β2 receptor is known to assemble in two functional subunit stoichiometries, 3α:2β and 2α:3β. The latter stoichiometry has an ~100-fold higher affinity for both acetylcholine and nicotine, lower single channel conductance and calcium permeability, and its expression is selectively upregulated by nicotine8–10. We used a small-scale fluorescence-based approach to optimize conditions for protein expression and purification that would yield the 2α:3β form11. Growth of well-diffracting crystals required deleting most of the intracellular domain between transmembrane spans M3 and M4 in both subunits (Extended Data Figs. 1–2). This crystallized receptor construct, referred to here as α4β2, retains function comparable to full-length protein, as discussed below. The best-diffracting crystals were obtained by co-crystallization with nicotine and a cholesterol analog, and allowed for collection of a complete dataset to 3.9 Å resolution (see Methods and Extended Data Table 1).
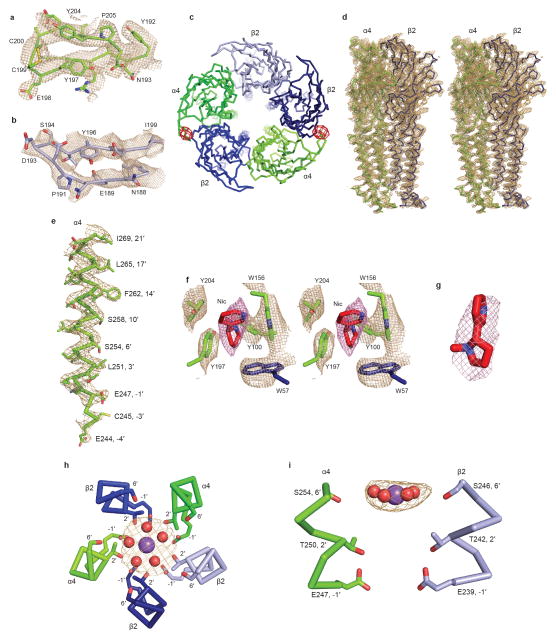
The structure of the α4β2 receptor was solved by molecular replacement (see Methods). Subunit identities were initially assigned based on features in electron density maps from the vicinity of the neurotransmitter binding pocket (Extended Data Figs. 3a, b). To further interrogate subunit identity, we co-crystallized the receptor with 5-Iodo-A-85380, a potent agonist that, like acetylcholine and nicotine, is expected to bind only at α-β interfaces12. From a low-resolution isomorphous dataset we observed iodine anomalous signal in only the two assigned α-β interfaces (Extended Data Fig. 3c). After finalizing subunit assignment, electron density maps were of sufficient quality to build and refine nearly all of the extracellular and transmembrane domains, as well as a portion of the intracellular domain (Extended Data Figs. 1 and 3).
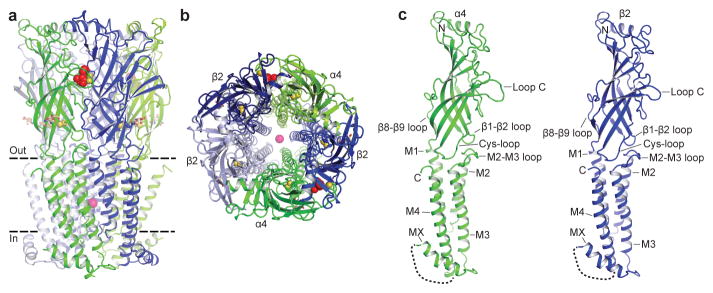
The α4β2 receptor resembles a cylinder formed from 5 subunits in a pseudo-symmetric arrangement about the channel axis. The crystal structure reveals a subunit ordering of α-β-β-α-β around the pentameric ring (Figs. 1a, b), consistent with functional studies of concatameric receptors13. The α4 and β2 subunits share 59% amino acid sequence identity and adopt similar backbone conformations (Fig. 1c and Extended Data Figs. 4a, b). Each subunit comprises a large extracellular domain with an amino-terminal α-helix and 10 β-strands that wrap inward to form a sandwich. The C-terminal bundle comprises 3 transmembrane α-helices (M1–M3), an amphipathic or intracellular MX helix, and a final transmembrane α-helix (M4). The overall architecture is similar to that found in the other Cys-loop receptor family members of known structure (Extended Data Fig 4c and Extended Data Table 2)7. The MX helix, about which comparatively little structural information is available, closely resembles the conformation observed in the 5-HT3 receptor (5-HT3R) structure (Extended Data Fig. 4c)14. The Cys-loop receptor superfamily takes its name from a conserved disulfide bond linking the β6 and β7 strands in the extracellular domain. A second disulfide bond is formed between adjacent cysteines at the tip of Loop C in the α4 subunits (Extended Data Fig. 3a,b and 5g–i), a feature that defines nicotinic receptor α subunits and is absent in all other Cys-loop receptors15. Electron density was observed for nicotine at the two α-β interfaces in the extracellular domain and for a single N-acetylglucosamine residue linked to a conserved asparagine in the Cys-loop of each subunit (Extended Data Figs. 3f, g). The interior surface of the receptor begins at a large extracellular vestibule that narrows into a funnel-shaped transmembrane channel defined by the pore-lining M2 α-helices; mutations in this region are linked to autosomal dominant nocturnal frontal lobe epilepsy (Extended Data Fig. 1)4. A strong electron density peak in the pore was modeled speculatively as a combination of Na+ ion and water in an arrangement similar to that seen in a prokaryotic pentameric receptor, GLIC16 (see Methods and Extended Data Figs. 3h, i). The channel is in a desensitized, non-conducting conformation most similar to that observed in the GABAAR structure17, however the overall receptor conformation is distinct.
Figure 1. Architecture of the α4β2 nicotinic receptor.
a, View parallel to the plasma membrane. α4 subunits are in green and β2 in blue. Nicotine (red) and sodium (pink) are represented as spheres. The Cys-loop and Loop C disulfide bonds are shown as yellow spheres. N-linked glycans (brown) are shown as sticks. Dashed lines indicate approximate membrane position. b, View perpendicular to the plasma membrane looking from the extracellular side. c, Orientation as in a of the individual subunits. Unmodeled residues from the intracellular domain are represented as a dashed line.
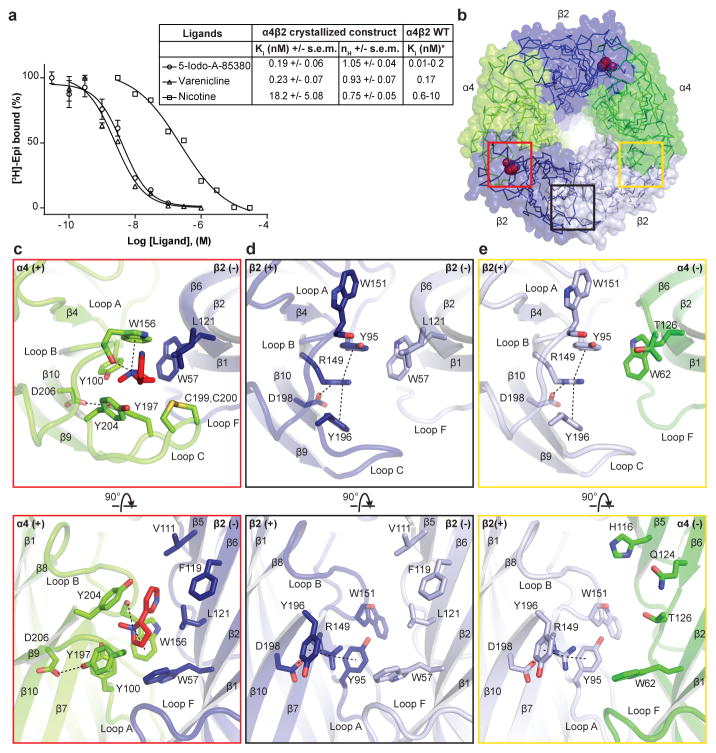
Nicotine activity in the brain, including its reinforcing properties that lead to addiction, is mediated principally by α4β2 receptors18,19. To validate the receptor constructs used in crystallization, we quantified the binding affinities of a panel of ligands for the purified receptor (Fig. 2a and Extended Data Fig. 2d). Among the three classes of subunit interfaces, we observed electron density for nicotine only at the α-β interfaces (Fig. 2b). The ligand was positioned based on the strong omit electron density (6.8–8.0 σ, Extended Data Figs. 3f, g) and comparison with the high resolution structure of the acetylcholine binding protein (AChBP) in complex with nicotine (Extended Data Fig. 6)6. We first analyzed interactions of nicotine with the receptor and then compared the positions of corresponding residues at non α-β interfaces to understand principles of binding selectivity.
