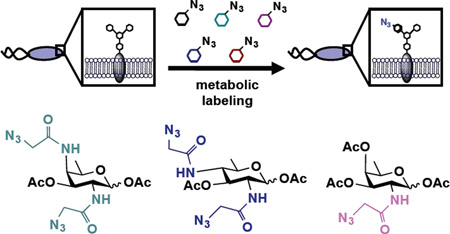
Abstract
Bacterial glycans contain rare, exclusively bacterial monosaccharides that are frequently linked to pathogenesis and essentially absent from human cells. Therefore, bacterial glycans are intriguing molecular targets. However, systematic discovery of bacterial glycoproteins is hampered by the presence of rare deoxy amino sugars, which are refractory to traditional glycan-binding reagents. Thus, the development of chemical tools that label bacterial glycans is a crucial step toward discovering and targeting these biomolecules. Here we explore the extent to which metabolic glycan labeling facilitates the studying and targeting of glycoproteins in a range of pathogenic and symbiotic bacterial strains. We began with an azide-containing analog of the naturally abundant monosaccharide N-acetylglucosamine and discovered that it is not broadly incorporated into bacterial glycans, thus revealing a need for additional azidosugar substrates to broaden the utility of metabolic glycan labeling in bacteria. Therefore, we designed and synthesized analogs of the rare deoxy amino d-sugars N-acetylfucosamine, bacillosamine, and 2,4-diacetamido-2,4,6-trideoxygalactose and established that these analogs are differentially incorporated into glycan-containing structures in a range of pathogenic and symbiotic bacterial species. Further application of these analogs will refine our knowledge of the glycan repertoire in diverse bacteria and may find utility in treating a variety of infectious diseases with selectivity.
Graphical Abstract
INTRODUCTION
One widespread challenge in the healthcare industry is the increasing ineffectiveness of existing antibiotics due to the evolution of microbial resistance. Even when existing antibiotics are effective at treating bacterial disease, these treatments are often broad-spectrum and suffer from deleterious effects on our beneficial bacteria.1 Such disruptions to the normal gut microbiome can precipitate long-term health problems2 including obesity, autoimmune disorders, allergies, malnutrition, and expansion of enteric pathogens in the gut.3,4 Thus, there is an urgent need to develop novel antibiotics that target specific bacterial populations while leaving the rest of the microbiome intact.
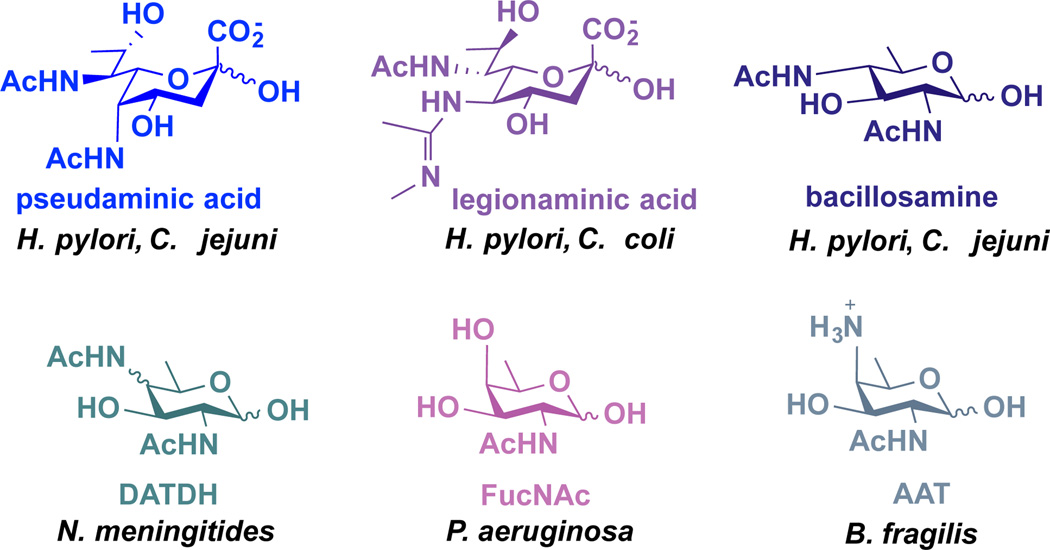
Bacterial glycoproteins serve as attractive targets of therapeutic intervention because they are often linked to pathogenesis, their structures differ markedly from those found in eukaryotes, and they contain distinctive monosaccharide building blocks that are exclusively present on a small number of bacterial pathogens.5,6 For example, the gastric pathogen Helicobacter pylori synthesizes glycoproteins that contain the rare amino- and deoxy-monosaccharides pseudaminic acid, legionaminic acid, and bacillosamine (Figure 1).7,8 These sugars have limited expression on pathogenic bacteria, and there are no reports of these three monosaccharides in nonpathogenic bacteria that dominate the human gut microbiome3 (e.g. Bacteroides spp., Prevotella spp.). More broadly, Neisseria meningitidis utilizes 2,4-diacetamido-2,4,6-trideoxyhexose (DATDH),9 Pseudomonas aeruginosa incorporates FucNAc residues,10 and the symbiotic Bacteroides fragilis appends 2-acetamido-4-amino-2,4,6-trideoxygalactose (AAT)11 into its cell surface polysaccharides (Figure 1). These distinctive building blocks appear to be exclusively present on a small number of bacterial species.6,12
Figure 1.
Structures of rare, exclusively bacterial deoxy amino monosaccharides and examples of bacteria that install them in their glycans.
Despite the potential of distinct monosaccharides within bacterial glycans to serve as molecular targets, the systematic discovery of bacterial glycoproteins is hampered by the presence of rare deoxy amino sugars, which are refractory to traditional glycan-binding reagents.13,14 In particular, lectins and glycosidases employed to detect and remove eukaryotic glycans, respectively, often cannot be utilized to study bacterial glycans due to their restricted binding specificity.15,16,17 The development of chemical tools that label bacterial glycans is a crucial step toward discovering and targeting these biomolecules.
