Abstract
Diverse glycans on proteins help impact cell and organism physiology along with drug activity. Since many protein-based biotherapeutics are glycosylated and these glycans have biological activity, there is a desire to engineer glycosylation for recombinant protein-based biotherapeutics. Engineered glycosylation can impact the recombinant protein efficacy and also influence many cell pathways by first changing glycan-protein interactions and consequently modulating disease physiologies. However, its complexity is enormous. Due to recent advances in glycoengineering, modulating protein-glycan interactions become more amenable to therapeutic approaches. Here, we discuss how engineered glycans contribute to therapeutic monoclonal antibodies (mAbs) in the treatment of cancers, how these glycoengineered therapeutic mAbs affect the transformed phenotypes and downstream cell pathways, and how systems biology can help in the next generation mAb glycoengineering process by aiding in data analysis and guiding engineering efforts to tailor mAb glycan and ultimately drug efficacy, safety and affordability.
Graphical Abstract
Introduction
Monoclonal antibodies (mAbs) are the major category of glycoprotein-based therapeutic drugs, approved by the US Food and Drug Administration (FDA) [1]. Furthermore, they have attained considerable success in many therapies, including cancer, over the past three decades [2]. Despite remarkable advances in contemporary biopharmaceutical technologies, many challenges remain in efficiently manufacturing effective and affordable antibody-based drugs. Since glycosylation essentially impacts the therapeutic efficacies of mAbs [3], it is desirable to control glycoforms on therapeutic mAbs for the next generation mAb development. With fast growing innovative engineering and cutting-edge technologies, glycoengineering provides a promising method to tune the activities of therapeutic mAbs [4,5]. An effective glycoengineered mAb usually modulates specific interactions between designed glycans and target proteins, thereby impacting the activity of downstream pathways that control cancer physiology. Conversely, a wrong glycan can induce unwanted side effects and even adverse immunogenic response [6]. For example, some colorectal cancer patients develop hypersensitivity to the FDA approved mAb cetuximab [7].
Several intriguing and unsolved questions in mAb glycoengineering include the following. Which glycan structures will provide the optimal mAb? How can we efficiently and reliably engineer a consistent glycoform on mAbs? Challenges in answering these questions stem from our limited understanding regarding the intricate relationships between glycans, proteins, and host cell physiologies. Furthermore, even when desired glycoforms are known, it has been difficult to unravel all of the factors that influence glycosylation and to control the complex system.0020Systems biology provides a powerful toolbox for integrating heterogeneous omics data and for deciphering the mechanisms and interactions between molecules and pathways, using network analysis, mathematical modeling, and simulation [8,9]. An abundance of omics technologies have been developed to aid in studying glycoengineering and expression systems (e.g., [10]), but the application of omics data and systems biology in glycoengineering is still in its infancy. Here we review the state-of-art knowledge of glycan-protein interactions in the context of FDA-approved therapeutic mAbs and then summarize several innovative technologies that can help control the glycoforms on mAbs. Finally, systems biology-based glycoengineering approaches are explored with an emphasis on how systems biology can be used to advance anti-tumor mAb development toward a predictable glycoengineering era.
Glycan-protein interaction, therapeutic antibody, and cancer physiology
Glycosylation helps to modulate interactions between mAbs and antigens or Fcγ receptors (Figure 1A), and impact the efficacy and safety of a biotherapeutic drug. The glycan-protein interactions of FDA-approved therapeutic mAbs in various cancer settings and their subsequent effects reported in the literature are summarized in Table 1.