Figure 2. Neurotransmitter binding site.
a, Competition experiments against [3H]-epibatidine. Calculated inhibition constant (Ki) values assume a Kd for [3H]-epibatidine of 96 pM (Extended Data Fig. 2d). n = 4 independent experiments. Error bars are s.e.m. and nH is the Hill coefficient. *Published range of the Ki of the ligands against WT α4β2. b, Extracellular view, with colored boxes indicating the three different interface classes. c–e, Architectural details of interfaces boxed in b. The top row is from the same orientation as in b. Nicotine and interacting residues are shown as sticks. Potential hydrogen bonding and cation-π interactions are represented as dashed lines (2.7–5 Å). In the lower row, the loop C backbone is hidden to aid in clarity.
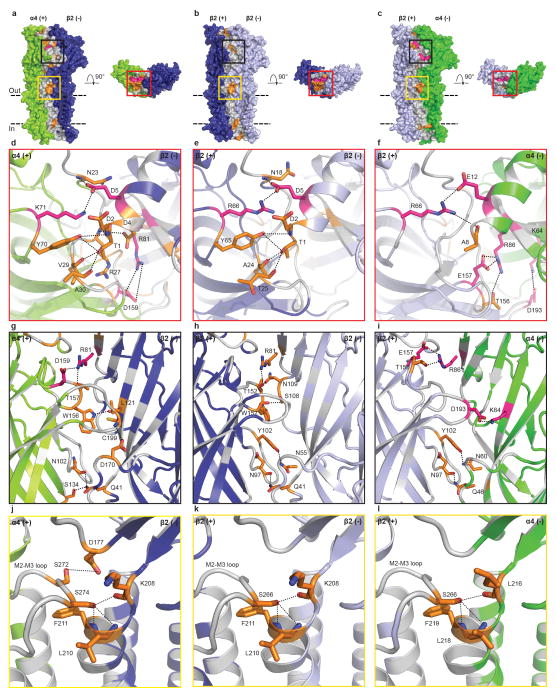
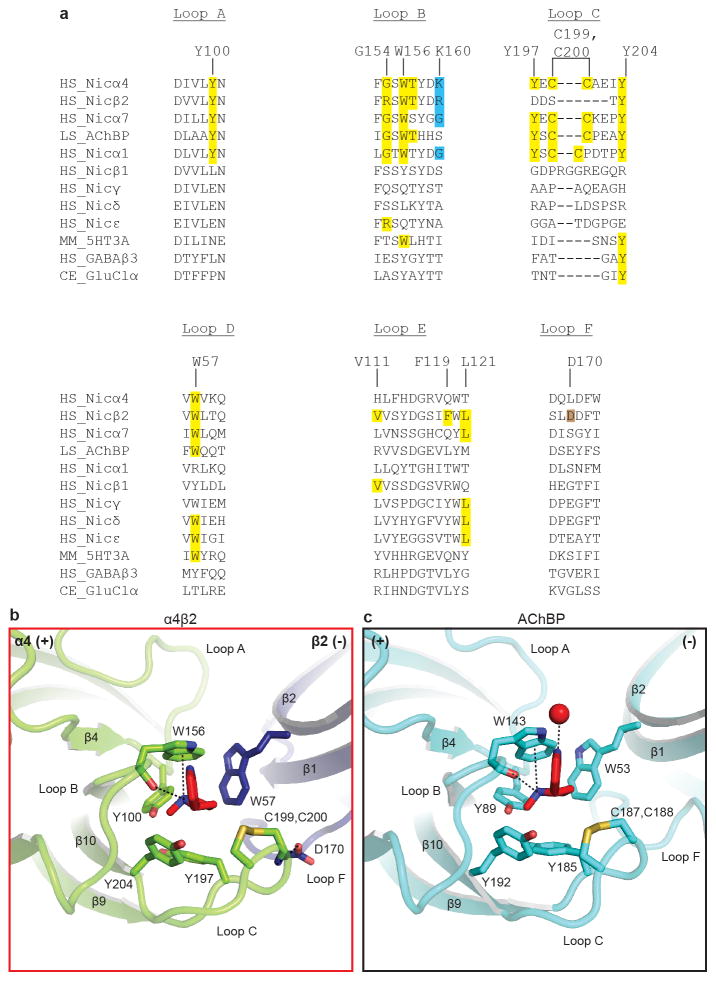
Nicotine binds in the classical neurotransmitter site at the α-β interface, almost fully buried from solvent. The α4 subunit forms the (+) side of the binding pocket and the β2 subunit forms the (−) side (Figs. 2b, c). Three loops from each side of the interface contribute to binding of orthosteric ligands, A, B and C from the (+) side, and D, E and F from the (−) side. Residues from loops A–E form a tightly-packed aromatic box surrounding nicotine, with the floor formed by Y100 on Loop A and W57 on the β2 strand in Loop D. The back walls are defined by W156 in Loop B and L121 on the β6 strand in Loop E. The front wall of the pocket is formed by Loop C, which packs tightly onto the ligand, contributing interactions from the vicinal cysteines and from Y197 and Y204. The hydrophobic top of the pocket is formed by V111 and F119 in Loop E. In addition to the aromatic and hydrophobic interactions with these side chains, nicotine is poised to form a hydrogen bond between its electropositive pyrrolidine nitrogen and the backbone carbonyl oxygen of W156. The pyrrolidine nitrogen is also well-oriented to form a cation-π interaction with the indole ring of W156, a recurring ligand-receptor interaction in the superfamily, though not always to this tryptophan20. Residues in Loop F do not contribute directly to nicotine binding, however D170 on Loop F likely stabilizes loop C via a hydrogen bond to the backbone nitrogen of C199 (Extended Data Figs. 5, 6).
To date all high resolution structural information for Cys-loop receptors has come from homopentameric assemblies, leaving many questions unanswered regarding architecture of the non-canonical interfaces. The α4β2 crystal structure reveals a surprising reorganization of the conserved aromatic residues in the β-β and β-α interfaces that precludes nicotine binding. The source of the reorganization appears to be the identity of the residue that precedes the Loop B tryptophan by two positions. In the α4 subunit, this residue is a glycine (G154); in β2, it is an arginine (R149). When the β2 subunit contributes to the (+) side of the interface (Figs. 2d, e), this R149 orients longitudinally into the base of the binding pocket. The second tyrosine on Loop C is not present in the β2 subunit, which allows Y196 to change its rotameric position, orienting toward the membrane. A second tyrosine, Y95 in Loop A, rotates away from the membrane. The result of the switch in conformations of these two tyrosines is that the positively charged guanidinium group of R149 is sandwiched between their two aromatic rings, in a sense satisfying the electron-rich π system as the pyrrolidine nitrogen of nicotine does in the α-β interfaces. A consequence of the reorganization around the arginine is that W151 in Loop B must move; its side chain rotates out of the binding pocket completely. The conformations of these residues on the (+) side are similar between the β-β and β-α interfaces; the differences between them arise from the (−) side of the interface, where three hydrophobic groups on the (−) side of the β2 subunit are replaced by polar side chains on the (−) side of the α4 subunit (Fig. 2e). This difference in chemical environment may affect nicotine binding to α4-α4 interfaces in the 3α:2β stoichiometry21. The polar environment on the (−) face of the α4 subunit may be less favorable for nicotine binding in the orientation we observe at the α-β interfaces, wherein the pyridine ring packs against the hydrophobic (−) face of the β subunit. By comparison, the homopentameric α7 nicotinic receptor preserves two of the three hydrophobic residues in Loop E (Extended Data Fig. 6a) and maintains nicotine binding, albeit with lower affinity.