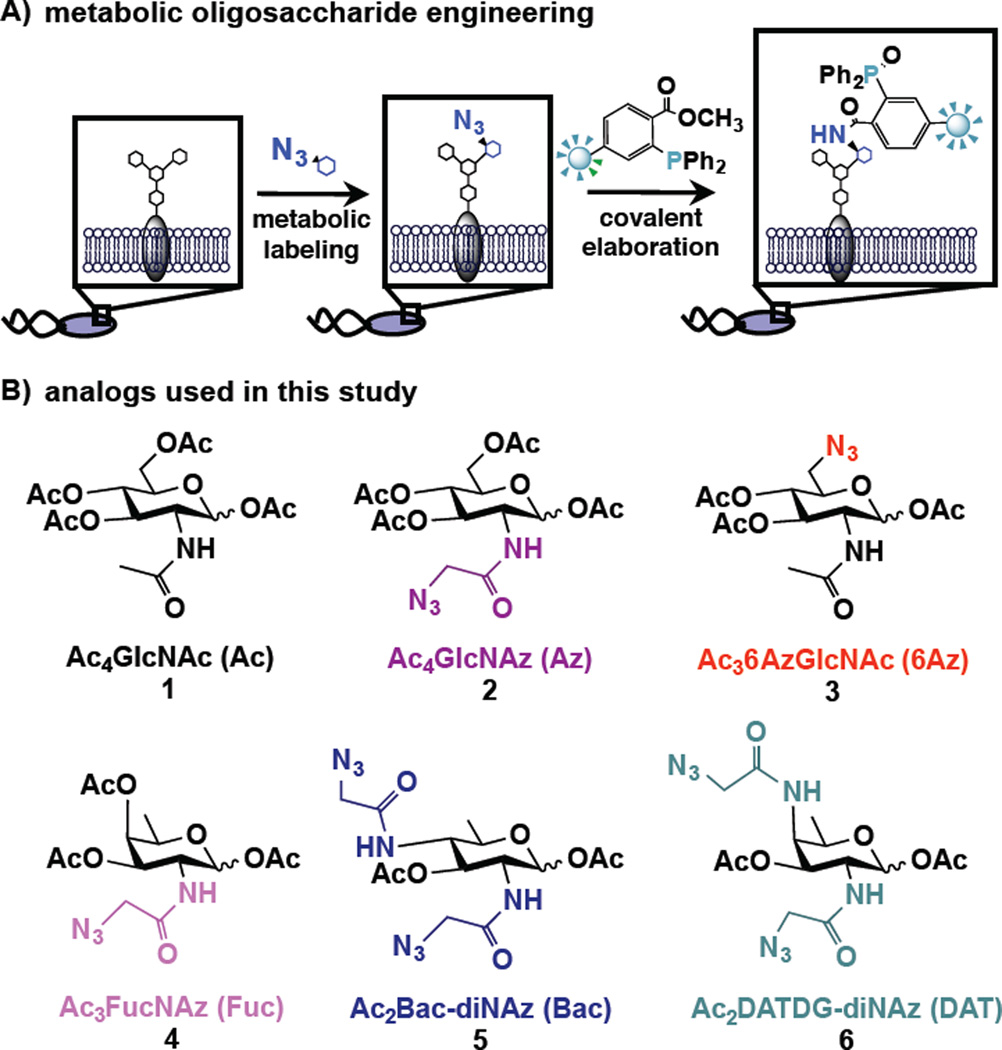
The chemical technique known as metabolic oligosaccharide engineering (MOE), which was pioneered by Bertozzi,18,19 Reutter20 and others for the study of mammalian glycoproteins, offers a powerful approach to label and detect bacterial glycoproteins.21 In MOE, cells are supplemented with an unnatural sugar that is taken up, processed by endogenous carbohydrate biosynthetic pathways, and installed into glycans in place of natural monosaccharides. Using this approach, bio-orthogonal chemical reporters, such as azides,22 can be incorporated into cellular glycans (Figure 2A). Once incorporated into cellular glycoproteins, the azide acts as a chemical handle that undergoes covalent elaboration with exquisitely selective reaction partners, including triarylphosphines23 and cyclooctynes,24 for the attachment of biological probes25 (Figure 2A). Incorporation of azide-containing sugars into bacterial glycans thus facilitates detection, enrichment, and identification of bacterial glycoproteins, and enables targeting with covalent therapeutics to incite cell damage.26
Figure 2.
Schematic overview of metabolic oligosaccharide engineering (MOE) experiments performed, and structures of monosaccharide analogs used in this study. A) In MOE experiments, cells are first metabolically labeled with an unnatural azide-containing sugar. Azide-bearing glycans then undergo Staudinger ligation with phosphine probes to enable their detection on cells or in cell lysates. B) Metabolic incorporation of azide-containing analogs of the naturally abundant monosaccharide N-acetylglucosamine (GlcNAc) as well as of the rare bacterial monosaccharides FucNAc (4), bacillosamine (5), and DATDG (6) were explored in this work.
MOE has been utilized to incorporate an azide-bearing analog of the common monosaccharide N-acetylglucosamine (GlcNAc) into glycoproteins on the gastric pathogen Helicobacter pylori.21 In particular, the GlcNAc analog peracetylated-N-azidoacetylglucosamine (Ac4GlcNAz (2), Figure 2B) was robustly incorporated into H. pylori’s glycoproteins and facilitated the identification of 125 candidate H. pylori glycoproteins.27 In addition to enabling bacterial glycoprotein discovery, Ac4GlcNAz could form the basis of a bacterial glycan targeting strategy as it is minimally presented onto the surfaces of mammalian cells,28 despite its robust incorporation into mammalian O-GlcNAc modified nuclear and cytosolic proteins.29 Indeed, our work led to the development of immune-stimulant bearing phosphines that catalyzed the destruction of azide-covered H. pylori in an immune-cell dependent manner.30 Thus, Ac4GlcNAz is a reasonable choice from the perspective of discovering and targeting H. pylori glycoproteins and may have broad utility for probing glycoproteins or other glycan-containing structures in diverse bacteria.
Here, we explored the extent to which MOE facilitates the study of glycoproteins in a range of pathogenic and symbiotic bacterial strains. Our results demonstrate that relatively few bacterial strains are azide-labeled upon treatment with Ac4GlcNAz, and thus suggest that differential labeling of bacterial populations is possible. However, these results reveal a need for additional azidosugar substrates to broaden the utility of MOE in bacteria. To meet this need, we synthesized azide-containing analogs of rare bacterial monosaccharides (Figure 2B; compounds 4, 5, 6) and established that pathogenic bacteria effectively process these distinctive sugars. This work sets the stage for facilitating the study of glycoproteins in diverse bacterial species and providing a means to inactivate select bacteria based on their distinctive glycan coating.
RESULTS AND DISCUSSION
To facilitate the studying and targeting of glycans in a range of pathogenic bacteria, we set out to assess the metabolic incorporation of a range of azide-bearing monosaccharides in diverse bacteria. Since incorporation of azide-containing sugars into a wide range of bacterial glycoproteins is beneficial from the perspective of glycoprotein discovery but detrimental for creating narrow-spectrum glycan-targeted therapeutics,6 we explored the incorporation of azide-bearing analogs of both common and rare monosaccharide substrates. We reasoned that rare monosaccharides with restricted metabolic fates (e.g. conversion to a single product) would be processed into glycans on fewer bacterial strains than common monosaccharides with broad metabolic fates.
Differential labeling of bacterial glycoproteins with Ac4GlcNAz
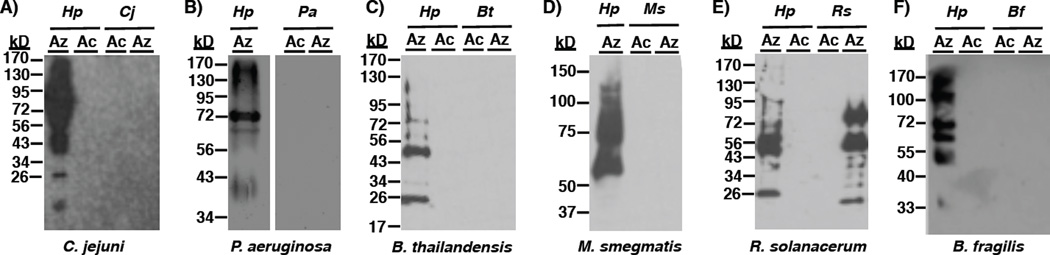
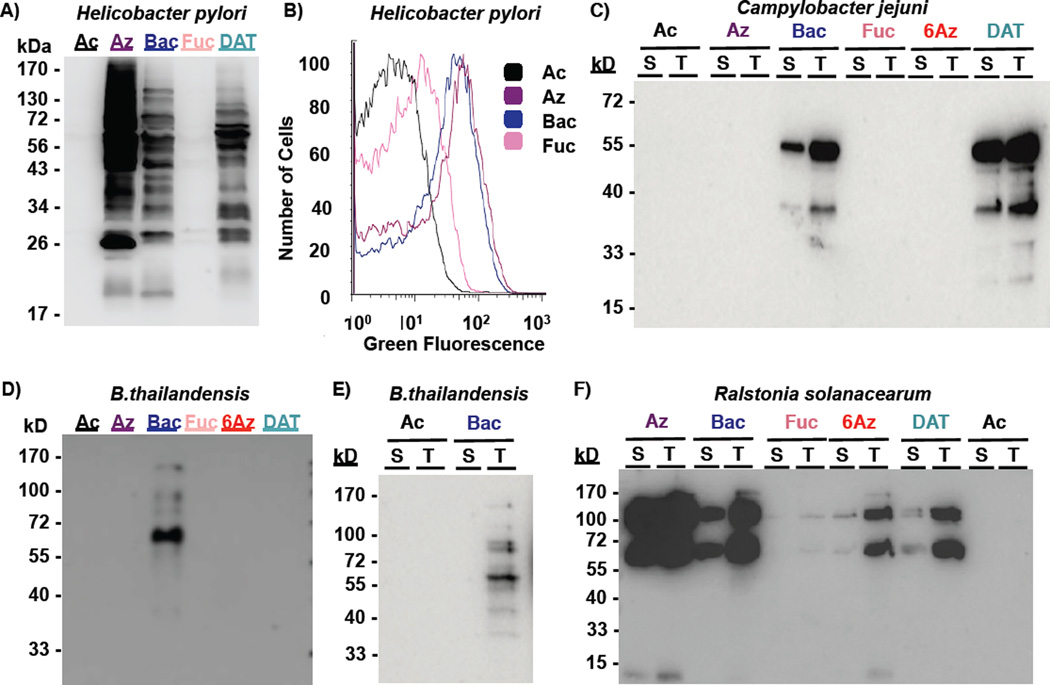