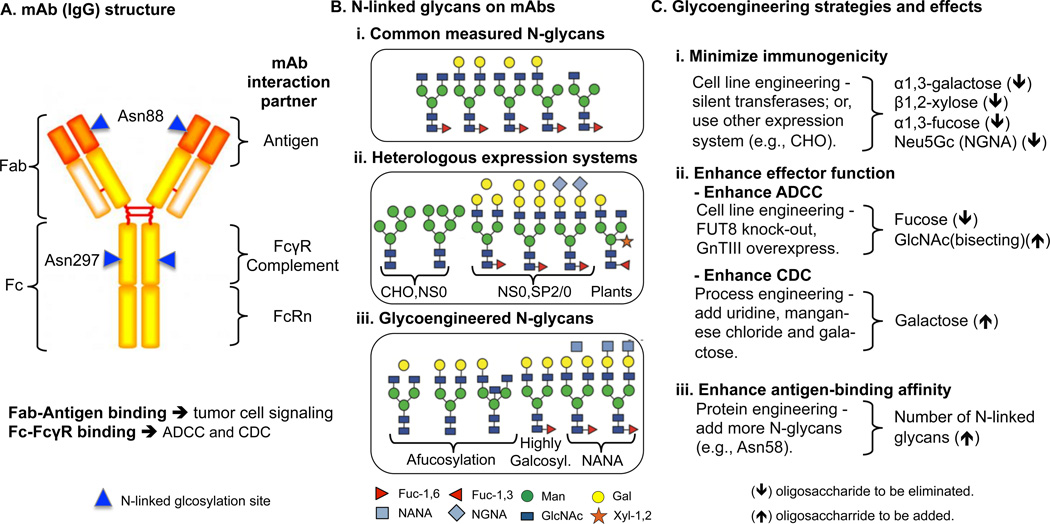
Figure 1. Therapeutic mAb structure, glycoforms, and glycoengineering strategies for generating desired glycoforms.
(A) The structure of an IgG with interaction-partner binding regions and N-linked glycosylation sites (highlighted in blue triangles) are annotated. (B) The dominant N-linked glycans on mAbs can vary depending on the host and product. However, (i) common glycans on therapeutic mAbs have been measured. (ii) MAbs expressed in heterologous expression systems introduce non-human compatible sugars and linkages, leading to immunogenicity and low serum half-life. (iii) Glycoengineering aims to make mAbs with N-glycans that are human compatible and exhibit enhanced mAb efficacy and safety. (C) Many glycoengineering efforts aim to enhance the drugs and achieve any of the three effects (i-iii) by modifying glycans on mAbs. NANA: N-glycolylneuraminic acid (hyper-sialylation). Data of (B) in this figure was adapted from [34].
Table 1.
Cancer physiology impacted by the FDA approved therapeutic mAbs through glycan-protein interactions
| Targeting mechanism |
Impacted physiologies in tumors |
State-of-art knowledge of glycan-protein interactions |
Examples of FDA-approved product (Receptor; First approved indications) |
|---|---|---|---|
| Fab-antigen binding | Tumor cell proliferation reduced (PI3K-AKT, MAPK); Apoptosis of tumor cells. |
|
Cetuximab (EGFR; Head and neck, colorectal cancer) Alemtuzumab (CD52; Chronic myeloid leukemia) Epratuzumab (CD22; Acute lymphocytic leukemia) |
| Angiogenesis inhibited |
|
Bevacizumab (VEGF; Colorectal cancer) | |
| T cell activation and tumor cell killed |
Fab-antigen binding affinity is affected by N- glycans on receptors (e.g., Asn16, Ans25, Asn41, and Asn83 at PD1). |
Pembrolizumab (PD1; Ipi-refractory melanoma) Nivolumab (PD1; Metastatic melanoma) |
|
| Fc-FcγR binding | Tumor cell killed by immune cells (ADCC/CDC) |
|
Mogamulizumab (CCR4; T-cell leukemia/lymphoma) Rituximab (CD20; Chronic lymphocytic leukemia) Obinutuzumab (CD20; Chronic lymphocytic leukemia) |
Fab-antigen interaction
Fab glycans have several roles in modulating interactions with receptors and glycoengineering can help reduce negative interactions leading to immunogenicity. Early research demonstrated that engineering N-linked oligosaccharides on the Fab region can enhance the antigen-binding affinity of mAbs [11]. However, a comprehensive understanding of the glycans on Fab region is still lacking. One example of their role is the allergenic responses to therapeutic antibody cetuximab [7]. It was produced by murine myeloma cells (SP2/0), which adds an additional α1,3-galactose (the α-Gal epitope) on the N-linked oligosaccharide at Asn88 of the Fab region (Figure 1A). Unfortunately, the human IgE recognizes the non-human α-Gal epitope and leads to downstream immune responses, such as hypersensitivity reactions (anaphylaxis) after drug treatments. Therefore, extra efforts must be made to glycoengineer the drug to reduce α-Gal content and solve the immunogenicity problem (e.g., [12]). Moreover, the Fab-antigen binding affinity could be affected by targeting receptors. Recent mutagenesis studies showed that the potential glycosylation sites (Asn16, Ans25, Asn41, and Asn83) on Programmed death 1 (PD1) could impact antigen-binding affinity (e.g., nivolumab and pembrolizumab) by influencing its local structure [13]. Physiologically, PD1 is an inhibitory receptor that suppresses T cell responses to avoiding auto-immunity. Indeed, many factors can affect Fab-antigen binding in cancer treatment, and glycoengineering can be applied in mAb design either to optimize Fab-antigen binding affinity or to eliminate immunogenicity problem.