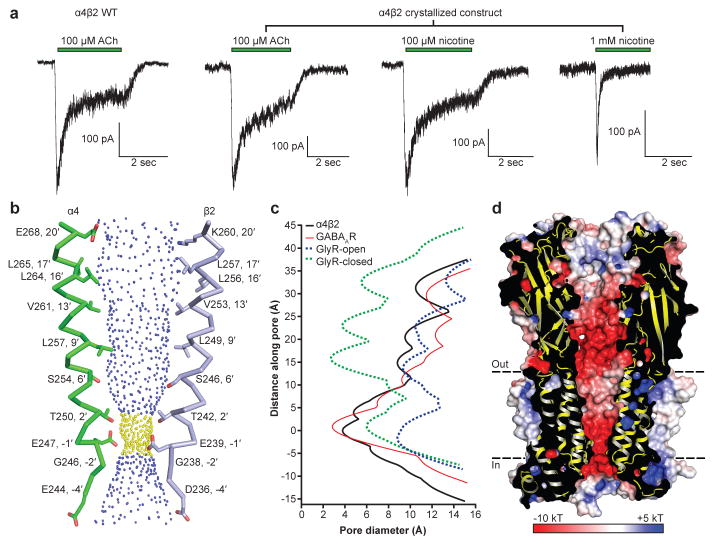
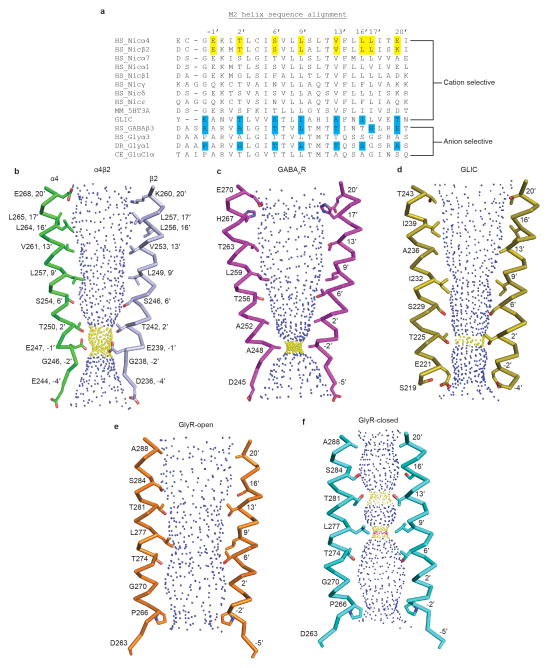
After prolonged exposure to agonist, nicotinic receptors desensitize, adopting a high-affinity and agonist-bound, non-conducting conformation7. We performed patch clamp electrophysiology experiments comparing responses of full-length and crystallized α4β2 receptor constructs to acetylcholine and found them to behave similarly (Fig. 3a). We next measured responses to 1 mM nicotine, as was used throughout purification and for crystallization, and observed that the receptor desensitized profoundly within a few milliseconds. This functional result predicts that we would observe a desensitized, non-conducting conformation in the structure. The receptor structure reveals the transmembrane channel tapering to a constriction point at the interface with the cytosol (Fig. 3b). The narrowest point in the pore is defined by glutamate side chains at the −1′ position of the M2 α-helices, which give rise to a constriction of 3.8 Å in diameter (Figs. 3b, c). The consensus on minimum pore diameter among cation-selective Cys-loop receptors is in the range of ~6–8 Å22,23, consistent with the permeant ion being at least partially hydrated. The α4β2 receptor is a non-selective cation channel, being permeable to Na+, K+ and Ca2+. Na+ is the smallest, with an ionic diameter of 1.90 Å. Adding a single equatorial water molecule (2.8 Å diameter) would put the diameter of the permeant species above the observed constriction size. We compared the α4β2 receptor pore conformation to those from recent structures that likely represent the three principal receptor states: resting-closed (glycine receptor + strychnine24; GlyR-closed), activated-open (glycine receptor + glycine24; GlyR-open) and desensitized-closed (GABAAR17) (Fig. 3c and Extended Data Fig. 7). The pore conformation of the α4β2 receptor most closely resembles the desensitized GABAAR, where the gate is at the cytosolic end of the pore. Functional studies also suggest that the desensitization gate is located at the cytosolic side of the pore25. Thus, structural and functional analyses are consistent with the α4β2 receptor structure representing a desensitized, non-conducting state.
Figure 3. Ion permeation pathway.
a, Patch-clamp recordings of the wild type (WT) and crystallized α4β2 receptor. ACh, acetylcholine. b, M2 α-helices from opposing α4 and β2 subunits with side chains shown for pore-lining residues. Blue spheres indicate pore diameters >5.6 Å; yellow are >2.8 Å and <5.6 Å. c, Pore diameter for the α4β2 receptor and representative Cys-loop receptors in distinct functional states: desensitized-closed (GABAAR + benzamidine; PDB:4COF), activated-open (GlyR + glycine; PDB:3JAE) and resting-closed (GlyR + strychnine; PDB:3JAD). Structures were aligned using the M2 helix 9′ leucine, which occurs at y = ~15 Å. The zero value along the Y-axis in the plot is aligned with the α-carbon of the M2 helix −1′ glutamate residue in α4β2. d, Cutaway of the receptor showing the permeation pathway colored by electrostatic potential.
To probe mechanisms of ion selectivity, we analyzed the electrostatic properties of the permeation pathway of the α4β2 receptor (Fig. 3d). The surface of the extracellular vestibule is strongly electronegative, which likely serves to increase the local concentration of cations near the channel mouth. The electrostatic potential becomes more neutral at the extracellular end of the pore, where the 20′ glutamate side chains from the two α4 subunits are offset by the 20′ lysine side chains from the three β2 subunits. This 20′ position is the only site in the pore where the α4 and β2 subunits contribute opposing charges to the electrostatic surface, and thus is where alternate subunit stoichiometries would be expected to most strongly influence permeation properties. Indeed, the higher Ca2+ permeability of the 3α:2β stoichiometry of this receptor has been shown to result from the swap of lysine to glutamate at the 20′ position in that assembly9. Approaching the constriction point in the pore, the surface becomes strongly electronegative, dominated by the five glutamate side chains that form the selectivity filter at the base of the pore. The side chains are folded toward the pore axis with their carboxylates likely stabilized through hydrogen bonding with the −2′ backbone carbonyl oxygens from adjacent subunits.
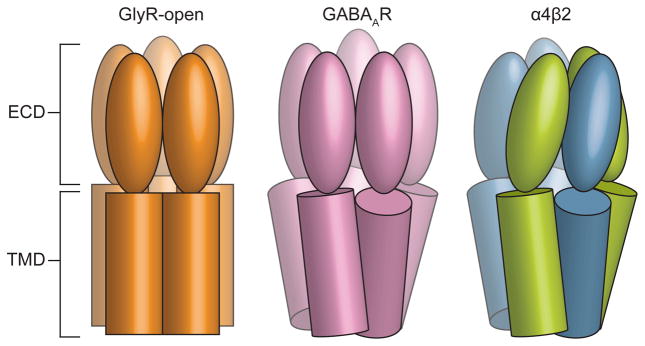
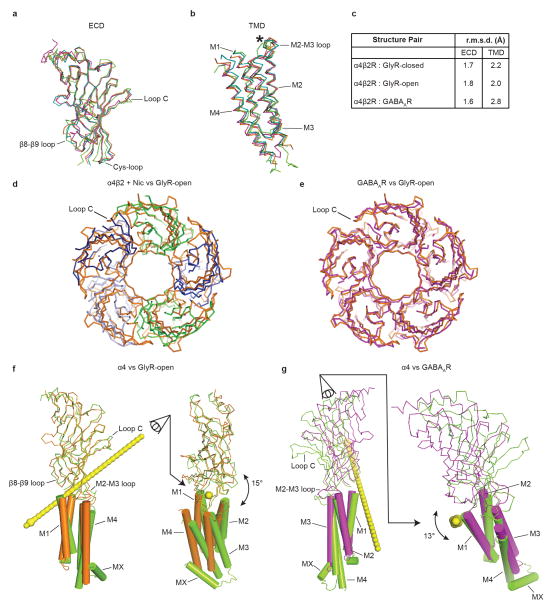
To move beyond the local conformation observed in the pore, and to place the α4β2 receptor structure in the context of the resting-activated-desensitized gating cycle, we next compared the overall conformation of the α4β2 receptor to the reference structures for distinct conformations. Structures of GluCl26 and the glycine receptor24, each in multiple conformations, suggest that within an individual subunit, the extracellular (ECD) and transmembrane subdomains (TMD) behave in large part as rigid bodies during state transitions. Thus we initially compared the extracellular and transmembrane subdomains of an α4 subunit with the analogous subdomains from the open and desensitized structures described in the previous section (Extended Data Figs. 8a–c). We found that the Cα backbones from these subdomains superimpose well (Cα r.m.s.d 1.6–2.8 Å), with noteworthy differences in loops at the extracellular-transmembrane interface thought to be involved in signal transduction. These loops include the β1-β2, M2–M3 and Cys-loops from the (+) subunit and the β8-β9 loop and the β10-M1 helix junction in the (−) subunit. To understand how the reorganization of these interfacial loops relates to global conformational changes, we superimposed whole receptors based on alignment of their pentameric transmembrane domains, and examined corresponding differences in the extracellular domains. We were surprised to find that while the GABAAR pore is tightly closed, more so even than α4β2 (Fig. 3c), the conformation of the GABAAR extracellular domain much more closely resembles the open GlyR structure than the α4β2 receptor structure (Extended Data Figs. 8d, e).
