Abstract
Glycosylation is a ubiquitous mammalian post-translational modification that both decorates a majority of expressed proteins and regulates their function. Cellular glycan biosynthesis is facilitated by a few hundred enzymes that are collectively termed ‘glycoenzymes’. The expression and activity of these enzymes is controlled at the transcription, translation and post-translation levels. New wet-lab advances are providing analytical methods to collect large-scale data at these multiple levels, relational databases are starting to collate these results, and computer models are beginning to integrate this information across scales in order to gain new knowledge. These activities are likely to enable the qualitative and quantitative mapping of pathways regulating glycan production and function in proteins, cells and tissue.
Graphical Abstract
Introduction
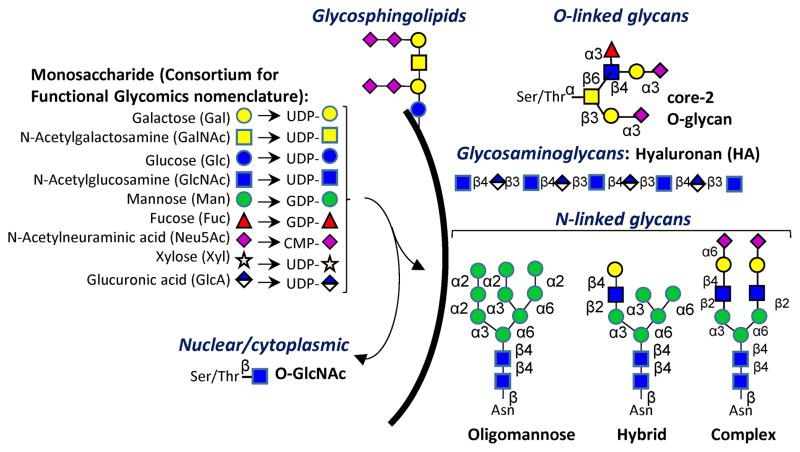
Cell surface, secreted and intracellular proteins and lipids are extensively glycosylated. These modifications affect a range of molecular recognition and signaling events [1]. Whereas intracellular glycoproteins often contain short carbohydrate structures, commonly bearing a single O-linked GlcNAc (N-Acetyl glucosamine) residue, extracellular glycans bear more elaborate structures [2]. On human cells, these include branched O- and N-linked glycans, glycolipids and repeating glycosaminoglycans (Figure 1). These sugars are commonly made up of nine different monosaccharides. Altogether, human glycoproteins and glycolipids are estimated to contain a minimum of 3,000 unique 2–5 monosaccharide determinants [3]. Individual cell and tissue only have a subset of the glycoenzymes and thus individual glycome typically have fewer members. Similarly, glycosaminoglycans are estimated to contain up to ~4,000 possible pentasaccharide sequences [3]. These determinants provide the recognition motifs underlying the biological functions of glycans.
Figure 1. Glycosylation overview.
Commonly, nine types of monosaccharides in humans form corresponding sugar-nucleotides (UDP-Gal etc.). A variety of glycoenzymes (particularly the glycosyltransferases and glycosidases) enable the transfer of these monosaccharides to lipid and protein scaffolds. Additional monosaccharides can form, e.g. Iduronoic acid (IdoA), by the epimerization of Glucuronic acid (GlcA) on glycosaminoglycan chains. Carbohydrate structures thus formed decorate the cell surface, and they are also found in intra-cellular compartments. Most glycan families have a limited number of core-structures that are elaborated by repeat extensions (like N-Acetyllactosamine repeats) and terminal modifications.
Classical studies into the biosynthesis of glycan epitopes use reductionist, biochemical methods to elaborate individual biosynthetic steps. In recent years, however, newer ‘omics’ approaches are rapidly bridging gaps in our knowledge of glycan biosynthesis, especially with respect to cell-type and context-specific glycosylation [4–7]. These newer experimental modalities often use mass spectrometry and lectin microarray based technologies [8]. Integrating these glycomics data sets with high-throughput genomics, transcriptomics, enzymology and proteomics methods is necessary as it can lead to a ‘systems level’ understanding of glycan biosynthetic pathways [9]. This review describes progress and challenges in the development of such systems-based understanding, with emphasis on computer based modeling and pathways regulating canonical O- and N-linked glycosylation in humans.