Beyond immunogenicity, Fab-binding can target cancer by modulating glycan-protein interactions. This occurs by either directly targeting glycans (Fab-glycan interaction) or indirectly modulating downstream protein-glycan interactions to treat cancer. At least four types of glycan-protein interactions involving the Fab have been explored. First, the Fab can interact with a target glycan, such as seen with Alemtuzumab which directly targets a glycosylphosphatidylinositol (GPI) anchor on the CAMPATH-1 (CD52) antigen to treat chronic myeloid leukemia. CD52 plays a dominant role in mediating cell depletion via apoptosis [14]. Second, the Fab can target siglec-sialic acid interactions of inhibitory siglecs (e.g., CD22) that normally prevent B-cell over-stimulation by inhibiting B-cell receptor signaling [15]. For example, the anti-CD22 antibody (epratuzumab) targets siglec-sialic acid interactions between B cells with endothelial cells, leading to downstream B-cell receptor signaling suppression [16]. Third, the Fab can interfere with angiogenesis by blocking interactions between glycans and VEGF, a key inducer for several downstream signaling pathways (e.g., MAPK and PI3K/AKT [17]) that lead to vascular and stromal cell ablation [18]. Bevacizumab binds VEGF to interrupt interactions between VEGF and heparin sulfate proteoglycans (HSPGs), leading to VEGF-VEGFR interaction blockade [19]. Fourth, mAbs can modulate Gal1-N-linked glycan interactions. Specifically, anti-VEGF refractory tumors can secrete excess Galectin-1 (Gal1). This enables tumor immune escape and metastasis by interacting with the β1,6-GlcNAc branching N-glycans on the endothelial cell receptors [20]. Anti-Gal1 antibodies can block the interaction between Gal1 and the N-glycan on VEGFR, leading to angiogenesis inhibition. While some mAbs might not require a glycosylation on the Fab-antigen binding site, to successfully interrupt downstream glycan-protein interactions, engineering glycosylation can improve their pharmacological properties (e.g., stability and effector functions) for maintaining optimal efficacy (see discussion below).
Fc-FcγR interaction
Fc binding with the Fcγ receptor (FcγR) on immune cells is crucial to mAb efficacy in governing downstream immunological responses of complement-dependent cytotoxicity (CDC) [21] and antibody-dependent cellular cytotoxicity (ADCC) [22] activities, which are immune mechanisms for killing target cells. In essence, the interaction between Fc and FcγR is a glycan-glycan interaction that is significantly affected by Fc-glycosylation (at Asn297). Indeed, mAbs with aglycosylated Fc regions lose effector functions, and mAbs with glycosylated Fc can activate downstream effector mechanisms. However, different engineered glycoforms significantly alter the efficacies of effector functions; for example, removal of the α1,6-linked core fucose from IgG-Fc glycans results in better effector functions compared to the IgG with fucosylated Fc glycans [23]. The N-glycans attached to the FcγRIIIa mediate the interaction with nonfucosylated IgG1-Fc, thereby stabilizing the Fc-FcγRIIIa complex, while fucosylation of the Fc N-glycans otherwise inhibits the interaction due to the steric hindrance. NMR analysis shows that defucosylation facilitates the active conformation of the Fc Tyr296 and the formation of a high-affinity complex; however, presence of fucose inhibits complex formation by reducing the flexibility of Tyr296. Other modifications to glycans further impact function. For example., bi-antennary N-glycans that are terminated with alpha-2,6-linked sialic acids is an optimal structure to enhance effector functions by strengthening interactions with the FcγRIIIa [24]. Intriguingly, effector functions are highly correlated with structural stability [25,26]. Glycosylated mAbs exhibit better structural stability and aglycosylated mAbs tend to unfold [27]. Indeed, a recent study indicated that an Fc Nglycan substantially stabilized the conformation