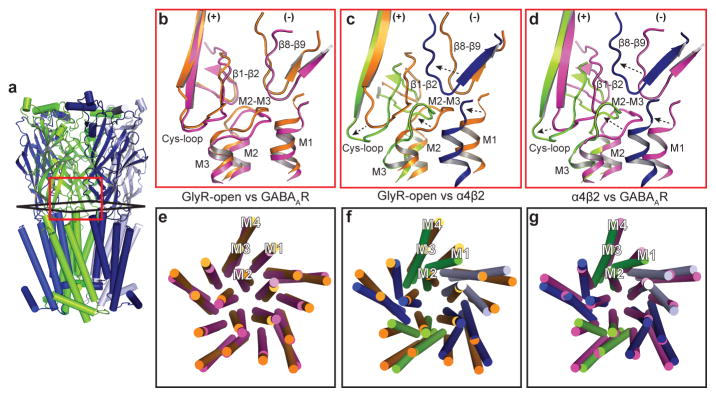
Examination of the interactions between the extracellular and transmembrane domains further illustrates the differences between the open and the two desensitized conformations (Figs. 4a–d). At the ECD-TMD interface, local loop conformations are similar between the GlyR-open and the GABAAR structures (Fig. 4b). Comparison of α4β2 with both the GlyR-open (Fig. 4c) and the GABAAR (Fig. 4d) structures reveals concerted displacements in α4β2 of the β1-β2, M2–M3 and Cys-loops on the (+) subunit and the β8-β9 loop and the β10-M1 helix on the (−) subunit. These displacements are maximal at the Cys-loop, with differences between reference Cα atoms of 6.5 Å for α4β2 vs. GABAAR and 7.4 Å for α4β2 vs. GlyR-open. Analysis of the conformational differences at the subunit level between α4β2 and GlyR-open that generate these displacements suggests a 15° rotation around an axis passing through the Cys-loop (Extended Data Fig. 8f). This rotation results in closure of the ion channel and necessitates reorganization of the ECD-TMD interface. In contrast, analysis of the conformational differences between α4β2 and GABAAR suggests a 13° tilting of the ECD (Extended Data Fig. 8g). As a result, from α4β2 to the GABAAR, the pore remains similarly closed, but the ECD-TMD interface is different. In both cases, the resulting displacement of the Cys-loop at the pivot point coincides with a major alteration in the conformation of the M1 helix of α4β2 relative to GlyR-open and to GABAAR (Figs. 4e–g).
Figure 4. Rearrangements at the membrane interface underlie desensitization in the α4β2 receptor.
a, Reference orientation of the α4β2 receptor. b–d, Superimpositions of whole pentamers based on alignment of transmembrane domains, showing local structural differences at the membrane interface. e–g, Superimpositions of whole pentamers based on alignment of extracellular domains, showing global differences in transmembrane domains. b,e, GlyR-open (orange) vs. GABAAR (magenta). c,f, GlyR-open vs. α4β2 structure (green, blue). d,g α4β2 vs. GABAAR.
Our structural analysis suggests that the α4β2 and GABAAR structures represent distinct desensitized states. Kinetically-distinct desensitized states are well described for both GABAA and nicotinic receptors27,28. The electrophysiology data for nicotine at the α4β2 receptor, and other studies of nicotine at the rat α4β2 receptor29, are consistent with a desensitized receptor; those presented with the GABAAR structure are potentially consistent with an intermediate or transitional state stabilized by the novel agonist benzamidine. We speculate that the extensive conformational rearrangements observed in the α4β2 receptor ECD-TMD interface further stabilize the receptor and thereby contribute to the increased affinity for agonist in the desensitized state7. This progression of quaternary rearrangements is illustrated in Figure 5. These interpretations are tentative as both of these structures were determined in the presence of detergent, removed from the native membrane environment known to be important for pentameric receptor function30. Additional Cys-loop receptor structures in desensitized states, and of nicotinic receptors in additional states, will help elucidate the detailed structural changes underlying desensitization.
Figure 5. Conformational changes underlying desensitization.
Cartoon illustrates the relative positions of ECD and TMDs in the α4β2 receptor compared to the open conformation of the glycine receptor and the desensitized conformation of the GABAA receptor.
Here we describe the X-ray structure of a nicotinic acetylcholine receptor, the heteropentameric α4β2 receptor. This structure of a heteromeric Cys-loop receptor sheds light on the architecture of the neurotransmitter site with bound nicotine and illustrates why the two other classes of binding sites are unable to bind classical nicotinic agonists. The receptor is locked in a non-conducting, desensitized conformation by the agonist nicotine. The α4β2 receptor conformation is strikingly distinct from prior structural information on a desensitized GABAA receptor, and thereby provides an important addition toward mapping the structural basis of allosteric gating in Cys-loop receptors.
Methods
Protein expression and purification
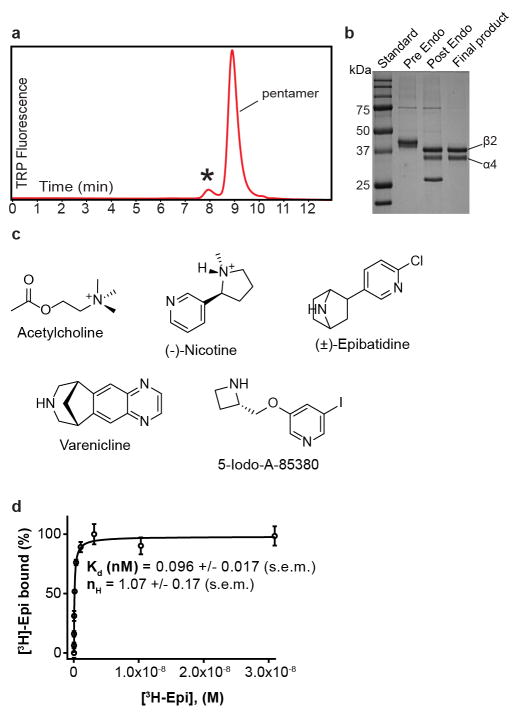
The human α4 and β2 nicotinic receptor genes were provided by Dr. Jon Lindstrom at the University of Pennsylvania. For the purposes of small-scale biochemical screening, a synthesized EGFP gene was spliced into the M3–M4 loop of each subunit and the genes were subcloned into the pEZT bacmam expression vector11. The EGFP fusion to one subunit was co-transfected into GnTI- HEK cells (ATCC CRL-3022) with a panel of deletion constructs for the partner subunit; a large number of constructs were screened in this manner for expression and pentameric monodispersity by Fluorescence-detection Size Exclusion Chromatography (FSEC)31. The final expression constructs for crystallization included the native signal peptides and residues 1–338 and 556–601 in the α4 subunit and residues 1–330 and 417–477 in the β2 subunit (residue numbering here is for the wild-type mature, signal-peptide-cleaved protein sequence). Deletion of the M3–M4 loop has been shown to not affect function in other Cys-loop receptor family members32. To promote crystallization a Glu-Arg linker was inserted in the MX-M4 junction, between Phe559-Ser560 in the α4 subunit and between Gln420-Ser421 in the β2 subunit. For purification purposes a Strep-tag was inserted at the C-terminus of the β2 subunit preceded by a Ser-Ala linker. Previously identified expression conditions resulted in a homogenous receptor subunit stoichiometry of two α4 and three β2 subunits11. For large-scale expression, 1.6 L of suspension GnTI- cells were transduced with multiplicities of infection (MOIs) of 0.25:0.5 for the α4 and β2 subunits, respectively. Nicotine (Sigma-Aldrich) and sodium butyrate (Sigma-Aldrich) were added at the time of transduction to 0.1 mM and 3 mM, respectively. At the time of transduction, suspension cells were moved to 30 °C and 8% CO2. After 72 hours, cells were collected by centrifugation, resuspended in 20 mM Tris, pH 7.4, 150 mM NaCl (TBS buffer), 1 mM nicotine and 1 mM phenylmethanesulfonyl fluoride (Sigma-Aldrich), and disrupted using an Avestin Emulsiflex. Lysed cells were centrifuged for 15 minutes at 10,000 g; supernatants containing membranes were centrifuged 2 hours at 186,000 g. Membrane pellets were mechanically homogenized and solubilized for 1 hour at 4 °C, in a solution containing TBS, 40 mM n-dodecyl-β-D-maltopyranoside (DDM; Anatrace), 1 mM nicotine and 0.2 mM cholesteryl hemisuccinate (CHS; Anatrace). Solubilized membranes were centrifuged for 40 minutes at 186,000 g then passed over high capacity Strep-Tactin (IBA) affinity resin. The resin was washed with Size Exclusion Chromatography (SEC) buffer containing TBS, 1 mM DDM, 1 mM nicotine, 0.2 mM CHS and 1 mM TCEP (Thermo Fisher Scientific) and eluted in the same buffer containing 5 mM desthiobiotin (Sigma-Aldrich). Peak elution fractions were concentrated and digested with Endoglycosidase H overnight in a 1:8 w:w ratio at 4 °C. This material was then injected over a Superose 6 10/300 GL column equilibrated in SEC buffer wherein DDM was replaced with 2 mM n-undecyl-β-D-maltopyranoside (Anatrace). Peak fractions were assayed by FSEC, monitoring tryptophan fluorescence, before pooling and concentrating for crystallization.