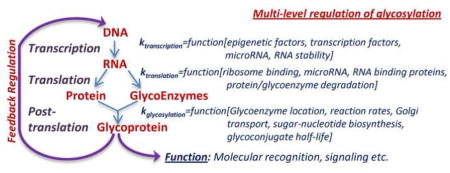
Multi-level regulation of glycosylation
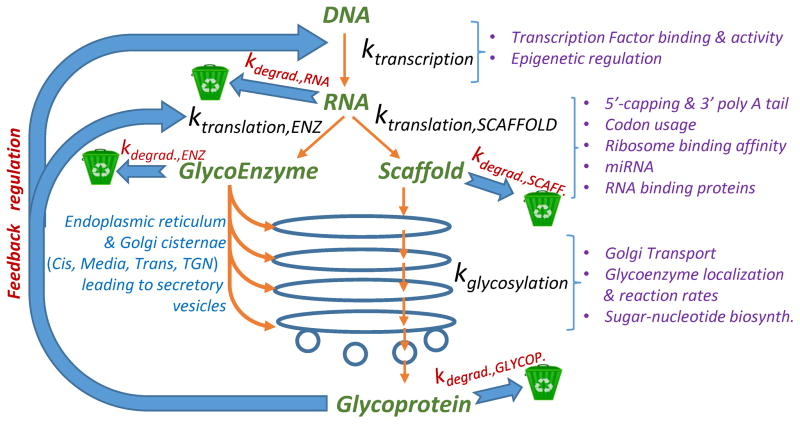
The regulation of glycoproteoforms (i.e. the glycosylation variants of a protein) in metazoans is complex. It involves not only the biosynthesis of the underlying protein scaffold built on its mRNA template, but also non-template driven post-translational modifications that is driven by the glycosylation-related enzymes or ‘glycoenzymes’. In humans, ~500 glycoenzymes participate in this process including 209 glycosyltransferases, 76 glycosidases, 114 enzymes involved in sugar metabolism & transport, 54 sulfation-related enzymes and 31 enzymes regulating additional lipid and GPI-anchor biosynthesis pathways [10,11]. Regulation of protein scaffolds and glycoenzymes (themselves proteins) underlying glycoproteoforms occurs at multiple steps, including transcriptional, translational and post-translational levels (Figure 2).
Figure 2. Regulation of glycosylation.
The central dogma of molecular biology states the passage of information from DNA to RNA to protein in all living organisms. These proteins can be further glycosylated by the glycoenzymes, mainly in the endoplasmic reticulum and Golgi compartments. RNA and protein degradation rates, shown using recycle bins, are important checkpoints that control cell surface carbohydrate structures. Additional parameters that track the process of information transfer from genes to RNA to proteins (including glycoenzymes) to glycoproteins are shown using purple text. Glycoproteins may provide feedback and feedforward control to regulate cellular transcription and translation. The parameter ‘k’ (e.g. ktranscription and kdegrad.) is used to denote lumped rate constants for individual steps.
Transcription is perhaps the most recognized regulator of protein, and thus glycoproteoform, expression. Transcription of a glycosylation-related gene (glycogene) is controlled by both transcription factors binding to gene promoter and enhancer elements, and by epigenetic regulation of gene accessibility [12]. Transcription factors, which can act as promoters or repressors, are often glycosylated by O-GlcNAc which can regulate the transcriptional machinery [2]. In general, there is little available information about transcription factor-mediated control of glycogene expression, with only ~2 dozen glycogene-transcription factor interactions known to date and mechanistic work available for ~15 glycogenes [13–16]. The importance of transcription factors in control of glycosylation is exemplified by recent work on the transcription factor XBP1s, a major regulator of the unfolded protein response (UPR) [15]. Here, the artificial activation of XBP1s induced expression of a series of N-glycan biosynthetic enzymes, shifting the glycosylation profile of a secreted glycoprotein from oligomannose towards more processed hybrid and complex epitopes, tying together the UPR with changes in the glycome [15]. In another example, the phosphorylated form of the transcription factor ATF2, which has a role in therapeutic resistant melanoma, was recently found to directly repress expression of fucokinase (FUK). This enzyme is a key component of the fucose salvage pathway and its repression by activated ATF2 resulted in ~40% loss of cellular fucosylation [16]. Beyond transcription factors, access to the genome, which is controlled by epigenetic modifications such as acetylation, methylation and ubiquitinylation also regulate glycogene transcription [12]. This is even less studied than transcription factor binding to glycogenes, although a recent study analyzing DNA methylation of 86 glycogenes suggests a positive correlation between the promoter methylation status of glycogenes and expected tumor-associated glycan structures [17]. A handful of studies have also shown a direct interaction between the methylation status of specific glycogenes and glycogene expression [18–21]. In this regard, the methylation of the α(2,3)sialyltransferase ST6Gal1 promoter was shown to silence expression of the ST6Gal1 transcript in bladder cancer [18], confirming an earlier report in breast cancer showing methylation-dependent silencing of this glycogene [22]. Mapping transcription factors and epigenetic regulation onto glycogene regulatory networks will require a combination of methods from traditional Chromatin Immunoprecipitation (ChIP) assays to more sophisticated promoter-reporter assays that can reveal indirect and direct effects, to more sophisticated analysis of genome accessibility and silencing [15,23]. All of this is important to our understanding of the glycome and its regulation.