of the Fc C’E loop by interacting with self-protein. Interestingly, the C’E loop stability is correlated with the Fc-binding affinity [28]. Moreover, an Fc with bisecting or high galactosylation N-glycans may open up the horseshoe-shaped conformation of the Fc fragment, which is favored by Fc-FcγR interaction, resulting in the enhanced effector functions [29]. In addition, different glycoforms also impact the pharmacokinetic behavior of mAbs [30]. Indeed, glycosylated mAbs can significantly increase their serum half-life, due to the prevention of proteolytic degradation [31]. Indeed, conformational changes of aglycosylated mAbs increase protease accessibility to the hinge region [32]. Notably, terminal sugars affect proteolytic resistance; glycoengineered mAb terminated with GlcNAc (G0 glycoforms) showed at least two times greater resistance to papain digestion than those terminated with sialic acid (G2S2 glycoform) and beta-galactose (G2 glycoform) [33].
Two glycoengineered mAbs have been approved by the FDA — mogamulizumab and obinutuzumab. Mogamulizumab targets CC chemokine receptor 4 (CCR4) to inhibit CCR4-mediated signal transduction pathways for T-cell leukemia/lymphoma treatment [34]. The antitumor activity strongly relies on ADCC, which is enhanced by glycoengineered afucosylated-Fc [35]. Obinutuzumab targets CD20 to treat chronic lymphocytic leukemia by directly inducing tumor cell apoptosis. Obinutuzumab demonstrated superior antitumor activity than rituximab, a leading FDA-approved CD20-targeting mAb. The superiority of obinutuzumab comes from the enhanced ADCC by glycoengineering afucosylated N-glycans with bisecting GlcNAc [36]. Inspired by these successful examples, more than 20 glycoengineered mAbs are currently in clinical trials.
Remarkably, the structure of glycans and their locations are the two most prominent properties governing the interaction between mAb and receptor, three generally desired properties of glycosylation on mAbs are summarized. First, removal of the N-glycan on Fab (Asn88) could avoid immunogenicity. Second, modulation of N-glycans on the Fc (Asn297) can enhance structural stability and Fc binding affinity. Third, three structures for the N-glycan on Fc (Asn297) have shown to enhance effector functions (ADCC/CDC): the bi-antennary N-glycans terminated with two alpha-2,6-linked sialic acids (ADCC) [24], the afucosylated Fc (ADCC) [23], and the high galactosylated Fc (CDC) [42]. These properties have been considered extensively while glycoengineering mAbs, but it is possible that many more desirable structures and locations on mAbs remain undiscovered.
Glycoengineering to control glycoforms on therapeutic proteins
While there is a vast diversity of glycans in mammalian cells, far fewer are commonly on mAbs (Figure 1B). Novel technologies and knowledge allow us to modify the glycan for more effective mAbs for cancer treatments. Glycoengineered mAbs can enhance therapeutic efficacy (antigen binding affinity and effector functions) and safety (immunogenicity). They also influence other properties (e.g., pharmacokinetics (PK) and pharmacodynamics (PD)) on biotherapeutics, which has been covered elsewhere recently (see [37–39]).
There are three major considerations when glycoengineering mAbs for cancer therapy. First, modified glycans should minimize immunogenicity. Most protein expression systems are nonhuman, and if the non-human sugars and linkages are added to the mAbs (e.g., fucose or xylose in plants [40]), severe adverse drug reactions can occur (e.g., "Cytokine Storm" of TGN142 [41] and anaphylaxis of cetuximab [7]). Indeed, several glycoforms from heterologous expression systems (see Figure 1B (ii)) have been identified to be immunogenic to human. Thus, incompatible expression systems need to be humanized by knocking out relevant transferases (Figure 1C (i)), or human-compatible expression systems (e.g., Chinese hamster ovary cells) must be selected to create safe and bioactive glycoforms.