Crystallization, X-ray Data Collection and Structure Solution
Purified α4β2 was concentrated to 1.5–2.5 mg/mL in SEC buffer and crystallized by hanging drop vapor diffusion. The best-diffracting crystals of the nicotine-bound receptor were obtained after mixing protein with reservoir solution containing 0.05 M ADA pH 6.8, 12.5% PEG 1500 and 10% PEG 1000 in a 1:1 ratio and incubating over sealed wells containing 0.5 mL reservoir, at 14 °C. The crystals were cryoprotected with additional PEG 1000, PEG 1500 and ethylene glycol before flash freezing in liquid nitrogen. Crystals of the 5-Iodo-A-85380 (IA)33-bound receptor were obtained using the same approach, however the protein was purified in the absence of ligand, with IA added after SEC to a concentration of 0.5 mM. The best diffracting crystals of the IA complex were obtained at 14 °C using a reservoir solution of 0.05 M ADA pH 6.5 and 24% PEG 400; crystals were cryoprotected with additional PEG 400 before flash freezing in liquid nitrogen. X-ray data were collected at the 24-ID-C beamline at the Advanced Photon Source (Argonne, IL). Both datasets were collected from single crystals. The dataset from the IA complex was collected at low energy (7300 eV) to maximize anomalous signal from iodine in the ligand.
Diffraction datasets were integrated and scaled using HKL200034. The “Auto Corrections” option was used to assess anisotropic signal to noise, determine the resolution to use in refinement, and perform ellipsoidal truncation of the data as well as anisotropic B factor sharpening. The data from the nicotine complex were highly anisotropic, extending to ~3.6 Å in the best direction and ~4.5 Å in the worst. Electron density maps using the auto-corrected data contain far more features than the unmodified data and thus were used for all of the manual model building. However, truncated data from “auto corrections” suffer from low completeness in the high resolution shells. We thus used the UCLA diffraction anisotropy server35 to perform more conservative truncation and sharpening of the data; the deposited model underwent a final round of refinement against this truncated dataset to generate the statistics shown in Extended Data Table 1. The deposited structure factors include both sets of these truncated, sharpened data.
The structure of the nicotine-bound α4β2 receptor was solved by molecular replacement using a pentameric homology model based on the desensitized GABAA β3 receptor structure (PDB: 4COF)17, with models of the acetylcholine receptor α4 and β2 subunits generated using Swissmodel36. A panel of homology models was made comprising different orderings of subunits around the pentameric ring; the best molecular replacement search model had an ordering of α-β-β-α-β. Distinct electron density features, mainly in Loop C, provided the first convincing clues into subunit identity. Swapping positions of α4 and β2 subunits in the pentamer, followed by monitoring of R factors after refinement, supported the subunit assignment, however we sought additional validation. The potent agonist IA is expected to bind only in the canonical neurotransmitter site found at α-β interfaces. We exploited anomalous signal in a low resolution dataset of the α4β2-IA complex to independently validate subunit assignment. After rigid body refinement of the nicotine-bound model in this IA-complex dataset, strong anomalous difference peaks were observed: one in each of the two binding pockets that we had assigned as α4-β2 interfaces (4.5σ and 5.8σ) and similarly strong peaks near Cys-loop disulfides where four sulfur atoms are in close proximity. No anomalous difference signal was observed at the corresponding position in the β-α or β-β interfaces. Once the subunit arrangement was confirmed, iterative cycles of manual rebuilding in Coot37, jelly body refinement in Refmac38 and further restrained refinement in Phenix39 were performed. The Fitmunk server40 was used to identify improved side chain rotamers. Torsion-angle non-crystallographic symmetry restraints (α4 subunits and β2 subunits as separate groups), group B factors (one per residue) and TLS parameters (two groups per subunit) were used in refinement with Phenix.
The ECDs and TMDs were modeled with a high degree of confidence, with electron density visible for most side chains, one GlcNAc residue per subunit and two molecules of nicotine. One exception to the overall well-ordered ECD is the distal end of Loop C in the β2 subunits, which exhibited weak electron density in two of the three β subunits, and thus its modeling is tentative. A pancake-shaped difference electron density peak midway along the ion channel was modeled as a sodium ion coordinated by water molecules mediating H-bonds to the proximal threonine side chains. The sodium ion and water assignments are speculative; they were based on NaCl being the only salt present in purification and crystallization, the channel being selective for cations, B factors after refinement, and a similar arrangement of sodium and water in the high resolution structure of the bacterial pH-gated cation channel GLIC16. The register matches that of the AChBPs in the extracellular domain and the 5-HT3R, GABAAR, GlyR and GluCl structures in the transmembrane domain. Comparisons were also made with the Torpedo ACh receptor structure and were found to be different in register throughout much of the TMD, as previously described17,41–43. There was no observable electron density for 7 residues in the N-terminus of the α4 subunit, 11 and 15 residues linking the MX helix (following M3) to the M4 helix of the α4 and β2 subunits and 5 and 30 residues from the C-termini of the α4 and β2 subunits. While there was clear electron density for the MX helix, the observable density between M3 and M4 was disordered relative to the rest of the receptor leading to some ambiguity in modeling, in particular in the linker between the M3 helix and the MX helix. In the final refined model the MX helix register matches that observed in the 5-HT3R structure14. The five glutamate residues that define the pore constriction were not all well resolved. We modeled all five side chains in the same rotameric conformation based on convincing electron density for a subset. In the open state these glutamates are likely highly dynamic with heterogeneous conformations affecting conductance44.
Sequence alignments were made using PROMALS3D45. Ligand-receptor interactions were analyzed with areaimol in the CCP4 suite46,47 and the CaPTURE program48. Structural superpositions were made using Superpose49 in the CCP4 suite. Subunit interfaces were analyzed using the PDBe-PISA server50. Pore diameters were calculated using HOLE51. Structural figures were made with PyMOL (Schrödinger, LLC) including the APBS electrostatics plugin52. Crystallographic software packages were compiled by SBGrid53. Domain movements were analyzed using DynDom, http://fizz.cmp.uea.ac.uk/dyndom/.
Radioligand Binding
Experiments to measure binding of [3H]-epibatidine (PerkinElmer, 32.46 Ci/mmol) to the α4β2 receptor, as well as competition with other ligands, were performed with protein purified as for crystallization but in the absence of ligands. The concentration of binding sites was kept at 0.1 nM after a preliminary experiment to determine optimal receptor concentration. In addition to the protein, the binding assay conditions included 20 mM Tris pH 7.4, 150 mM NaCl, 1 mM DDM, and 1 mg/mL streptavidin-YiSi scintillation proximity assay beads (SPA; GE Healthcare Life Sciences). Non-specific signal was determined in the presence of 100 μM [1H]-nicotine; all data shown are from background-subtracted measurements. For competition assays [3H]-epibatidine concentration was fixed at 1 nM. All data were analyzed using Prism 6 software (GraphPad) with variable Hill slope. Ki values were calculated based on the experimentally determined Kd of 96 pM for [3H]-epibatidine.
Electrophysiology
To test the α4β2 receptor channel function, adherent GnTI- HEK cells were transfected with 0.5 μg of plasmid DNA for each subunit and 0.2 μg of a GFP expression plasmid using Lipofectamine 2000 (Thermo Fisher Scientific). The GFP expression plasmid was included to identify the cells for recording. After incubating for 72 hours at 30°C and 5% CO2 the cells were patched using the whole-cell configuration and clamped at a membrane potential of −90 mV. The recordings were made with an Axopatch 200B amplifier, low-pass filtered at 5 kHz and digitized at 10 kHz using the Digidata 1440A and pClamp 10 software (Molecular Devices). Borosilicate glass pipettes (King Precision Glass, Inc) were pulled and polished to 2–4 MΩ resistance. The bath solution contained (in mM): 140 NaCl, 2.4 KCl, 4 CaCl2, 4 MgCl2, 10 Hepes pH 7.3 and 10 glucose. The pipette solution contained (in mM): 150 CsF, 10 NaCl, 10 EGTA, 20 Hepes pH 7.3. The acetylcholine chloride (Sigma-Aldrich) and nicotine solutions were prepared in bath solution. Solution exchange was achieved using a gravity driven RSC-200 rapid solution changer (Bio-Logic).
Extended Data
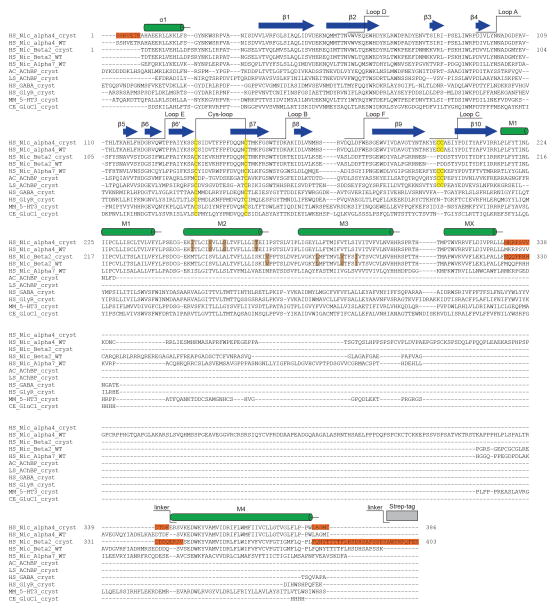
Extended Data Figure 1. Sequence alignment of α4β2 receptor with other Cys-loop receptors and AChBPs.