Although transcript (i.e. cellular mRNA) levels are often used as a key indicator of what glycogenes are being translated and the levels of biosynthetic enzymes and glycans that result, the relationship is not straightforward. A number of investigators have attempted to relate transcript levels to glycoenzyme activity and carbohydrate structures, including the response of the system to perturbations [11,24–26]. Such studies have been conducted using human promyelocytic leukocytes differentiated to primary neutrophils [24], a limited panel of mouse tissue [11], mouse embryonic stem cells differentiated to embryoid bodies and endodermal cells [25], and epithelial ovarian cancer cell types [26]. The broad conclusion of these studies is that glycogene, and more specifically glycosyltransferase, expression measurements in a number of cases can semi-quantitatively predict corresponding enzyme activity and cell-surface glycan expression. However, the relationship is often non-linear and in some instances, changes in glycan structures upon perturbation do not correlate with changes in glycogene mRNA expression levels. Although part of this discrepancy may be due to difficulties with quantitatively measuring low-abundance glycogene transcript levels [11], the post-transcriptional regulation of glycogene expression may also control carbohydrate structures.
MicroRNAs (miRNAs), small non-coding RNAs, are major post-transcriptional regulators of protein expression, acting as rheostats to maintain key proteins critical to a cell state at set levels [27,28]. Genes with lower expression levels, such as glycogenes, are both preferentially targeted by miRNA and more impacted by miRNA binding [28]. Recent work has shown that miRNA are indeed a major regulator of the human glycome and the impact of miRNA on the glycome via alteration of glycosyltransferase protein levels may explain discrepancies with the transcriptome [4,29]. For example, miR-34a, a microRNA associated with hepatocellular carcinoma, targets FUT8. This miRNA did no impact FUT8 transcripts in human HepG2 cells, although it concomitantly altered FUT8 protein expression and core fucosylation levels [30]. miRNA regulate biological pathways by regulating sets of gene transcripts. Multiple laboratories have demonstrated that downregulation of single glycogene (or other gene) targets of miRNA mimic the biological effects of the miRNA [31–33]. Thus, miRNAs could be used as a proxy to identify glycosylation enzymes involved in the pathways they regulate (miRNA proxy approach) [32,33]. The miRNA proxy approach has been applied to identify functionally relevant glycogenes impacting cell states like epithelial-to-mesenchymal transition (EMT) [32] and the cell cycle [33]. For example, miR-200b, a regulator of EMT, was found to regulate a set of glycogenes from hard to analyze cohorts those involved in glycolipid (ST3GAL5 and ST6GALNAC5) and non-canonical O-glycan biosynthesis (B3GLCT). Downregulation of these glycogenes using shRNA phenocopied the miRNA, showing a role for these genes in the control of EMT [32]. This work opens up the possibility of leveraging post-transcriptional regulation of glycosylation as a means to reveal critical functional changes in the glycome that underlie biological processes. This may be especially important for the portions of the glycome, such as glycosylaminoglycans and glycolipids, which are overlooked in most analysis due to analytical challenges.
In addition to transcription, the measurement of glycoenzyme translation, turnover rates and activity are key parameters controlling glycan biosynthesis. In this regard, newer technologies like ribosome profiling are enabling a more nuanced understanding of protein synthesis [31]. This technology uses deep sequencing to identify stretches of mRNA that are actively bound to the ribosome during in vivo translation. Such quantitative estimation of protein biosynthesis rates may provide a new avenue for evaluating translation, not just transcription, as a measure of glycoenzyme expression, taking into account posttranscriptional regulation by miRNA.
Beyond expression, the direct measurement of glycoenzyme activity in situ is important. However, currently there is no established method for this measurement in the milieu of the Golgi. In complex mixtures, enzyme activity is typically followed in reaction vessels by assaying the conversion of a synthetic acceptor to product using cell/tissue lysates as the enzyme source. Product quantitation methods range from radioactivity based chromatography assays [34], to fluorescence based strategies that quantify glycan biosynthesis in an array format [35,36] and mass spectrometry [37]. Here, solution based enzymology assays are more sensitive compared to studies performed in 2D-array format, since only limited amounts of the carbohydrate acceptor can be immobilized on an array spot. As an alternative, mass spectrometry can be used to follow glycan biosynthesis in cells by feeding them small molecule acceptors, such as per-acetylated Benzyl-α-GalNAc, and subsequently measuring carbohydrate products formed on the synthetic substrate [38]. Data from such experiments, in tandem with genome editing methods (TALEN, CRISPR-Cas9 etc.), may enable estimation of glycosyltransferase activities and dissection of the interdependencies among glycogenes in the same biosynthetic pathway [5,39].
Overall, high-throughput analysis of the transcriptome is starting to provide clues to the nature of the cellular glycome and its response to system perturbation. Integration of additional data including miRNA, glycoenzyme expression and activity is necessary in order to obtain a more complete picture of the glycan regulatory process.