A second consideration for glycoengineering in cancer therapeutics is the need to enhance effector functions, such as ADCC and CDC. Many studies have reported that N-linked glycans at Asn297 in the Fc region are crucial to ADCC. Glycoengineering by depleting fucosylation or increasing N-glycan branching (e.g. by over-expressing the GnTIII gene) can improve ADCC. These glycoforms increase binding affinity to Fc-FcγRIIIa [25]. The CDC effector function can also be modulated through glycoengineering. Specifically, high galactosylation can improve effector function of CDC, and this can be achieved by process engineering of adding uridine, manganese chloride, and galactose in media [42]. The increased galactosylation can improve the binding affinity between the mAb and complement (C1q) [43].
A third consideration is that glycoengineering can enhance antigen-binding affinity. mAb affinity has been widely studied, and efforts use tools like phage-display libraries, combined with point mutations, to engineer the protein for improving antigen-binding affinity [44]. However, glycoengineering mAbs by introducing more N-glycosylation sites using protein engineering can improve antigen-binding affinity. For example, adding an N-glycan at Asn58 on Fab region can increase antigen-binding affinity by 50 fold [11].
Systems biology for predictive and rational glycoengineering of mAbs
In the past decade, considerable efforts have aimed to control glycosylation through media optimization, chemical treatments, or genetic changes (e.g., [45]). Despite many successful examples, it is still challenging to obtain desired quality and quantity of mAbs. However, omics technologies and systems biology modeling promise to aid in the glycoengineering of improved anti-tumor mAbs [10] (Figure 2). Indeed, systems biology provides platforms to integrate omics data with a holistic perspective, thereby guiding glycoengineering while accounting for competing pathways and processes [46]. Moreover, computational models in systems biology can be used for predicting knock-in or knockout strategies to obtain desired glycoforms [47].
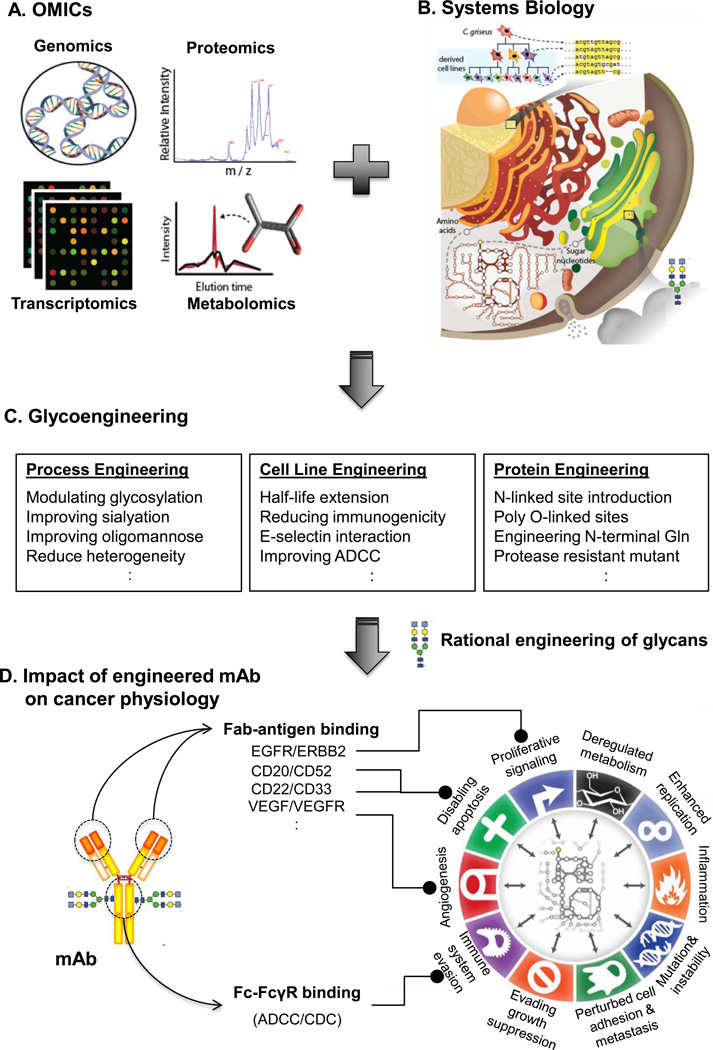
Figure 2. Omics data and systems biology provide novel tools for rational glycoengineering of therapeutic mAbs and to assess the impact of glycoengineered mAbs on cancer.