Sequences are numbered starting with the first amino acid in the mature protein. NCBI GI accession numbers are provided for full-length proteins and PDB codes for sequences from crystal structures. Human α4 nAChR (29891586), human β2 nAChR (29891594), human α7 nAChR (29891592), Aplysia californica AChBP (2WN9)54, Lymnaea stagnalis AChBP (1UW6)55, human GABAA β3 (4COF)17, human glycine α3 (5CFB)56, Mus musculus 5-HT3 receptor (4PIR)14 and Caenorhabditis elegans α (3RHW)41. Secondary structure, binding pocket loops and other selected structural elements are labeled. Disulfide bonds are highlighted in yellow and residues that lacked electron density and are not present in the model are highlighted in orange. Residues with mutations linked to autosomal dominant nocturnal frontal lobe epilepsy are highlighted in brown.
Extended Data Figure 2. Biochemical analysis.
a, FSEC trace of the α4β2 nicotinic receptor. The protein sample used for crystallization was tested by FSEC using an SRT SEC-500 column (0.35 mL/min) monitoring tryptophan fluorescence. The receptor exhibited time-dependent oligomerization/aggregation indicated by an asterisk. Pentamer indicates the elution peak of the heteropentameric assembly. b, SDS-PAGE stained with coomassie of the stages of receptor purification. c, Chemical structures of ligands used in crystallization, electrophysiology and binding assays. d, Saturation binding experiments with [3H]-epibatidine. Binding affinity (Kd) was calculated using the one site binding with variable slope equation in Graphpad Prism. The published range for epibatidine Ki, for reference, is 0.042–0.150 nM (all published values in paper are from a pharmacological review57). The experiment was performed in triplicate. Error bars are s.e.m. and nH is the Hill coefficient.
Extended Data Figure 3. Electron density quality.
a and b, 2Fo-Fc electron density maps of Loop C from an α4 and β2 subunit, respectively (contoured at 1 σ), with reference residues indicated. Perspective is from inside binding pocket looking toward receptor periphery. c, View down the channel axis toward the cyotosol. Anomalous difference peaks from co-crystallization with 5-Iodo-A-85380 are shown as red mesh and contoured at 3 σ. No detectable anomalous signal was present in other interfacial pockets. d, Stereo pair of 2Fo–Fc electron density maps (contoured at 1.5 σ) from an interface of α4 and β2 subunits. e, 2Fo–Fc electron density map of an α4 subunit M2 α-helix (contoured at 1.5 σ). Reference residues in the M2 helix are indicated. f, Stereo pair of Fo–Fc omit maps (contoured at 2 σ) of selected residues and nicotine in the neurotransmitter binding pocket. Residues and ligand omitted from map calculation are labeled. g, Fo–Fc omit map (contoured at 2 σ) for nicotine in the α-β interface. h–i, Fo–Fc omit map (contoured at 2 σ) of the ion and waters in the pore. The Na+ ion (purple) and water (red) are represented as spheres. The nearest residues on the M2 α-helices are indicated.
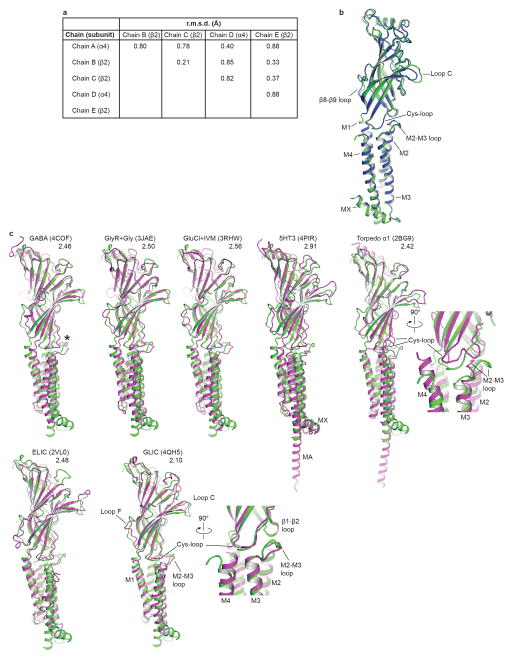
Extended Data Figure 4. Structural superimpositions.
a, Cα atom r.m.s.d. from pairwise superimpositions of all α4 and β2 chains. b, Backbone comparison of the α4 (green) and β2 (blue) subunits. c, Superimpositions of subunits of representative pentameric ligand gated ion channel structures (magenta) on the chain A α4 subunit (green). PDB codes and Cα r.m.s.d. are listed. Asterisk indicates bulging caused by inserted leucine residue found in the M2–M3 loops of α4 and β2 subunits relative to other receptors shown here (this loop was unmodeled in the 5-HT3R structure, however that protein has the same loop length as α4 and β2). The most similar subunit structure overall to α4 is GLIC, which has been thought to represent an open state, however studies on its desensitization properties58–60 and comparison to the α4β2 receptor structure here and in Extended Data Fig. 8 suggest it may rather represent a desensitized conformation. Conversely, the Torpedo nicotinic receptor structure, while clearly adopting the same overall fold, aligns less well structurally with α4 than does GLIC. This difference may relate to the Torpedo receptor being in a closed-resting state; notable differences in the backbone conformation of the Torpedo M2–M3 and Cys-loops (inset) compared to all other structures are less straightforward to interpret.
Extended Data Figure 5. Detailed interface interactions.
a–c, Views parallel and perpendicular to the plasma membrane coloring potential van der Waals (gray), H-bonds (orange) and electrostatic (pink) interactions in the subunits interface. Parallel views are from periphery of receptor. d–f, Close-up of the red boxes on the apical receptor surface. g–i, Close-up of the black boxes in the view parallel to the plasma membrane. j–l, Close-up of the yellow boxes in the view parallel to the plasma membrane. Panels j–l highlight the N-capping of the M1 helix by a serine in the M2–M3 loop, an interaction seen in GlyR-closed, but absent in GlyR-open and GABAAR17,24. For simplicity, only the residues likely to be involved in forming H-bonds and electrostatic interactions are shown. These potential interactions are shown as dashed lines (2.4–3.9 Å). The subunit interfaces are predominantly stabilized through van der Waals interactions, with interspersed hot spots of hydrogen bonding and electrostatic interactions of known functional importance. The N-terminal helix of the receptor is important in pentameric assembly and mutations in this region of other pentameric receptors results in disease17. Loop C is essential for orthosteric ligand binding, the M2–M3 loop is critical for allosteric signal transduction7, and residues at the apex of M1 and at the intracellular base of the pore are known to affect desensitization25,61.
Extended Data Figure 6. Determinants of nicotine binding.
a, Sequence alignment of loops implicated in nicotine binding. The human nicotinic α1 (NCBI GI accession number:87567783), β1 (41327726), γ (61743914), δ (4557461) and ε (4557463) subunits were added to the sequence alignment. Residues making contact with nicotine or stabilizing the binding pocket indirectly are highlighted in yellow and brown, respectively. Determinants indirectly affecting the receptor-nicotine cation-π interaction are highlighted in blue. b, Close-up of the α4β2 nicotinic receptor binding pocket. c, Close-up of the corresponding region in AChBP (PDB: 1UW6)55. The water in the AChBP pocket is represented as a red sphere and forms a hydrogen bond between the pyridine nitrogen on nicotine and the protein backbone. Potential hydrogen bonding and cation-π interactions are represented as dashed lines (2.7–5 Å).
Extended Data Figure 7. Cys-loop receptor ion channel conformations.
a, Sequence alignment of the M2 α-helices. Residues lining the α4β2 receptor pore are highlighted in yellow and the residues lining the pores of GlyR (closed: 3JAD; open: 3JAE)24, GLIC (4QH5)62 and GABAAR (4COF)17 are highlighted in blue. b–e, View of the M2 α-helices from opposing subunits with side chains shown for pore-lining residues. The blue and yellow spheres represent the internal surface of the transmembrane ion channel. Blue spheres are pore diameters >5.6 Å; yellow are >2.8 Å and <5.6 Å and pink are <2.8 Å.