To build upon the established utility of Ac4GlcNAz (2),29 an azide-containing analog of the common monosaccharide GlcNAc, for discovering27 and targeting30 Helicobacter pylori’s glycoproteins, we first sought to investigate the utility of this substrate to study glycoproteins in bacteria beyond H. pylori. We began by assessing the levels of incorporation of Ac4GlcNAz into bacterial glycans in a range of species including pathogenic and symbiotic bacteria. In particular, we focused on incorporation into the gram-negative pathogens Campylobacter jejuni and Pseudomonas aeruginosa, close relatives of pathogens Burkholderia thailandensis and Mycobacterium smegmatis, the plant pathogen Ralstonia solanacerum, and the symbiotic bacterium Bacteroides fragilis. These bacteria were selected as their ability to produce glycosylated proteins has been previously established.31–37 To gauge metabolic incorporation, bacteria were treated with 1 mM Ac4GlcNAz (2) or the azide-free control sugar peracetylated N-acetylglucosamine (Ac4GlcNAc (1), Figure 2B), then their proteins were harvested by cell lysis and reacted with a phosphine probe comprising a FLAG peptide38 (Phos-FLAG). In all Western blot analyses, lysates from H. pylori treated with Ac4GlcNAz followed by Phos-FLAG were included as a positive control.21 Analysis of proteins from treated H. pylori and R. solanacerum yielded an array of azide-labeled glycan-containing structures that were detected by Western blot analysis with anti-FLAG antibody (Figure 3). In contrast, similar treatment of the other bacterial strains with Ac4GlcNAz led to no detectable azide-labeled glycoproteins (Figure 3). Control samples from cells treated with the azide-free compound peracetylated GlcNAc (Ac4GlcNAc, 1) yielded no azide-dependent signal, as expected. Zinc or Coomassie staining of electrophoresed samples confirmed that all samples contain proteins (Supplementary Figure 1), indicating that the lack of detectable azides in some samples was not due to low protein levels. Counter to our expectation, these data reveal that metabolic incorporation of azides into bacterial glycans upon treatment with Ac4GlcNAz is not a widespread phenomenon but rather appears limited to select bacterial strains.
Figure 3.
Western blot analyses indicate that Ac4GlcNAz (2) supplementation leads to installation of azides into glycoproteins in select bacterial pathogens. A) Helicobacter pylori (Hp), Campylobacter jejuni (Cj), B) Pseudomonas aeruginosa (Pa), C) Burkholderia thailandensis (Bt), D) Mycobacterium smegmatis (Ms), and E) Ralstonia solanacerum (Rs) were grown in media supplemented with 1 mM Ac4GlcNAz (2, Az) or the azide-free control sugar Ac4GlcNAc (1, Ac) for 5 days, then harvested by lysis. F) Due to its faster doubling time, Bacteroides fragilis (Bf) was grown in media supplemented with 1 mM Ac4GlcNAz (2, Az) or the azide-free control sugar Ac4GlcNAc (1, Ac) for 2 days, then harvested by lysis. The presence of azides in cellular glycoproteins was detected by reacting lysates with 250 µM Phos-FLAG for 6–12 hours at 37 °C, and then analyzing samples via Western blot with anti-FLAG antibody. The data shown are representative of replicates (n≥3).
Synthesis of azide-containing variants of rare bacterial monosaccharides
Due to the observed narrow incorporation of Ac4GlcNAz into bacterial glycans, we wondered whether analogs of rare bacterial monosaccharides might facilitate glycoprotein discovery while setting the stage for novel antibiotic strategies that interfere with select pathogenic strains but leave the microbiome largely intact. Therefore, we designed a panel of azide-containing variants of the rare bacterial sugars FucNAc, bacillosamine, and DATDG (Figure 2B, 4–6). Given that a variety of carbohydrate biosynthetic pathways are permissive of N-azidoacetyl substituents in place of the natural N-acetyl moieties,18,26 we reasoned that the requisite bacterial enzymes would likely process analogs bearing N-azidoacetyl groups. Further, we chose to temporarily mask the hydrophilic hydroxyl groups of these analogs with hydrophobic acetyl groups39 to maximize efficiency of uptake by bacterial strains.21
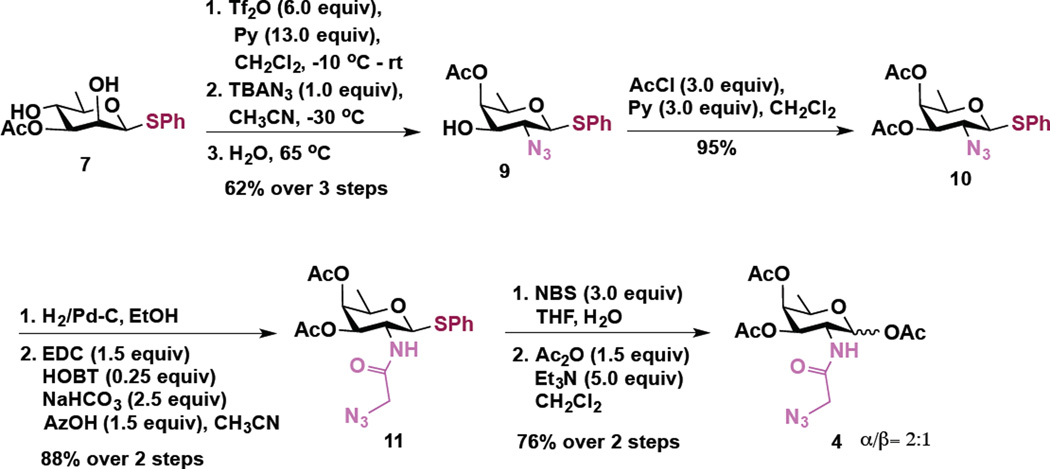
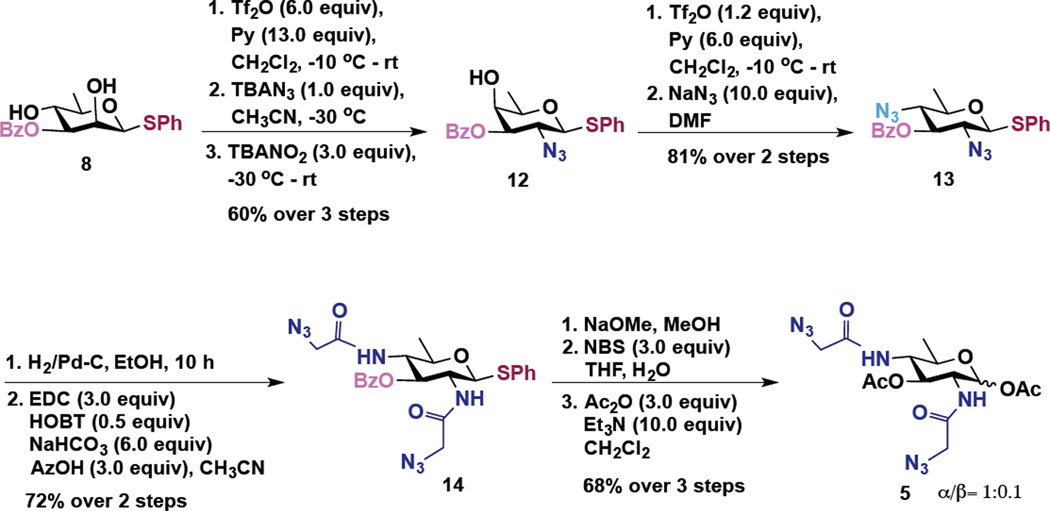
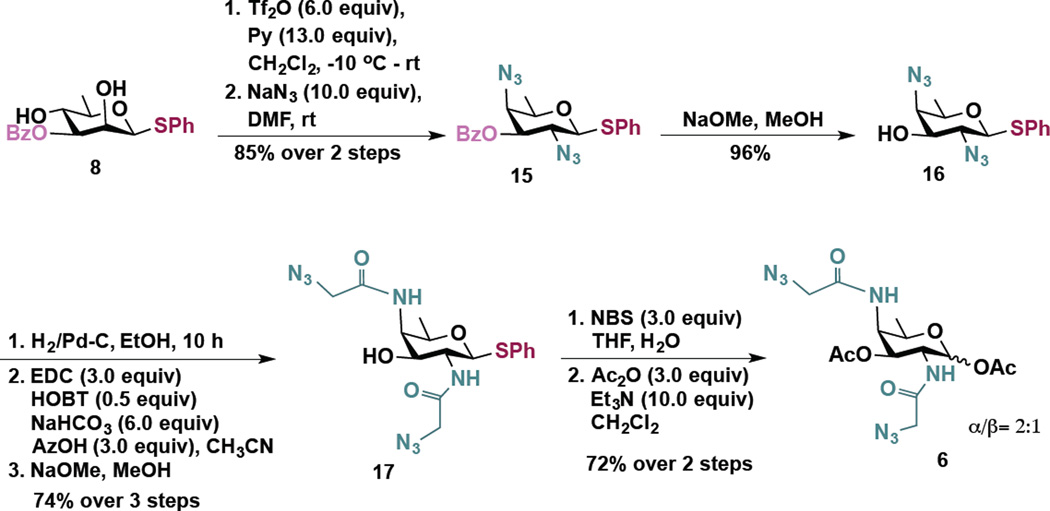
To access these compounds, we built upon a strategy developed by Kulkarni and coworkers to synthesize rare bacterial amino and deoxy monosaccharide building blocks including FucNAc, bacillosamine, and DATDG.40,41 In this strategy, changing the sequence of addition of nucleophiles in a series of one-pot double serial and double parallel displacement reactions using azide, phthalimide and nitrite ions as nucleophiles and mannose-derived 2,4 bis-trifluoromethanesulfonates (triflates) as electrophiles enabled rapid access to various orthogonally protected rare deoxy amino sugar building blocks.40,41 Accordingly, the 2,4-diol substrates (Scheme 1, compound 7 and Scheme 2, compound 8) for the one-pot double displacement reaction were first prepared starting from cheaply available d-mannose in six steps.41