(A) Omics data can be acquired to identify how to glycoengineer mAb production cells. (B) Computational models of the pathways influencing glycosylation and protein production can serve as a platform for interpreting omics data and (C) guiding the rational design of glycans on therapeutic mAbs. (D) The impact of engineered mAbs on cancer physiology can further be analyzed using systems biology techniques, especially assessing the glycan-protein interactions [9].
Systems biology holds great promise in advancing at least three glycoengineering processes. First, it can be used for reliable cell line development. Cell line development is often a bottleneck step in protein-based drug manufacturing (usually taking more than a year). Many cellular processes must be considered during cell line development, since they influence protein quality and glycosylation. These include metabolism, protein secretion, and cell growth/apoptosis [48]. Computational models can be developed to understand how to control these processes in expression systems, as we can predict growth characteristics and production capabilities of mAbs [49,50]. Second, with increased understanding of the molecular pathways influencing mAb production, models can be used for predictable glycoengineering. The complexity of glycosylation greatly hampers the glycoengineering process. Computational models have been developed that enables us to predict glycoforms under different glycoengineering strategies [47,51]. The third area where systems biology can aid in glycoengineering is for its use in assessing the physiological effects of glycoengineering. Some glycoengineering designs have proven toxic to host cell lines; for example, the OCH1 deletion [52] and double knockouts of PMT-family genes [53] in yeast were shown to significantly reduce host-cell fitness. However, there has not been a systematic study concerning how glycoengineering affects host cells and their protein secretion system. In the future, we can combine several omics data (e.g., transcriptomics [54,55] and glycomics [56]) and relevant phenotypes (e.g., [57]) to analyze them with systems biology approaches. These results can provide insights into how key metabolic and signaling pathways are modulated by alterations in glycosylation [58]. We are also hopeful that future research should be pursued in developing reliable prediction models for predicting outcomes of glycosylation alterations on mAbs. Ultimately, a clear blueprint of what glycosylation to be engineered on a mAb with desire properties of low cost, safety and effectiveness will be revealed.
Conclusions
While therapeutic effects of mAbs are profoundly influenced by how the glycans impact cancer physiology, the detailed mechanistic relationships are largely unknown. By analyzing the FDA-approved mAbs in oncology indications, deeper insights will be obtained into how glycosylation and glycan-protein interactions impact on cancer physiology. In the future, systems biology technologies will aid in glycoengineering as it will allow us to better control and engineer glycans at the system level, thus, facilitating the development of novel therapeutic mAbs with increased efficacy, safety, and affordability.
Highlights.
-
*
Glycan-protein interactions modulate tumor cell killing.
-
*
Different glycans can significantly alter the glycan-protein interactions.
-
*
Glycan structure and location on mAb are essential properties need careful control.
-
*
Glycoengineering can modify glycans on therapeutic mAbs with desired glycoforms.
-
*
Systems biology can advance mAb glycoengineering toward a rational design era.
Acknowledgments
This work was supported by generous funding from the Novo Nordisk Foundation provided to the Center for Biosustainability at the Technical University of Denmark (grant no. NNF16CC0021858), and from NIGMS (grant no. R35 GM119850). AWTC was also partially supported by a Postdoctoral fellowship from the Ministry of Science and Technology of Taiwan (grant no. 104-2917-I-564-028).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no conflict of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1.Walsh G. Biopharmaceutical benchmarks 2014. Nat. Biotechnol. 2014;32:992–1000. doi: 10.1038/nbt.3040. [DOI] [PubMed] [Google Scholar]
- 2.Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat. Rev. Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 3.Jefferis R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat. Rev. Drug Discov. 2009;8:226–234. doi: 10.1038/nrd2804. [DOI] [PubMed] [Google Scholar]
- 4. Herter S, Birk MC, Klein C, Gerdes C, Umana P, Bacac M. Glycoengineering of therapeutic antibodies enhances monocyte/macrophage-mediated phagocytosis and cytotoxicity. J. Immunol. 2014;192:2252–2260. doi: 10.4049/jimmunol.1301249. ** This paper demonstrated that glycoengineered antibodies could enhance effector functions through enhanced binding to Fcγ receptors.