Integrating knowledge using computer models of glycosylation reaction networks
The analysis of large-scale experimental datasets available from the above technologies may be streamlined by developing computational tools that can integrate results collected using these diverse methods. While such computational tools have been widely developed for genomics and proteomics research, they lag behind in the glycosciences [9,40]. This is in part due to the greater complexity of this research field, since glycoconjugate biosynthesis is regulated by multiple pathways including: i. sugar-nucleotide donor biosynthesis (e.g. UDP-Galactose) in the cellular cytoplasmic and nuclear compartments; ii. glycosylation reactions in the endoplasmic reticulum (ER) and Golgi cisternae; iii. transport processes that result in the sub-cellular localization of glycoenzymes, glycoconjugates and other reaction components in specific compartments; iv. trafficking of glycoproteins and lipids through the ER and Golgi which is impacted by cellular lectins and other transport mechanisms; and v. salvage pathways that aid the recycling of glycoconjugates back to their basic building blocks (Figure 2).
Integration of data across scales requires a robust modeling framework to formulate glycosylation reaction networks, integrate data from different relational databases, and enable code sharing among laboratories. Due to the last requirement, the Systems Biology Markup Language provides the ideal schema for model building [41,42]. Although this modeling language does not have specific facilities to handle glycan structures and glycosylation related reaction pathways, the annotation fields of the eXtensible Markup Language (XML) files can be easily modified to store pertinent information. For example, the annotation element of the Species field could store glycan structure information in XML format, the Reaction field can hold glycoenzyme and carbohydrate transport definitions/data, and Compartment is available to describe the physical features and reaction mechanisms in the ER and Golgi cisternae [42,43].
Some initial examples of glycosylation pathway modeling have appeared, and these investigations suggest that quantitative analysis of glycan structure data using computer modeling can yield novel biological insight. Notably, the computer models of Lau et al. showed that the activities of various N-acetylglucosaminyltransferase enzymes (MGAT1–5) together with the intracellular concentrations of UDP-GlcNAc regulate the pattern of N-glycan branching on cell surface proteins, especially growth factor receptors [44]. Higher levels of N-glycan-branching results in more robust receptor cross-linking (or lattice formation) using multivalent galectin-3, longer receptor retention/lifetimes on the cell-surface, and consequently more pronounced cell signaling. In another study of O-linked glycosylation, Liu et al. studied the critical enzymes (fucosyltransferases and sialyltransferases) regulating microheterogeneity in the leukocyte cell surface protein P-selectin glycoprotein ligand-1 and leukocyte cell adhesion function [45]. Predictions from such modeling have been validated by developing RNA-interference and CRISPR-Cas9 based knockouts [39,46]. In another example, Bennun et al. modeled cancer cell transcriptome and glycome data to demonstrate augmented α(1,2)fucosylation on prostate cancer cells during their transition from androgen-dependent to androgen-independent lymph node carcinoma [47]. Besides these studies that are of biomedical relevance, several studies have modeled the glycosylation status of recombinant proteins produced using CHO cell lines as this is of biopharmaceutical relevance (reviewed in [48]).
While some examples of glycosylation networks have appeared as illustrated above, additional development of machine-readable glycoenzyme definitions can accelerate such modeling efforts as this will enable: i) model-sharing among investigators, and ii) streamlined in silico description of different biological aspects such as sugar-nucleotide biosynthesis, species transport and rate expressions. In this regard, XML-based definitions for glycosyltransferases and glycosidases have appeared for O- and N-glycosylation pathways [42,43]. Succinct, computationally efficient definitions for the specificity of enzymes involved in N-linked glycosylation have also been described, though these definitions appear in string rather than XML format [47]. McDonald et al. also describe a similar modeling language specifically for O-glycosylation [49]. It seems possible that the merits of the above approaches may be combined to develop a more universally acceptable and comprehensive glycoenzyme database. Besides being linked to the GenBank for gene-level information and the carbohydrate-enzyme database CAZY for glycoenzyme structure and carbohydrate-binding module information [50], such a resource may also be parameterized using enzyme catalytic activity and rate constant data stored in BRENDA. Additionally, the glycoenzyme definitions may include reaction descriptions from the IUBMB ExploreEnz database, and pathway data from KEGG GLYCAN, an extension of the Kyoto Encylopedia of Genes and Genomes (KEGG) [51] (Table 1). Such efforts can enable the seamless integration of computational models with a variety of glycosylation related databases.
In addition to model formulation, advanced computer simulation methods are also needed in the field. In this regard, glycosylation reaction networks typically have a large number of reactions and reactants, but relatively few model parameters due to the limited number of glycoenzymes in a given system. Fitting such over-determined systems using deterministic ordinary differential equation (ODE) networks is complicated, though this is commonly done [44,45,47,52]. In such studies, optimization approaches like genetic algorithms are sometimes used to determine the bounds of the enzyme rate constants rather than exact solutions [45]. Another approach uses stochastic Markov chain methods for data fitting [53]. This method is computationally efficient but empirical (i.e. not mechanism based) in that similar reactions catalyzed by the same enzyme may have different rate constants. Besides these approaches, there are few examples of other computational techniques in the field like Boolean networks, agent-based models and statistical modeling [40]. Overall, streamlined methods to formulate and solve glycosylation-related mathematical models are awaited.