Scheme 1.
Synthesis of Ac3FucNAz (Fuc) 4
Scheme 2.
Synthesis of Ac2Bac-diNAz (Bac) 5
Transformation of d-rhamnosyl 2,4-diol 7 into d-fucosamine derivative 4 is shown in Scheme 1. Following our earlier established procedure, compound 7 was converted to the corresponding 2,4-bistriflate derivative, which upon a brief work-up was subjected to a one-pot, regioselective, nucleophilic displacement reaction. Accordingly, a sequential C2-OTf displacement of the crude bistriflate with a stoichiometric amount of tetrabutylammonium azide (TBAN3) at −30 °C followed by water mediated intramolecular displacement of the C4-OTf by C3-OAc, via the ring opening of the corresponding orthoester intermediate, resulted in the formation of the C4-OAc derivative of d-fucosamine 9 in 62% yields over three steps.41 The remaining 3-OH group was capped as an acetate to form 10 (95%), which upon azide reduction under hydrogenation conditions, followed by EDC mediated coupling of the so-formed amine with azidoacetic acid,19 afforded the azido derivative 11 in 88% yield over two steps. Finally, hydrolysis of the thioglycoside by using NBS and water generated the corresponding hemiacetal which upon subsequent treatment with acetic anhydride in the presence of trielthylamine afforded triacetate compound 4 (α/β =2:1)(Ac3FucNAz, Fuc) in 76% yield over 2 steps.
For the synthesis of Ac2Bac-diNAz (Bac) 5, we started from 3-OBz derivative 8 (Scheme 2). Diol 8 upon similar triflation followed by sequential, one pot SN2 reactions with TBAN3 and with tetrabutyl ammonium nitrite (TBANO2), at C2 and C4 respectively, afforded d-fucosamine derivative 12 in 60% overall yield.41 Concomitant triflation of the axial 4-OH group and its displacement with NaN3 furnished bacillosamine 13.41 Reduction of 2,4-azido groups in 13 followed by EDC coupling with azidoacetic acid afforded 14 in 72% yield over two steps. Debenzoylation, conversion of anomeric thioglycoside to the corresponding hemiacetal with NBS/water and acetylation afforded the target molecule Ac2Bac-diNAz (Bac) 5 (α/β = 1:0.1) in 68% yield over three steps.
The synthesis of an analog of the rare sugar DATDG is outlined in Scheme 3. A double displacement of the 2,4-bistriflate, obtained from 8, with excess NaN3 cleanly afforded di-azido derivative 15 in excellent overall yield.41 The 3-O-benzoate group was cleaved using NaOMe to obtain 16 (96%). This step was deemed necessary in order to avoid the possible migration of benzoate (C3→C4) during the ensuing reduction of azides to corresponding amines. The azides were then reduced under hydrogen atmosphere and so formed amines were coupled with the azidoacetic acid using EDC as the coupling reagent. In this reaction, along with the desired di-N-acylated product, we also obtained fully acylated product. Treatment of the mixture of acylated products with NaOMe in MeOH gave the desired product 17 in 74% yield over 3 steps. Thioglycoside was treated with NBS/water and the so formed hemi acetal was acetylated to give the desired derivative Ac2DATDG-diNAz (DAT) 6 (α/β = 2:1) in 72% yield over 2 steps.
Scheme 3.
Synthesis of Ac2DATDG-diNAz (DAT) 6
Rare bacterial monosaccharide analogs are incorporated into diverse bacterial pathogens
With access to these rare bacterial monosaccharide analogs, we performed metabolic labeling experiments to assess incorporation into diverse bacterial species. We first evaluated metabolic incorporation into bacterial pathogens or close relatives of pathogens. For these experiments, bacteria were grown in media supplemented with one of each of the panel of rare bacterial monosaccharide analogs (Ac3FucNAz (4), Ac2Bac-diNAz (5), or Ac2DATDG-diNAz (6)), or with one of three GlcNAc analogs (Ac4GlcNAz (2),28 the peracetylated 6-azido GlcNAc analog Ac36AzGlcNAc (3),42 or the azide-free control Ac4GlcNAc (1); Figure 2B) that are not appreciably incorporated onto mammalian cell surfaces.28,42 After metabolic labeling, proteins were harvested by cell lysis and reacted with Phos-FLAG for Western blot analysis with anti-FLAG antibody to yield a snapshot of all (“total”) azide-labeled glycans from the bacteria. Alternatively, intact cells were rinsed and reacted with the cell impermeable compound Phos-FLAG to probe for the presence of surface accessible (“surface”) azides on the bacteria;30 these surface accessible azide-labeled species were then detected by flow cytometry or Western blot analysis with an anti-FLAG antibody.