Extended Data Figure 8. Comparison of Cys-loop receptor conformational states.
a, View parallel to the plasma membrane of a superposition of the α4 subunit (green) ECD with the GABAAR (magenta) and GlyR-open (orange) and GlyR-closed (cyan). b, View parallel to the plasma membrane of a superposition of the TMDs. Asterisk indicates an inserted leucine in the M2–M3 loop of α4β2, which is conserved in 5-HT3 receptors. In the high-resolution structure of the 5-HT3R, the majority of the M2–M3 loop including the leucine of interest is not modeled, precluding comparison of the two structures for this analysis. c, Table of Cα r.m.s.d. values between isolated regions of one subunit per structure. d–e, View down the channel axis from the synaptic cleft toward the cyotosol of a superposition of the receptors based on alignments of the TMDs. f–g, Analysis of intrasubunit rotation angles between different conformational states. Rotation axes indicated by yellow bar. In f, the ECD of GlyR-open was superposed on the ECD of α4 and relative displacement of the TMD is shown. In g, the TMD of GABAAR was superposed on the TMD of α4 and relative displacement of the ECD is shown.
Extended Data Table 1.
Data collection and refinement statistics.
| Dataset | Nicotine | 5-lodo-A-85380# |
|---|---|---|
| Data collection | ||
|
| ||
| Space group | P212121 | P212121 |
| Resolution (Å)* | 40.00–3.94(4.01–3.94) | 30.00–6.50(6.61–6.50) |
| Wavelength (Å) | 0.9791 | 1.6984 |
| Cell dimensions a, b ,c (Å)¥ | 127.1, 132.6,202.4 | 128.1, 133.6, 205.6 |
| Number of unique reflections | 30759 | 7259 |
| Completeness (%)* | 99.5 (97.8) | 99.2 (100) |
| Redundancy* | 9.1 (7.5) | 6.3 (6.5) |
| I/σ(I)* | 14.9(1.1) | 19.4(1.5) |
| CC1/2 in the last shell | 0.547 | 0.528 |
|
| ||
| Refinement | ||
|
| ||
| Resolution (Å)* | 25.00–3.94 (4.08–3.94) | |
| Number of reflections (test set) | 26,718(1,330) | |
| Completeness (%)* | 86.8 (33) | |
| Rwork/Rfree (%) | 28.5/30.7 | |
| Number of non-H atoms | 14,805 | |
| Mean B factors (Å2) | ||
| Protein | 170 | |
| Ligand/carbohydrate | 147 | |
| Water/ion | 74 | |
| r.m.s.d. values | ||
| Bond lengths (Å) | 0.003 | |
| Bond angles (°) | 0.745 | |
| Ramachandran analysis | ||
| Favored (%) | 93.8 | |
| Outliers (%) | 0 | |
| Molprobity score | 2.47 (99th percentile) | |
This dataset is of low resolution and was only used to generate anomalous difference maps.
Values in parentheses are for the highest resolution shell.
All angles = 90°
Extended Data Table 2.
Surface areas buried at subunit interfaces. a,Buried area at subunit interfaces in the α4β2 receptor and other pentameric receptors. The 5-HT3R structure contains an extra section of the intracellular domain (Extended Data Fig. 4c), which accounts for its larger subunit interface area. Glycine receptor structures include two from cryo-EM studies (3JAE and 3JAD in the open and resting states, respectively)24 and one from X-ray crystallography in the resting state (5CFB)56. b, Surface areas buried by only Loop C. We analyzed inter-subunit interactions in the α4β2 receptor to investigate mechanisms underlying heteromeric receptor assembly. The crystal structure of the receptor reveals three classes of subunit interfaces: α-β, β-β and β-α. All three interface types in the receptor are comparable in terms of surface area buried to the most tightly packed Cys-loop receptor structures. Of the three interface classes in the α4β2 receptor, the α-β interface is the most extensive; the majority of this difference is provided by Loop C, which is significantly longer in the α subunit and forms extensive contacts with the neighboring β subunit (Extended Data Fig. 5g–i). Among the pentameric receptors of known structure, the α4β2 nicotinic receptor is closest in sequence and function to the Torpedo nicotinic receptor5. We compared backbone conformations and inter-subunit interactions between these two structures (Extended Data Figs. 4c). We found that the α4β2 receptor conformation is more similar to other eukaryotic receptors and the bacterial receptor GLIC than to the Torpedo receptor. We additionally observed that subunit interfaces are much more loosely packed in the Torpedo receptor structure. Due to these differences and to a previously-described register inconsistency in its transmembrane domain17,24,41–43 we limited our further structural comparisons with the Torpedo nicotinic receptor.
| a | ||
|---|---|---|
| Structure (PDB ID) | Interface; area (Å2) | |
|
| ||
| (+) subunit | (−) subunit | |
|
| ||
| α4β2 [α-β interface] | 2820 | 2806 |
| α4β2 [β-β interface] | 2501 | 2575 |
| α4β2 [β-α interface] | 2544 | 2561 |
| nAChR [α-γ interface] (2BG9) | 1665 | 1658 |
| nAChR [α-δ interface] (2BG9) | 1308 | 1300 |
| nAChR [β-α interface] (2BG9) | 1426 | 1401 |
| nAChR [γ-α interface] (2BG9) | 1684 | 1714 |
| nAChR [δ-β interface] (2BG9) | 1858 | 1842 |
| 5-HT3R (4PIR) | 3125 | 3012 |
| GABAAR (4COF) | 2560 | 2621 |
| GlyR + gly (3JAE) | 1708 | 1760 |
| GlyR + strychnine (3JAD) | 2155 | 2137 |
| GlyR + strychnine (5CFB) | 2214 | 2273 |
| GluCI (3RHW) | 2231 | 2298 |
| GLIC (4HFI) | 2215 | 2121 |
| ELIC (2VL0) | 2593 | 2474 |
| b | ||
|---|---|---|
| Structure (PDB ID) | Loop C interface area (Å2) | |
|
| ||
| (+) subunit | (−) subunit | |
|
| ||
| α4β2 [α-β interface] | 234 | 249 |
| α4β2 [β-β interface] | 31 | 34 |
| α4β2 [β-α interface] | 31 | 34 |
Acknowledgments
We thank Jon Lindstrom at the University of Pennsylvania for providing the α4 and β2 receptor genes, Dominika Borek and Zbyszek Otwinowski for guidance in structural analyses, José Cabrera for assistance with illustrations, and members of the Hibbs lab for comments on the manuscript. X-ray diffraction experiments at the Argonne National Laboratory’s Advanced Photon Source 24-ID-C beamline were supported by the NIH (GM103403 and RR029205) and the DOE (DE-AC02-06CH11357). This research project was supported by an NIH training grant (T32 NS069562) and an HHMI Gilliam Fellowship to C.L.M.P.; R.E.H. is supported by a McKnight Scholar Award, a Klingenstein-Simons Fellowship Award in the Neurosciences, The Welch Foundation (I-1812), The Friends of the Alzheimer’s Disease Center and the NIH (DA037492, DA042072, and NS077983).
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions: CLMP, CMN and REH contributed to all aspects of the project.
Atomic coordinates and structure factors are deposited with the Protein Data Bank under the code 5KXI for the α4β2 + nicotine structure.
We declare no competing financial interests.