Besides data collected by individual laboratories, relational databases with systems-level information necessary for mathematical modeling of the glycome are starting to emerge (Table 1). Some of these repositories are a results of efforts by large consortia supported in the USA (Consortium of Functional Glycomics [CFG], www.functionalglycomics.org) [54], Europe (EuroCarbDB) [55] and Japan (Japan Consortium for Glycobiology and Glycotechnology Database [JCGGDB]). Among these, at the transcript level, the CFG had developed glycogene specific microarrays and deposited related experimental data for two model organisms (human and mouse) under a limited range of experimental conditions [54]. These entries complement more generic transcriptomics data stored in the GEO (Gene Expression Omnibus) database. The JCGGDB also maintains a glycosylation-related disease database, which is an abridged form of the OMIM (Online Mendelian Inheritance in Man) catalog. MALDI-TOF mass spectrometry experiments that profile the N- and O-glycome of cells and tissue by analyzing the distribution of carbohydrates released from their underlying scaffolds are stored at the CFG database. The Unicarb-KB database catalogs intact glycoproteomics analysis mostly from literature [56]. While there is no specific database for the storage of lectin array results, the CFG has developed glycan microarrays that complement these efforts. Here, the binding specificity and affinity of glycan binding proteins (lectins) and antibodies to carbohydrates printed on chips is provided [4,57]. Complementing this, the JCGGDB has mapped the binding specificity of a range of glycan related antibodies (Table 1). The application of computational modeling concepts to quantitatively analyze genomics and proteomics resources stored in these databases is an important future step for our understanding of glycosylation processes.
Conclusions and outlook
The review highlights the manner in which glycans are synthesized and the myriad of control mechanisms regulating this process. The development of computational models to relate data available from different experimental modalities can provide a platform, initially for the qualitative understanding of these glycosylation reaction networks and subsequently for providing quantitative testable hypotheses.
Table I.
Databases for the Glycosciences*
| Level | Description and unique feature | Website |
|---|---|---|
| Glycan entries | GlyTouCan | glytoucan.org |
| UnicarbKB | unicarbkb.org | |
|
| ||
| Enzyme data | CAZY (Carbohydrate-Active Enzyme) | www.cazy.org |
| IUBMB ExploreEnz database | enzyme-database.org | |
| BRENDA (Comoprehensive enzyme info. system) | brenda-enzymes.org | |
|
| ||
| Transcript | Glycogene microarray data - for human/mouse samples (CFG) | www.functionalglycomics.org |
| Gene Expression Omnibus (GEO)- functional genomics repository | www.ncbi.nlm.nih.gov/geo | |
|
| ||
| Glycomics/Glycoproteomics | GlycoBase | glycobase.nibrt.ie |
| UniCarb-KB | www.unicarbkb.org | |
| CFG MALDI-TOF/TOF datasets | www.functionalglycomics.org | |
|
| ||
| Glycan binding data | CFG glycan array entries | www.functionalglycomics.org |
| LfDB (Lectin Frontier Database) & GlycoEpitope | jcggdb.jp/index_en.html | |
|
| ||
| Pathway | KEGG GLYCAN | www.genome.jp/kegg/glycan |
This table only lists relational databases that would be useful for computational modeling of cellular glycosylation biosynthetic pathways. For a more comprehensive list of all glycan-related databases, please see ref. [9].
HIGHLIGHTS.
Glycosylation is regulated at the transcriptional, translational and post-translational levels
Advances in sequencing and structural analysis enable data collection at all levels
Relational databases that store glycosylation-related information are starting to emerge
Computational models can integrate knowledge across different experimental scales
Acknowledgments
SN is supported by NIH/NHLBI grants HL103411 and the Program for Excellence in Glycosciences award HL107146. LKM is supported by the NIH/NIAID grant U01AI111598.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Varki A. Biological roles of oligosaccharides: All of the theories are correct. Glycobiology. 1993;3(2):97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between o-glcnacylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cummings RD. The repertoire of glycan determinants in the human glycome. Mol Biosyst. 2009;5(10):1087–1104. doi: 10.1039/b907931a. [DOI] [PubMed] [Google Scholar]