As depicted in Figure 4, the gastric pathogen H. pylori robustly incorporates Ac4GlcNAz (2), Ac2Bac-diNAz (5), and Ac2DATDG-diNAz (6) into surface glycans, as revealed by Western blot (Figure 4A) and flow cytometry (Figure 4B; Supplemental Figure S2B) analysis. In contrast, Ac3FucNAz (4) is minimally incorporated into H. pylori’s glycans and H. pylori treated with Ac4GlcNAc (1) remain azide-free (Figure 4A, B). The gut pathogen C. jejuni displays a different metabolic incorporation phenotype; Western blot analysis reveals robust processing of only Ac2Bac-diNAz (5) and Ac2DATDG-diNAz (b) into select primarily surface-accessible glycans in C. jejuni (Figure 4C). The opportunistic pathogen B. thailandensis incorporates Ac2Bac-diNAz (5) but no other azidosugars into its glycan-containing structures (Figure 4D); follow up analysis of the Ac2Bac-diNAz (5) labeled species from B. thailandensis indicates that they are not surface accessible (Figure 4E). Finally, the plant pathogen R. solanacearum exhibits the broadest incorporation profile, with treatment with Ac4GlcNAz (2), Ac2Bac-diNAz (5), Ac2DATDG-diNAz (6), Ac36AzGlcNAc (3) and, to a lesser extent, Ac3FucNAz (4) yielding surface accessible azides detectable by Western blot (Figure 4F) and flow cytometry (Supplementary Figure 3). In all cases Coomassie staining of electrophoresed samples confirmed equivalent protein levels (Supplementary Figure 2), indicating that the lack of detectable azides in some samples was not due to low protein content. Further, the negative control samples from cells treated with Ac4GlcNAc (1) consistently lacked azide-dependent signal. Taken together, these data suggest that the extent of incorporation into bacterial glycans is dependent upon the structure of the metabolic input and upon the bacteria under study. Relative to Ac4GlcNAz, analogs of rare bacterial monosaccharides appear better suited to study bacterial glycans.
Figure 4.
Metabolic incorporation of azide-containing rare bacterial monosaccharide analogs onto bacterial pathogens. H. pylori, C. jejuni, B. thailandensis, and R. solanacerum were grown in media supplemented with 1 mM Ac4GlcNAz (2, Az), Ac2Bac-diNAz (5, Bac), Ac3FucNAz (4, Fuc), Ac36AzGlcNAc (3, 6Az), Ac2DATDG-diNAz (6, DAT), or the azide-free control sugar Ac4GlcNAc (1, Ac) for 5 days, then probed for the presence of azide-labeled glycans on cells or within lysates by reaction with 250 µM Phos-FLAG and subsequent detection with anti-FLAG antibody. A) The presence of azides in H. pylori’s lysates was detected via Western blot analysis with anti-FLAG-HRP. B) Presentation of azides on H. pylori cell surfaces was assessed via flow cytometry with anti-FLAG-FITC. C) Incorporation of azides into C. jejuni’s surface-accessible (“S”) versus total (“T”) glycoproteins was probed by reacting intact cells or cell lysates, respectively, with Phos-FLAG, then analyzing samples via Western blot analysis. D, E) Display of azides in B. thailandensis lysates (D) or in surface-accessible versus total glycoproteins (E) was analyzed via Western blot. F) Incorporation of azides into R. solanacearum surface-accessible versus total glycoproteins was assessed via Western blot analysis. All lanes contain equivalent protein levels. The data shown are representative of replicate experiments (n≥3).
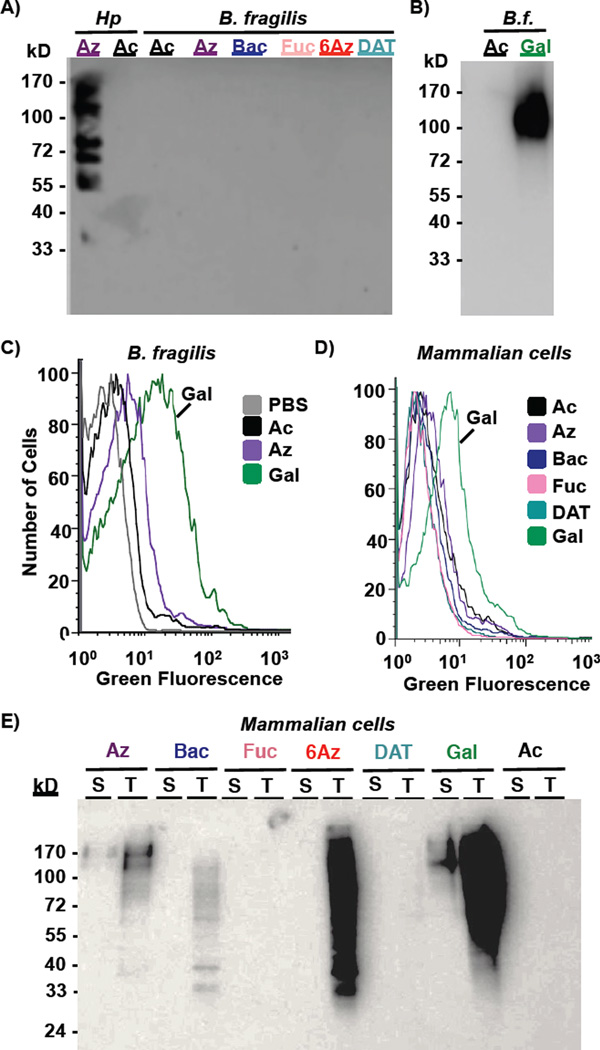
Rare bacterial monosaccharide analogs are not incorporated onto symbiotic or mammalian cells
With compelling evidence that this panel of azidosugars could be used to study or target glycans in a variety of bacterial pathogens, we next explored whether these azidosugars are incorporated into the beneficial bacteria Bacteroides fragilis. Such information is critical to assess potential undesired impacts on innocent bystanders of a rare bacterial monosaccharide-based antibiotic strategy. B. fragilis supplemented with the panel of azide-containing sugars did not incorporate azides into glycoproteins at a detectable level by Western blot analysis (Figure 5A) despite high protein content in these samples (Supplementary Figure 4A). In contrast, Western blot and flow cytometry analyses revealed that the azidosugar peracetylated N-azidoacetylgalactosamine (Ac4GalNAz),43 an analog of N-acetylgalactosamine, was robustly displayed in B. fragilis surface glycans (Figure 5B,C; Supplementary Figure 4B); this result is consistent with a recent demonstration by Kasper and co-workers that Ac4GalNAz is processed into polysaccharide A in B. fragilis.44 These data indicate that, though it is possible to incorporate azides into B. fragilis surface glycans, such incorporation is negligible with the rare bacterial monosaccharide analogs reported here. Thus, narrow range antibiotics based on differential incorporation of azides onto the surface of pathogenic but not symbiotic bacteria may be possible.
Figure 5.
Metabolic incorporation of azide-containing rare bacterial monosaccharide analogs onto symbiotic Bacteroides fragilis and mammalian Madin Darby Canine Kidney (MDCK) cells. A–C) B. fragilis were grown in media supplemented with 1 mM Ac4GlcNAz (2, Az), Ac2Bac-diNAz (5, Bac), Ac3FucNAz (4, Fuc), Ac36AzGlcNAc (3, 6Az), Ac2DATDG-diNAz (6, DAT), or the azide-free control sugar Ac4GlcNAc (1, Ac) for 2 days, then probed for the presence of azide-labeled glycans on cells or within lysates by reaction with 250 µM Phos-FLAG and subsequent detection with anti-FLAG antibody via Western blot (A, B) or by reaction with DIBO-488 (20 µM, 1 hour) followed by flow cytometry (C). D, E) MDCK cells were cultured with 0.25 mM azidosugar-supplemented media for 3 days, then probed for the presence of azide-labeled glycans using Phos-FLAG and anti-FLAG antibody. D) Analysis of intact MDCK was performed by flow cytometry with anti-FLAG-FITC. E) Incorporation of azides into MDCK surface-accessible (“S”) versus total (“T”) glycoproteins was probed by reacting intact cells or cell lysates, respectively, with Phos-FLAG, then analyzing samples via Western blot analysis. Data are representative of replicate experiments (n≥3).