References
- 1.Langley JN. On the reaction of cells and of nerve-endings to certain poisons, chiefly as regards the reaction of striated muscle to nicotine and to curari. The Journal of physiology. 1905;33:374–413. doi: 10.1113/jphysiol.1905.sp001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Changeux JP, Edelstein SJ. Nicotinic Acetylcholine Receptors: From Molecular Biology to Cognition. Odile Jacob Publishing Corporation; 2005. [Google Scholar]
- 3.Engel AG, Shen XM, Selcen D, Sine SM. Congenital myasthenic syndromes: pathogenesis, diagnosis, and treatment. The Lancet. Neurology. 2015;14:420–434. doi: 10.1016/S1474-4422(14)70201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becchetti A, Aracri P, Meneghini S, Brusco S, Amadeo A. The role of nicotinic acetylcholine receptors in autosomal dominant nocturnal frontal lobe epilepsy. Frontiers in physiology. 2015;6:22. doi: 10.3389/fphys.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. Journal of molecular biology. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- 6.Rucktooa P, Smit AB, Sixma TK. Insight in nAChR subtype selectivity from AChBP crystal structures. Biochem Pharmacol. 2009;78:777–787. doi: 10.1016/j.bcp.2009.06.098. [DOI] [PubMed] [Google Scholar]
- 7.Nemecz A, Prevost MS, Menny A, Corringer PJ. Emerging Molecular Mechanisms of Signal Transduction in Pentameric Ligand-Gated Ion Channels. Neuron. 2016;90:452–470. doi: 10.1016/j.neuron.2016.03.032. [DOI] [PubMed] [Google Scholar]
- 8.Nelson ME, Kuryatov A, Choi CH, Zhou Y, Lindstrom J. Alternate stoichiometries of alpha4beta2 nicotinic acetylcholine receptors. Molecular pharmacology. 2003;63:332–341. doi: 10.1124/mol.63.2.332. [DOI] [PubMed] [Google Scholar]
- 9.Tapia L, Kuryatov A, Lindstrom J. Ca2+ permeability of the (alpha4)3(beta2)2 stoichiometry greatly exceeds that of (alpha4)2(beta2)3 human acetylcholine receptors. Molecular pharmacology. 2007;71:769–776. doi: 10.1124/mol.106.030445. [DOI] [PubMed] [Google Scholar]
- 10.Lester HA, et al. Nicotine is a selective pharmacological chaperone of acetylcholine receptor number and stoichiometry. Implications for drug discovery. The AAPS journal. 2009;11:167–177. doi: 10.1208/s12248-009-9090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morales-Perez CL, Noviello CM, Hibbs RE. Manipulation of Subunit Stoichiometry in Heteromeric Membrane Proteins. Structure. 2016 doi: 10.1016/j.str.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zwart R, et al. 5-I A-85380 and TC-2559 differentially activate heterologously expressed alpha4beta2 nicotinic receptors. European journal of pharmacology. 2006;539:10–17. doi: 10.1016/j.ejphar.2006.03.077. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y, et al. Human alpha4beta2 acetylcholine receptors formed from linked subunits. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23:9004–9015. doi: 10.1523/JNEUROSCI.23-27-09004.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassaine G, et al. X-ray structure of the mouse serotonin 5-HT3 receptor. Nature. 2014;512:276–281. doi: 10.1038/nature13552. [DOI] [PubMed] [Google Scholar]
- 15.Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nature reviews. Neuroscience. 2002;3:102–114. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- 16.Sauguet L, et al. Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels. The EMBO journal. 2013;32:728–741. doi: 10.1038/emboj.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller PS, Aricescu AR. Crystal structure of a human GABAA receptor. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picciotto MR, et al. Acetylcholine receptors containing the beta2 subunit are involved in the reinforcing properties of nicotine. Nature. 1998;391:173–177. doi: 10.1038/34413. [DOI] [PubMed] [Google Scholar]
- 19.Tapper AR, et al. Nicotine activation of alpha4* receptors: sufficient for reward, tolerance, and sensitization. Science. 2004;306:1029–1032. doi: 10.1126/science.1099420. [DOI] [PubMed] [Google Scholar]
- 20.Dougherty DA. The cation-pi interaction. Accounts of chemical research. 2013;46:885–893. doi: 10.1021/ar300265y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mazzaferro S, et al. Additional acetylcholine (ACh) binding site at alpha4/alpha4 interface of (alpha4beta2)2alpha4 nicotinic receptor influences agonist sensitivity. The Journal of biological chemistry. 2011;286:31043–31054. doi: 10.1074/jbc.M111.262014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang J. Ion permeation through 5-hydroxytryptamine-gated channels in neuroblastoma N18 cells. The Journal of general physiology. 1990;96:1177–1198. doi: 10.1085/jgp.96.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dwyer TM, Adams DJ, Hille B. The permeability of the endplate channel to organic cations in frog muscle. The Journal of general physiology. 1980;75:469–492. doi: 10.1085/jgp.75.5.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du J, Lu W, Wu S, Cheng Y, Gouaux E. Glycine receptor mechanism elucidated by electron cryo-microscopy. Nature. 2015;526:224–229. doi: 10.1038/nature14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gielen M, Thomas P, Smart TG. The desensitization gate of inhibitory Cys-loop receptors. Nature communications. 2015;6:6829. doi: 10.1038/ncomms7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Althoff T, Hibbs RE, Banerjee S, Gouaux E. X-ray structures of GluCl in apo states reveal a gating mechanism of Cys-loop receptors. Nature. 2014;512:333–337. doi: 10.1038/nature13669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Celentano JJ, Wong RK. Multiphasic desensitization of the GABAA receptor in outside-out patches. Biophysical journal. 1994;66:1039–1050. doi: 10.1016/S0006-3495(94)80885-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lena C, Changeux JP. Allosteric modulations of the nicotinic acetylcholine receptor. Trends Neurosci. 1993;16:181–186. doi: 10.1016/0166-2236(93)90150-k. [DOI] [PubMed] [Google Scholar]
- 29.Paradiso KG, Steinbach JH. Nicotine is highly effective at producing desensitization of rat alpha4beta2 neuronal nicotinic receptors. The Journal of physiology. 2003;553:857–871. doi: 10.1113/jphysiol.2003.053447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Labriola JM, et al. Structural sensitivity of a prokaryotic pentameric ligand-gated ion channel to its membrane environment. The Journal of biological chemistry. 2013;288:11294–11303. doi: 10.1074/jbc.M113.458133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawate T, Gouaux E. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 2006;14:673–681. doi: 10.1016/j.str.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 32.Jansen M, Bali M, Akabas MH. Modular design of Cys-loop ligand-gated ion channels: functional 5-HT3 and GABA rho1 receptors lacking the large cytoplasmic M3M4 loop. The Journal of general physiology. 2008;131:137–146. doi: 10.1085/jgp.200709896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukhin AG, et al. 5-Iodo-A-85380, an alpha4beta2 subtype-selective ligand for nicotinic acetylcholine receptors. Molecular pharmacology. 2000;57:642–649. doi: 10.1124/mol.57.3.642. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 35.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biasini M, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research. 2014;42:W252–258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica. Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta crystallographica. Section D, Biological crystallography. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Porebski PJ, Cymborowski M, Pasenkiewicz-Gierula M, Minor W. Fitmunk: improving protein structures by accurate, automatic modeling of side-chain conformations. Acta crystallographica. Section D, Structural biology. 2016;72:266–280. doi: 10.1107/S2059798315024730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hibbs RE, Gouaux E. Principles of activation and permeation in an anion-selective Cys-loop receptor. Nature. 2011;474:54–60. doi: 10.1038/nature10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mnatsakanyan N, Jansen M. Experimental determination of the vertical alignment between the second and third transmembrane segments of muscle nicotinic acetylcholine receptors. Journal of neurochemistry. 2013;125:843–854. doi: 10.1111/jnc.12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corringer PJ, et al. Atomic structure and dynamics of pentameric ligand-gated ion channels: new insight from bacterial homologues. The Journal of physiology. 2010;588:565–572. doi: 10.1113/jphysiol.2009.183160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cymes GD, Grosman C. The unanticipated complexity of the selectivity-filter glutamates of nicotinic receptors. Nature chemical biology. 2012;8:975–981. doi: 10.1038/nchembio.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pei J, Grishin NV. PROMALS3D: multiple protein sequence alignment enhanced with evolutionary and three-dimensional structural information. Methods in molecular biology. 2014;1079:263–271. doi: 10.1007/978-1-62703-646-7_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee B, Richards FM. The interpretation of protein structures: estimation of static accessibility. Journal of molecular biology. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 47.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta crystallographica. Section D, Biological crystallography. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 50.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 51.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. Journal of molecular graphics. 1996;14:354–360. 376. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 52.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morin A, et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hibbs RE, et al. Structural determinants for interaction of partial agonists with acetylcholine binding protein and neuronal alpha7 nicotinic acetylcholine receptor. The EMBO journal. 2009;28:3040–3051. doi: 10.1038/emboj.2009.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Celie PH, et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- 56.Huang X, Chen H, Michelsen K, Schneider S, Shaffer PL. Crystal structure of human glycine receptor-alpha3 bound to antagonist strychnine. Nature. 2015;526:277–280. doi: 10.1038/nature14972. [DOI] [PubMed] [Google Scholar]
- 57.Wonnacott S, Barik J. Nicotinic ACh Receptors. Tocris Reviews. 2007;28:1–20. [Google Scholar]
- 58.Gonzalez-Gutierrez G, Grosman C. Bridging the gap between structural models of nicotinic receptor superfamily ion channels and their corresponding functional states. Journal of molecular biology. 2010;403:693–705. doi: 10.1016/j.jmb.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parikh RB, Bali M, Akabas MH. Structure of the M2 transmembrane segment of GLIC, a prokaryotic Cys loop receptor homologue from Gloeobacter violaceus, probed by substituted cysteine accessibility. The Journal of biological chemistry. 2011;286:14098–14109. doi: 10.1074/jbc.M111.221895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laha KT, Ghosh B, Czajkowski C. Macroscopic kinetics of pentameric ligand gated ion channels: comparisons between two prokaryotic channels and one eukaryotic channel. PLoS One. 2013;8:e80322. doi: 10.1371/journal.pone.0080322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouzat C, Bartos M, Corradi J, Sine SM. The interface between extracellular and transmembrane domains of homomeric Cys-loop receptors governs open-channel lifetime and rate of desensitization. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2008;28:7808–7819. doi: 10.1523/JNEUROSCI.0448-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fourati Z, Sauguet L, Delarue M. Genuine open form of the pentameric ligand-gated ion channel GLIC. Acta crystallographica. Section D, Biological crystallography. 2015;71:454–460. doi: 10.1107/S1399004714026698. [DOI] [PubMed] [Google Scholar]