- 4**.Agrawal P, Kurcon T, Pilobello KT, Rakus JF, Koppolu S, Liu Z, Batista BS, Eng WS, Hsu KL, Liang Y, Mahal LK. Mapping posttranscriptional regulation of the human glycome uncovers microrna defining the glycocode. Proc Natl Acad Sci U S A. 2014;111(11):4338–4343. doi: 10.1073/pnas.1321524111. Maps the lectin binding profile of the NCI-60 cell lines. Identifies microRNA as a key regulator of cellular glycosylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KT, Lavrsen K, Dabelsteen S, Pedersen NB, Marcos-Silva L, Gupta R, et al. Precision mapping of the human o-galnac glycoproteome through simplecell technology. EMBO J. 2013;32(10):1478–1488. doi: 10.1038/emboj.2013.79. Analyzed the spread of O-glycosylation sites using 12 different human cell types that lack the ability to extend O-GalNAc type carbohydrates (SimpleCells). The study identifies 3000 O-glycosites on 600 glycoproteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6*.Zielinska DF, Gnad F, Wisniewski JR, Mann M. Precision mapping of an in vivo n-glycoproteome reveals rigid topological and sequence constraints. Cell. 2010;141(5):897–907. doi: 10.1016/j.cell.2010.04.012. Describes the use of ‘filter aided sample preparation’ to map 6367 N-glycosylation sites on 2352 proteins in mouse tissue and blood plasma. Besides the consensus N-X-S/T (X≠P) sequon for N-glycosylation, the study also shows rare N-X-C sites for carbohydrate modification. [DOI] [PubMed] [Google Scholar]
- 7.Sun S, Shah P, Eshghi ST, Yang W, Trikannad N, Yang S, Chen L, Aiyetan P, Hoti N, Zhang Z, Chan DW, et al. Comprehensive analysis of protein glycosylation by solid-phase extraction of n-linked glycans and glycosite-containing peptides. Nat Biotechnol. 2016;34(1):84–88. doi: 10.1038/nbt.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakus JF, Mahal LK. New technologies for glycomic analysis: Toward a systematic understanding of the glycome. Annual review of analytical chemistry. 2011;4:367–392. doi: 10.1146/annurev-anchem-061010-113951. [DOI] [PubMed] [Google Scholar]
- 9.Liu G, Neelamegham S. Integration of systems glycobiology with bioinformatics toolboxes, glycoinformatics resources, and glycoproteomics data. Wiley Interdiscip Rev Syst Biol Med. 2015;7(4):163–181. doi: 10.1002/wsbm.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings RD, Pierce JM. The challenge and promise of glycomics. Chem Biol. 2014;21(1):1–15. doi: 10.1016/j.chembiol.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nairn AV, York WS, Harris K, Hall EM, Pierce JM, Moremen KW. Regulation of glycan structures in animal tissues: Transcript profiling of glycan-related genes. J Biol Chem. 2008;283(25):17298–17313. doi: 10.1074/jbc.M801964200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152(6):1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo H, Pierce JM. Transcriptional regulation of glycan expression. In: Taniguchi N, Endo T, Hart GW, Seeberger PH, Wong C-H, editors. Glycoscience: Biology and medicine. Springer Japan; Japan: 2015. p. 1568. [Google Scholar]
- 14.Taniguchi N, Honke K, Fukuda M, Narimatsu H, Yamaguchi Y, Angata TE. Handbook of glycosyltransferases and related genes. Springer; Tokyo, Japan: 2014. [Google Scholar]
- 15*.Dewal MB, DiChiara AS, Antonopoulos A, Taylor RJ, Harmon CJ, Haslam SM, Dell A, Shoulders MD. Xbp1s links the unfolded protein response to the molecular architecture of mature n-glycans. Chem Biol. 2015;22(10):1301–1312. doi: 10.1016/j.chembiol.2015.09.006. An example of how transription factor activity can change the patter of N-linked glycosylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau E, Feng Y, Claps G, Fukuda MN, Perlina A, Donn D, Jilaveanu L, Kluger H, Freeze HH, Ronai ZA. The transcription factor atf2 promotes melanoma metastasis by suppressing protein fucosylation. Sci Signal. 2015;8(406):ra124. doi: 10.1126/scisignal.aac6479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vojta A, Samarzija I, Bockor L, Zoldos V. Glyco-genes change expression in cancer through aberrant methylation. Biochim Biophys Acta. 2016 doi: 10.1016/j.bbagen.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Antony P, Rose M, Heidenreich A, Knuchel R, Gaisa NT, Dahl E. Epigenetic inactivation of st6gal1 in human bladder cancer. BMC Cancer. 2014;14:901. doi: 10.1186/1471-2407-14-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun Q, Zhang J, Zhou N, Liu X, Shen Y. DNA methylation in cosmc promoter region and aberrantly glycosylated iga1 associated with pediatric iga nephropathy. PLoS One. 2015;10(2):e0112305. doi: 10.1371/journal.pone.0112305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mi R, Song L, Wang Y, Ding X, Zeng J, Lehoux S, Aryal RP, Wang J, Crew VK, van Die I, Chapman AB, et al. Epigenetic silencing of the chaperone cosmc in human leukocytes expressing tn antigen. J Biol Chem. 2012;287(49):41523–41533. doi: 10.1074/jbc.M112.371989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serpa J, Mesquita P, Mendes N, Oliveira C, Almeida R, Santos-Silva F, Reis CA, LePendu J, David L. Expression of lea in gastric cancer cell lines depends on fut3 expression regulated by promoter methylation. Cancer Lett. 2006;242(2):191–197. doi: 10.1016/j.canlet.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 22.Fleischer T, Edvardsen H, Solvang HK, Daviaud C, Naume B, Borresen-Dale AL, Kristensen VN, Tost J. Integrated analysis of high-resolution DNA methylation profiles, gene expression, germline genotypes and clinical end points in breast cancer patients. Int J Cancer. 2014;134(11):2615–2625. doi: 10.1002/ijc.28606. [DOI] [PubMed] [Google Scholar]