To explore whether these rare bacterial monosaccharide analogs may have the potential to underpin a covalent antibiotic strategy aimed at gastrointestinal pathogens, we set out to evaluate the incorporation of these analogs in mammalian cells. As a model system, we turned to the well-studied Madin Darby Canine Kidney (MDCK) epithelial cell line, as this cell line is an established model for epithelial cells that line the gastrointestinal tract. In experiments analogous to those described above, MDCK cells were treated with each of a panel of azidosugars, and metabolic incorporation of azides into cellular glycans and onto cell surfaces was assessed using Phos-FLAG and anti-FLAG antibody.19 Western blot and flow cytometry analyses revealed that only the positive control sugar Ac4GalNAz, which is robustly incorporated into mucin-type O-linked glycoproteins,43 led to significant delivery of azides onto the surface of MDCK cells (Figure 5D, E). In contrast, none of the other azidosugars resulted in appreciable display of surface-accessible azides (Figure 5E, “surface” lanes); Coomassie staining confirmed that the differential azide labeling was not due to appreciable differences in protein loading across samples (Supplementary Figure 4C). As expected based on previous literature reports, Ac4GlcNAz (2) and Ac36AzGlcNAc (3) were incorporated into internal MDCK glycoproteins, consistent with the metabolic incorporation of these sugars into nuclear and cytosolic O-GlcNAc residues (Figure 5E, “total” lanes).42 Unexpectedly, bacillosamine analog 5 was processed by MDCK cells and led to installation of azides into some internal MDCK structures (Figure 5E, “total lane”). This result is surprising due to the assumed lack of metabolic enzymes to process this exclusively bacterial sugar analog. Regardless, the finding that none of the rare bacterial monosaccharide analogs led to appreciable presentation of azides on the surface of MDCK cells bodes well for the potential of these sugars to form the basis for selective covalent antimicrobial applications.
Discussion
Here we explore and expand the scope of metabolic glycan labeling across diverse bacterial species. Presumably due to the distinctive monosaccharide building blocks employed by bacteria, the analog Ac4GlcNAz (2) appears to have some but limited utility for metabolic labeling of bacterial glycoproteins. To address this limitation, we produced a series of novel bacteria-specific azidosugar substrates that are robustly incorporated into glycans in a variety of pathogenic bacteria. Our data indicate that the choice of azidosugar substrate dictates which bacterial species process azides into their glycans.
The ability of bacterial strains to process monosaccharide analogs into their glycans could reflect underlying differences in glycosylation machinery present within each species. For example, H. pylori and C. jejuni have biosynthetic pathways that convert bacillosamine to pseudaminic acid. A likely metabolic fate of Ac2Bac-diNAz (5) in these bacteria, therefore, is conversion to azido-pseudaminic acid and installation onto flagellin glycoproteins. In contrast, B. fragilis does not have the pseudaminic acid biosynthetic pathway enzymes, nor does it process Ac2Bac-diNAz into its glycans. Thus, metabolic incorporation of azidosugars into bacterial glycans is linked to which carbohydrate active enzymes are present within each species. The differences in glycan labeling across bacterial species observed in this work could also be a reflection of the labeling conditions used. Due to the dependence of glycan expression on a variety of factors (e.g. cell cycle, the environment), bacterial strains might exhibit a different metabolic incorporation phenotype at different growth stages or in different environments. For example, under different environmental conditions bacterial strains might incorporate monosaccharide analogs that were not displayed in their glycans in the conditions used in this study. Moreover, exploring a range of sugar analog concentrations (e.g. higher and lower than 1 mM) has the potential to broaden or narrow metabolic incorporation into glycans. Even within the scope of this work, where fixed concentrations of sugars were utilized for fixed periods of time, subtle differences in glycoprotein fingerprint were observed across samples; Ac4GlcNAz-treated H. pylori displayed altered labeling profiles (Figure 3), likely due to phase variation causing changes in protein and glycan expression levels.
Metabolic incorporation sets the stage to use the azide as a chemical handle to enrich these labeled species, ultimately enabling glycoproteomic analyses.19 Indeed, Woo et al. recently reported an azido-glycoproteomic method that could be applied to identify which bacterial proteins are glycosylated and to elucidate the structures of the modifying glycans.45 Moreover, metabolic labeling of bacterial glycoproteins provides an approach to monitor bacterial glycan dynamics; time- and environment-dependent changes in glycan expression have the potential to yield a dynamic picture of bacterial glycosylation, ultimately providing insight into the functions of these critical biomolecules. Expanding the scope of metabolic glycan labeling to a range of bacterial species is therefore an important step to facilitate the study of bacterial glycoproteins.
In addition to easing bacterial glycoprotein discovery, this work sets the stage for covalent targeting of distinctive bacterial glycans with molecules that incite damage or permit visualization.6 Our data indicate that the choice of azidosugar substrate dictates which bacterial species become azide-covered. Differential incorporation of azides across bacterial cells could form the basis for narrow-spectrum antibiotics that induce killing of pathogenic bacteria yet leave symbiotic bacteria unharmed. For example, azide-coated bacteria could be targeted with immune stimulant-bearing phosphines via Staudinger ligation30 or with copper-free click-chemistry46 delivered payloads. Analogously, azide-reactive molecules bearing imaging agents47,48 could be employed to track bacterial infection in an animal model. Taken together, the work presented here provides expanded tools for metabolic labeling of bacterial glycans, which might be harnessed for glycoprotein discovery, targeted cell killing, or molecular imaging.
Conclusions
Metabolic labeling of bacterial glycoproteins is a relatively new strategy to study and target these biomolecules. Prior to this work, common monosaccharide precursors were employed to metabolically label glycoproteins in H. pylori27 and Bacteroidales species,49 and a bacillosamine analog was employed as a committed pseudaminic acid precursor to label flagellin glycoproteins in C. jejuni.50 The work reported here adds a new dimension to this field by generating analogs of rare bacterial monosaccharides that are processed by a variety of pathogenic but not symbiotic bacterial strains, thus offering the potential to exploit the incorporated azides for glycoprotein discovery, targeted cell killing, and molecular imaging. Although we focus here on a relatively small panel of pathogenic and symbiotic bacterial strains, this strategy has the potential to be applied to study, target, and track distinctive glycans in complex microbial communities present in an animal infection model. Thus, this strategy opens the door to discovering novel antibiotic targets and treating a wide range of bacterial diseases in a glycosylation-dependent manner.
METHODS
Materials and Chemical Synthesis
Organic chemicals and anti-FLAG antibodies were purchased from Sigma-Aldrich. H. pylori strain G2751 was a gift of Manuel Amieva (Stanford University), and M. smegmatis strain Mc2155::otsA treS treY52 was a gift from Peter Woodruff (University of Southern Maine). Bacterial cells (C. jejuni ATCC 33560; B. thailandensis ATCC 700388; R. solanacearum ATCC 33291* (*incorrectly cataloged as C. jejuni; species confirmed as R. solanacearum by 16S ribosomal RNA sequencing); B. fragilis ATCC 23745; P. aeruginosa ATCC 39324) and MDCK cells (ATCC CCL-34) were purchased from ATCC and grown according to the supplier’s instructions. Ac4GlcNAc (1), Ac4GlcNAz (2), Ac4GalNAz and Phos-FLAG were synthesized as previously described.19,53 Ac36AzGlcNAc (3)42 was kindly provided by Matthew Pratt (University of Southern California). Ac3FucNAz (4), Ac2Bac-diNAz (5), and Ac2DATDG-diNAz (6) were synthesized using standard organic chemistry procedures and characterized by standard techniques including 1H and 13C NMR spectroscopy and mass spectrometry. Ac3FucNAz (4), Ac2Bac-diNAz (5), and Ac2DATDG-diNAz (6) were purified using flash silica gel chromatography.