- 23.MacNeil LT, Pons C, Arda HE, Giese GE, Myers CL, Walhout AJ. Transcription factor activity mapping of a tissue-specific gene regulatory network. Cell Syst. 2015;1(2):152–162. doi: 10.1016/j.cels.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marathe DD, Chandrasekaran EV, Lau JT, Matta KL, Neelamegham S. Systems-level studies of glycosyltransferase gene expression and enzyme activity that are associated with the selectin binding function of human leukocytes. FASEB J. 2008;22(12):4154–4167. doi: 10.1096/fj.07-104257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25**.Nairn AV, Aoki K, dela Rosa M, Porterfield M, Lim JM, Kulik M, Pierce JM, Wells L, Dalton S, Tiemeyer M, Moremen KW. Regulation of glycan structures in murine embryonic stem cells: Combined transcript profiling of glycan-related genes and glycan structural analysis. J Biol Chem. 2012;287(45):37835–37856. doi: 10.1074/jbc.M112.405233. Maps glycosylation related transcript levels to corresponding glycan structures. Concludes that glycan structure changes generally (but not always) correlate with alterations in glycogene transcript abundance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anugraham M, Jacob F, Nixdorf S, Everest-Dass AV, Heinzelmann-Schwarz V, Packer NH. Specific glycosylation of membrane proteins in epithelial ovarian cancer cell lines: Glycan structures reflect gene expression and DNA methylation status. Mol Cell Proteomics. 2014;13(9):2213–2232. doi: 10.1074/mcp.M113.037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartel DP, Chen CZ. Micromanagers of gene expression: The potentially widespread influence of metazoan micrornas. Nat Rev Genet. 2004;5(5):396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 28.Schmiedel JM, Klemm SL, Zheng Y, Sahay A, Bluthgen N, Marks DS, van Oudenaarden A. Gene expression. Microrna control of protein expression noise. Science. 2015;348(6230):128–132. doi: 10.1126/science.aaa1738. [DOI] [PubMed] [Google Scholar]
- 29.Kasper BT, Koppolu S, Mahal LK. Insights into mirna regulation of the human glycome. Biochem Biophys Res Commun. 2014;445(4):774–779. doi: 10.1016/j.bbrc.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bernardi C, Soffientini U, Piacente F, Tonetti MG. Effects of micrornas on fucosyltransferase 8 (fut8) expression in hepatocarcinoma cells. PloS one. 2013;8(10):e76540. doi: 10.1371/journal.pone.0076540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324(5924):218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurcon T, Liu Z, Paradkar AV, Vaiana CA, Koppolu S, Agrawal P, Mahal LK. Mirna proxy approach reveals hidden functions of glycosylation. Proc Natl Acad Sci U S A. 2015;112(23):7327–7332. doi: 10.1073/pnas.1502076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaiana CA, Kurcon T, Mahal LK. Microrna-424 predicts a role for beta-1, 4 branched glycosylation in cell cycle progression. J Biol Chem. 2016;291(3):1529–1537. doi: 10.1074/jbc.M115.672220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patil SA, Chandrasekaran EV, Matta KL, Parikh A, Tzanakakis ES, Neelamegham S. Scaling down the size and increasing the throughput of glycosyltransferase assays: Activity changes on stem cell differentiation. Anal Biochem. 2012;425(2):135–144. doi: 10.1016/j.ab.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blixt O, Allin K, Bohorov O, Liu X, Andersson-Sand H, Hoffmann J, Razi N. Glycan microarrays for screening sialyltransferase specificities. Glycoconjugate Journal. 2008;25(1):59–68. doi: 10.1007/s10719-007-9062-z. [DOI] [PubMed] [Google Scholar]
- 36.Houseman BT, Mrksich M. Carbohydrate arrays for the evaluation of protein binding and enzymatic modification. Chem Biol. 2002;9(4):443–454. doi: 10.1016/s1074-5521(02)00124-2. [DOI] [PubMed] [Google Scholar]