Metabolic Labeling
Bacterial cells were grown in rich liquid media supplemented with 1 mM21 Ac4GlcNAz (2), Ac36AzGlcNAc (3), Ac3FucNAz (4), Ac2Bac-diNAz (5), Ac2DATDG-diNAz (6), Ac4GalNAz, or the azide-free control Ac4GlcNAc (1) for 2–5 days, depending on the doubling rate of the organism. H. pylori, C. jejuni, and R. solanacearum were metabolically labeled for 5 days under microaerophilic conditions (14% CO2, 37 °C), B. fragilis were metabolically labeled for 2 days under anaerobic conditions (created by a Becton Dickinson EZ anaerobe Gaspack in an airtight container; 37 °C), and M. smegmatis, B. thailandensis, and P. aeruginosa were grown in 1 mM sugar-containing media for 5 days under aerobic conditions, as appropriate for each bacterial strain. MDCK were grown in Dulbecco’s Modified Eagles Medium supplemented with 250 µM of Ac4GlcNAc (1), Ac4GlcNAz (2), Ac36AzGlcNAc (3), Ac3FucNAz (4), Ac2Bac-diNAz (5), Ac2DATDG-diNAz (6), or Ac4GalNAz for 3 days. Cells were then harvested, rinsed with phosphate buffered saline (PBS), and reacted with 250 µM Phos-FLAG for 5 hours at 37 °C for detection of cell surface azides; whole cells were then lysed for Western blot, as described below. Alternatively, metabolically labeled cells were harvested, rinsed with PBS, lysed, and then reacted with 250 µM Phos-FLAG overnight at 37 °C. Protein lysates were standardized (BioRad’s DC protein concentration assay) and analyzed by Western blot.
Western Blot
Reacted lysates from metabolically labeled cells were standardized to a protein concentration of ~3 mg mL−1, loaded onto a 4–15% Tris–HCl SDS-PAGE gel, separated by electrophoresis, and transferred to nitrocellulose paper. Anti-FLAG-HRP was employed to visualize FLAG-tagged proteins via chemiluminescence.
Flow cytometry
Metabolically labeled intact cells were reacted with Phos-FLAG (250 µM) for 1 hour, then rinsed and treated with anti-FLAG-FITC to visualize FLAG-covered cells. Alternatively, the presence of azides on cells was probed using Click-IT Alexa Fluor 488 DIBO Alkyne (20 µM) for 5 hours in the dark for copper free click chemistry detection of azides (ThermoFisher Scientific; ex: 488/em: 519). Cells were analyzed by flow cytometry on a FACSCalibur (BD Biosciences) instrument, with 20,000 live cells gated for each replicate experiment.
Supplementary Material
Acknowledgments
We thank K. Moulton and A. McBride for helpful comments, M. Pratt and K. Chuh for generously providing Ac36AzGlcNAc, and P. Woodruff and M. Amieva for sharing bacterial strains. E.L.C. acknowledges support from a Surdna Foundation Undergraduate Research Fellowship, K.L.K. thanks Maine-INBRE for a summer research fellowship, M.E. and A.R.P thank CSIR-New Delhi for fellowships, and E.L.C., K.L.K., and J.D.H. received Grua/O’Connell Research Awards. Research reported in this publication was supported by the National Institutes of Health under grant number R15GM109397, by a Henry Dreyfus Teacher-Scholar award to D.H.D., and by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM0103423, and by Science and Engineering Research Board, Department of Science and Technology, Government of India (Grant No. EMR/2014/000235).
Footnotes
ASSOCIATED CONTENT
Supporting Information Available:
Detailed procedures for chemical syntheses and biological experiments, scanned NMR data for synthesized compounds, and Supplementary Figures. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Cotter PD, Stanton C, Ross RP, Hill C. The impact of antibiotics on the gut microbiota as revealed by high throughput DNA sequencing. Disc. Med. 2012;13:193–199. [PubMed] [Google Scholar]
- 2.Blaser M. Stop the killing of beneficial bacteria. Nature. 2011;476:393–394. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- 3.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zanevald J, Turnbaugh P, Lozupone C, Ley R, Hamady M, Gordon J, Knight R. Host-bacterial coevolution and the search for new drug targets. Curr. Opin. Chem. Biol. 2008;12:109–114. doi: 10.1016/j.cbpa.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dube DH, Champasa K, Wang B. Chemical tools to discover and target bacterial glycoproteins. Chem. Commun. 2011;47:87–101. doi: 10.1039/c0cc01557a. [DOI] [PubMed] [Google Scholar]
- 6.Tra VN, Dube DH. Glycans in pathogenic bacteria -- potential for targeted covalent therapeutics and imaging agents. Chem. Commun. 2014;50:4659–4673. doi: 10.1039/c4cc00660g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schirm M, Soo EC, Aubry AJ, Austin J, Thibault P, Logan SM. Structural, genetic and functional characterization of the flagellin glycosylation process in Helicobacter pylori. Mol. Microbiol. 2003;48:1579–1592. doi: 10.1046/j.1365-2958.2003.03527.x. [DOI] [PubMed] [Google Scholar]
- 8.Hopf PS, Ford RS, Zebian N, Merkx-Jacques A, Vijayakumar S, Ratnayake D, Hayworth J, Creuzenet C. Protein glycosylation in Helicobacter pylori: beyond the flagellins? PLoS ONE. 2011;6:e25722–e25722. doi: 10.1371/journal.pone.0025722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartley MD, Morrison MJ, Aas FE, Borud B, Koomey M, Imperiali B. Biochemical characterization of the O-linked glycosylation pathway in Neisseria gonorrhoeae responsible for biosynthesis of protein glycans containing N,N'-diacetylbacillosamine. Biochemistry. 2011;50:4936–4948. doi: 10.1021/bi2003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horzempa J, Held TK, Cross AS, Furst D, Qutyan M, Neely AN, Castric P. Immunization with a Pseudomonas aeruginosa 1244 pilin provides O-antigen-specific protection. Clin. Vacc. Immunol. 2008;15:590–597. doi: 10.1128/CVI.00476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baumann H, Tzianabos AO, Brisson JR, Kasper DL, Jennings HJ. Structural elucidation of two capsular polysaccharides from one strain of Bacteroides fragilis using high-resolution NMR spectroscopy. Biochemistry. 1992;31:4081–4089. doi: 10.1021/bi00131a026. [DOI] [PubMed] [Google Scholar]
- 12.Dube DH, Champasa K, Wang B. Chemical tools to discover and target bacterial glycoproteins. Chem. Commun. 2011;47:87–101. doi: 10.1039/c0cc01557a. [DOI] [PubMed] [Google Scholar]
- 13.Balonova L, Hernychova L, Bilkova Z. Bioanalytical tools for the discovery of eukaryotic glycoproteins applied to the analysis of bacterial glycoproteins. Exp. Rev. Proteomics. 2009;6:75–85. doi: 10.1586/14789450.6.1.75. [DOI] [PubMed] [Google Scholar]
- 14.Longwell SA, Dube DH. Deciphering the bacterial glycocode: recent advances in bacterial glycoproteomics. Curr. Opin. Chem. Biol. 2013;17:41–48. doi: 10.1016/j.cbpa.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linton D, Allan E, Karlyshev AV, Cronshaw AD, Wren BW. Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol. Microbiol. 2002;43:497–508. doi: 10.1046/j.1365-2958.2002.02762.x. [DOI] [PubMed] [Google Scholar]
- 16.Balnova L, Hernychova L, Bilkova Z. Bioanalytical tools for the discovery of eukaryotic glycoproteins applied to the analysis of bacterial glycoproteins. Exp. Rev. 2009;6:75–85. doi: 10.1586/14789450.6.1.75. [DOI] [PubMed] [Google Scholar]
- 17.Sambri V, Stefanelli C, Cevenini R. Detection of glycoproteins in Borrelia burgdorferi. Arch. Microbiol. 1992;157:205–208. doi: 10.1007/BF00245150. [DOI] [PubMed] [Google Scholar]
- 18.Dube DH, Bertozzi CR. Metabolic oligosaccharide engineering as a tool for glycobiology. Curr. Opin. Chem. Biol. 2003;7:616–625. doi: 10.1016/j.cbpa.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Laughlin ST, Bertozzi CR. Metabolic labeling of glycans with azido sugars and subsequent glycan-profiling and visualization via Staudinger ligation. Nat. Prot. 2007;2:2930–2944. doi: 10.1038/nprot.2007.422. [DOI] [PubMed] [Google Scholar]
- 20.Keppler OT, Horstkorte R, Pawlita M, Schmidts C, Reutter W. Biochemical engineering of the N-acyl side chain of sialic acid: biological implications. Glycobiology. 2001;11:11R–18R. doi: 10.1093/glycob/11.2.11r. [DOI] [PubMed] [Google Scholar]
- 21.Koenigs MB, Richardson EA, Dube DH. Metabolic profiling of Helicobacter pylori glycosylation. Mol. Biosyst. 2009;5:909–912. doi: 10.1039/b902178g. [DOI] [PubMed] [Google Scholar]
- 22.Prescher JA, Bertozzi CR. Chemistry in living systems. Nat. Chem. Biol. 2005;1:13–21. doi: 10.1038/nchembio0605-13. [DOI] [PubMed] [Google Scholar]
- 23.Saxon E, Bertozzi CR. Cell surface engineering by a modified Staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 24.Agard NJ, Prescher JA, Bertozzi CR. A strain-promoted [3+ 2] azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 25.Sletten EM, Bertozzi CR. From mechanism to mouse: a tale of two bioorthogonal reactions. Acc Chem Res. 2011;44:666–676. doi: 10.1021/ar200148z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dube DH. Metabolic labeling of bacterial glycans with chemical reporters. In: Reid CW, Twine SM, Reid AN, editors. Bacterial glycomics: current research, technology and applications. Norfolk, UK: Caister Academic Press; 2012. pp. 229–242. [Google Scholar]
- 27.Champasa K, Longwell SA, Eldridge AM, Stemmler EA, Dube DH. Targeted identification of glycosylated proteins in the gastric pathogen Helicobacter pylori. Mol. Cell. Prot. 2013;12:2568–2586. doi: 10.1074/mcp.M113.029561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saxon E, Luchansky SJ, Hang HC, Yu C, Lee SC, Bertozzi CR. Investigating cellular metabolism of synthetic azidosugars with the Staudinger ligation. J. Am. Chem. Soc. 2002;124:14893–14902. doi: 10.1021/ja027748x. [DOI] [PubMed] [Google Scholar]
- 29.Vocadlo DJ, Hang HC, Kim E-J, Hanover JA, Bertozzi CR. A chemical approach for identifying O-GlcNAc-modified proteins in cells. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9116–9121. doi: 10.1073/pnas.1632821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaewsapsak P, Esonu O, Dube DH. Recruiting the host's immune system to target Helicobacter pylori's surface glycans. ChemBioChem. 2013;14:721–726. doi: 10.1002/cbic.201300006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szymanski CM, Yao R, Ewing CP, Trust TJ, Guerry P. Evidence for a system of general protein glycosylation in Campylobacter jejuni. Mol. Microbiol. 1999;32:1022–1030. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- 32.Castric P. pilO, a gene required for glycosylation of Pseudomonas aeruginosa 1244 pilin. Microbiology. 1995;141:1247–1254. doi: 10.1099/13500872-141-5-1247. [DOI] [PubMed] [Google Scholar]
- 33.Goon S, Kelly JF, Logan SM, Ewing CP, Guerry P. Pseudaminic acid, the major modification on Campylobacter flagellin, is synthesized via the Cj1293 gene. Mol. Microbiol. 2003;50:659–671. doi: 10.1046/j.1365-2958.2003.03725.x. [DOI] [PubMed] [Google Scholar]
- 34.VanderVen BC, Harder JD, Crick DC, Belisle JT. Export-mediated assembly of Mycobacterial glycoproteins parallels eukaryotic pathways. Science. 2005;309:941–943. doi: 10.1126/science.1114347. [DOI] [PubMed] [Google Scholar]
- 35.Scott AE, Twine SM, Fulton KM, Titball RW, Essex-Lopresti AE, Atkins TP, Prior JL. Flagellar glycosylation in Burkholderia pseudomallei and Burkholderia thailandensis. J. Bacteriol. 2011;193:3577–3587. doi: 10.1128/JB.01385-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elhenawy W, Scott NE, Tondo ML, Orellano EG, Foster LJ, Feldman MF. Protein O-linked glycosylation in the plant pathogen Ralstonia solanacearum. Glycobiology. 2016;26:301–311. doi: 10.1093/glycob/cwv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell. 2009;137:321–331. doi: 10.1016/j.cell.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. U.S.A. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarkar AK, Fritz TA, Taylor WH, Esko JD. Disaccharide uptake and priming in animal cells: inhibition of sialyl Lewis X by acetylated Gal beta 1,4-GlcNAc beta-O-naphthalenemethanol. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3323–3327. doi: 10.1073/pnas.92.8.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emmadi M, Kulkarni SS. Expeditious synthesis of bacterial, rare sugar building blocks to access the prokaryotic glycome. Org. Biomol. Chem. 2013;11:3098–3102. doi: 10.1039/c3ob40615f. [DOI] [PubMed] [Google Scholar]
- 41.Emmadi M, Kulkarni SS. Synthesis of orthogonally protected bacterial, rare-sugar and D-glycosamine building blocks. Nat. Prot. 2013;8:1870–1889. doi: 10.1038/nprot.2013.113. [DOI] [PubMed] [Google Scholar]
- 42.Chuh KN, Zaro BW, Piller F, Piller V, Pratt MR. Changes in metabolic chemical reporter structure yield a selective probe of O-GlcNAc modification. J. Am. Chem. Soc. 2014;136:12283–12295. doi: 10.1021/ja504063c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hang HC, Yu C, Kato DL, Bertozzi CR. A metabolic labeling approach toward proteomic analysis of mucin-type O-linked glycosylation. Proc. Natl. Acad. Sci. U.S.A. 2003;100:14846–14851. doi: 10.1073/pnas.2335201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Geva-Zatorsky N, Alvarez D, Hudak JE, Reading NC, Erturk-Hasdemir D, Dasgupta S, von Andrian UH, Kasper DL. In vivo imaging and tracking of host-microbiota interactions via metabolic labeling of gut anaerobic bacteria. Nat. Med. 2015;21:1091–1100. doi: 10.1038/nm.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woo CM, Iavarone AT, Spiciarich DR, Palaniappan KK, Bertozzi CR. Isotope-targeted glycoproteomics (IsoTaG): a mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat. Meth. 2015;12:561–567. doi: 10.1038/nmeth.3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jewett JC, Sletten EM, Bertozzi CR. Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc. 2010;132:3688–3690. doi: 10.1021/ja100014q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohen AS, Dubikovskaya EA, Rush JS, Bertozzi CR. Real-time bioluminescence imaging of glycans on live cells. J. Am. Chem. Soc. 2010;132:8563–8565. doi: 10.1021/ja101766r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jewett JC, Bertozzi CR. Synthesis of a fluorogenic cyclooctyne activated by Cu-free click chemistry. Org. Lett. 2011;13:5937–5939. doi: 10.1021/ol2025026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Besanceney-Webler C, Jiang H, Wang W, Baughn AD, Wu P. Metabolic labeling of fucosylated glycoproteins in Bacteroidales species. Bioorg. Med. Chem. Lett. 2011;21:4989–4992. doi: 10.1016/j.bmcl.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu F, Aubry AJ, Schoenhofen IC, Logan SM, Tanner ME. The engineering of bacteria bearing azido-pseudaminic acid-modified flagella. Chembiochem. 2009;10:1317–1320. doi: 10.1002/cbic.200900018. [DOI] [PubMed] [Google Scholar]
- 51.Baltrus DA, Amieva MR, Covacci A, Lowe TM, Merrell DS, Ottemann KM, Stein M, Salama NR, Guillemin K. The complete genome sequence of Helicobacter pylori strain G27. J. Bacteriol. 2009;191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woodruff PJ, Carlson BL, Siridechadilok B, Pratt MR, Senaratne RH, Mougous JD, Riley LW, Williams SJ, Bertozzi CR. Trehalose is required for growth of Mycobacterium smegmatis. J. Biol. Chem. 2004;279:28835–28843. doi: 10.1074/jbc.M313103200. [DOI] [PubMed] [Google Scholar]
- 53.Luchansky SJ, Hang HC, Saxon E, Grunwell JR, Yu C, Dube DH, Bertozzi CR. Constructing azide-labeled cell surfaces using polysaccharide biosynthetic pathways. Meth. Enz. 2003;362:249–272. doi: 10.1016/S0076-6879(03)01018-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.