- 37.Northen TR, Lee JC, Hoang L, Raymond J, Hwang DR, Yannone SM, Wong CH, Siuzdak G. A nanostructure-initiator mass spectrometry-based enzyme activity assay. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(10):3678–3683. doi: 10.1073/pnas.0712332105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kudelka MR, Antonopoulos A, Wang Y, Duong DM, Song X, Seyfried NT, Dell A, Haslam SM, Cummings RD, Ju T. Cellular o-glycome reporter/amplification to explore o-glycans of living cells. Nat Methods. 2016;13(1):81–86. doi: 10.1038/nmeth.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mondal N, Buffone A, Jr, Stolfa G, Antonopoulos A, Lau JT, Haslam SM, Dell A, Neelamegham S. St3gal-4 is the primary sialyltransferase regulating the synthesis of e-, p-, and l-selectin ligands on human myeloid leukocytes. Blood. 2015;125(4):687–696. doi: 10.1182/blood-2014-07-588590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neelamegham S, Liu G. Systems glycobiology: Biochemical reaction networks regulating glycan structure and function. Glycobiology. 2011;21(12):1541–1553. doi: 10.1093/glycob/cwr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, Cuellar AA, et al. The systems biology markup language (sbml): A medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 42.Liu G, Puri A, Neelamegham S. Glycosylation network analysis toolbox: A matlab-based environment for systems glycobiology. Bioinformatics. 2013;29(3):404–406. doi: 10.1093/bioinformatics/bts703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Liu G, Neelamegham S. A computational framework for the automated construction of glycosylation reaction networks. PLoS One. 2014;9(6):e100939. doi: 10.1371/journal.pone.0100939. Describes methods to integrate glycan structure information into SBML format pathway models. Also presents algorithms for the automated construction of glycosylation reaction networks using machine readable enzyme definitions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex n-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129(1):123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- 45.Liu G, Marathe DD, Matta KL, Neelamegham S. Systems-level modeling of cellular glycosylation reaction networks: O-linked glycan formation on natural selectin ligands. Bioinformatics. 2008;24(23):2740–2747. doi: 10.1093/bioinformatics/btn515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buffone A, Jr, Mondal N, Gupta R, McHugh KP, Lau JT, Neelamegham S. Silencing alpha1, 3-fucosyltransferases in human leukocytes reveals a role for fut9 enzyme during e-selectin-mediated cell adhesion. J Biol Chem. 2013;288(3):1620–1633. doi: 10.1074/jbc.M112.400929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bennun SV, Yarema KJ, Betenbaugh MJ, Krambeck FJ. Integration of the transcriptome and glycome for identification of glycan cell signatures. PLoS Comput Biol. 2013;9(1):e1002813. doi: 10.1371/journal.pcbi.1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puri A, Neelamegham S. Understanding glycomechanics using mathematical modeling: A review of current approaches to simulate cellular glycosylation reaction networks. Ann Biomed Eng. 2012;40(4):816–827. doi: 10.1007/s10439-011-0464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald AG, Tipton KF, Davey GP. A knowledge-based system for display and prediction of o-glycosylation network behaviour in response to enzyme knockouts. PLoS Comput Biol. 2016;12(4):e1004844. doi: 10.1371/journal.pcbi.1004844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (cazy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hashimoto K, Goto S, Kawano S, Aoki-Kinoshita KF, Ueda N, Hamajima M, Kawasaki T, Kanehisa M. Kegg as a glycome informatics resource. Glycobiology. 2006;16(5):63R–70R. doi: 10.1093/glycob/cwj010. [DOI] [PubMed] [Google Scholar]
- 52.Jedrzejewski PM, del Val IJ, Constantinou A, Dell A, Haslam SM, Polizzi KM, Kontoravdi C. Towards controlling the glycoform: A model framework linking extracellular metabolites to antibody glycosylation. Int J Mol Sci. 2014;15(3):4492–4522. doi: 10.3390/ijms15034492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spahn PN, Hansen AH, Hansen HG, Arnsdorf J, Kildegaard HF, Lewis NE. A markov chain model for n-linked protein glycosylation--towards a low-parameter tool for model-driven glycoengineering. Metab Eng. 2016;33:52–66. doi: 10.1016/j.ymben.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Raman R, Venkataraman M, Ramakrishnan S, Lang W, Raguram S, Sasisekharan R. Advancing glycomics: Implementation strategies at the consortium for functional glycomics. Glycobiology. 2006;16(5):82R–90R. doi: 10.1093/glycob/cwj080. [DOI] [PubMed] [Google Scholar]
- 55.von der Lieth CW, Freire AA, Blank D, Campbell MP, Ceroni A, Damerell DR, Dell A, Dwek RA, Ernst B, Fogh R, Frank M, et al. Eurocarbdb: An open-access platform for glycoinformatics. Glycobiology. 2011;21(4):493–502. doi: 10.1093/glycob/cwq188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campbell MP, Peterson R, Mariethoz J, Gasteiger E, Akune Y, Aoki-Kinoshita KF, Lisacek F, Packer NH. Unicarbkb: Building a knowledge platform for glycoproteomics. Nucleic Acids Res. 2014;42(Database issue):D215–221. doi: 10.1093/nar/gkt1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kletter D, Singh S, Bern M, Haab BB. Global comparisons of lectin-glycan interactions using a database of analyzed glycan array data. Mol Cell Proteomics. 2013;12(4):1026–1035. doi: 10.1074/mcp.M112.026641. [DOI] [PMC free article] [PubMed] [Google Scholar]