- 5.Elliott S, Lorenzini T, Asher S, Aoki K, Brankow D, Buck L, Busse L, Chang D, Fuller J, Grant J, et al. Enhancement of therapeutic protein in vivo activities through glycoengineering. Nat. Biotechnol. 2003;21:414–421. doi: 10.1038/nbt799. [DOI] [PubMed] [Google Scholar]
- 6.Hansel TT, Kropshofer H, Singer T, Mitchell JA, George AJT. The safety and side effects of monoclonal antibodies. Nat. Rev. Drug Discov. 2010;9:325–338. doi: 10.1038/nrd3003. [DOI] [PubMed] [Google Scholar]
- 7.Chung CH, Mirakhur B, Chan E, Le QT, Berlin J, Morse M, Murphy BA, Satinover SM, Hosen J, Mauro D, et al. Cetuximab-induced anaphylaxis and IgE specific for galactose-alpha-1,3-galactose. N. Engl. J. Med. 2008;358:1109–1117. doi: 10.1056/NEJMoa074943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis NE, Schramm G, Bordbar A, Schellenberger J, Andersen MP, Cheng JK, Patel N, Yee A, Lewis RA, Eils R, et al. Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat. Biotech. 2010;28:1279–1285. doi: 10.1038/nbt.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis NE, Abdel-Haleem AM. The evolution of genome-scale models of cancer metabolism. Front. Physiol. 2013;4:237. doi: 10.3389/fphys.2013.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kildegaard HF, Baycin-Hizal D, Lewis NE, Betenbaugh MJ. The emerging CHO systems biology era: Harnessing the ’omics revolution for biotechnology. Curr. Opin. Biotechnol. 2013;24:1102–1107. doi: 10.1016/j.copbio.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 11.Wright A, Tao MH, Kabat EA, Morrison SL. Antibody variable region glycosylation: position effects on antigen binding and carbohydrate structure. EMBO J. 1991;10:2717–2723. doi: 10.1002/j.1460-2075.1991.tb07819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi C, Ruan C, Wang H, Xu X, Zhao Y, Fang M, Ji J, Gu X, Gao C. Function characterization of a glyco-engineered anti-EGFR monoclonal antibody cetuximab in vitro. Acta Pharmacol. Sin. 2014;35:1439–1446. doi: 10.1038/aps.2014.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Schwartz JCD, Guo X, Bhatia S, Cao E, Chen L, Zhang ZY, Edidin MA, Nathenson SG, Almo SC. Structural and functional analysis of the costimulatory receptor programmed death-1. Immunity. 2004;20:337–347. doi: 10.1016/s1074-7613(04)00051-2. [DOI] [PubMed] [Google Scholar]
- 14.James LC, Hale G, Waldmann H, Bloomer AC. 1.9 Å structure of the therapeutic antibody Campath-1H Fab in complex with a synthetic peptide antigen. J. Mol. Biol. 1999;289:293–301. doi: 10.1006/jmbi.1999.2750. [DOI] [PubMed] [Google Scholar]
- 15.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat. Rev. Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 16.Chang CH, Wang Y, Gupta P, Goldenberg DM. Extensive crosslinking of CD22 by epratuzumab triggers BCR signaling and caspase-dependent apoptosis in human lymphoma cells. MAbs. 2015;7:199–211. doi: 10.4161/19420862.2014.979081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goel HL, Mercurio AM. VEGF targets the tumour cell. Nat. Rev. Cancer. 2013;13:871–882. doi: 10.1038/nrc3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Smith GA, Fearnley GW, Tomlinson DC, Harrison MA, Ponnambalam S. The cellular response to vascular endothelial growth factors requires co-ordinated signal transduction, trafficking and proteolysis. Biosci. Rep. 2015;35(5):e00253. doi: 10.1042/BSR20150171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Croci DO, Cerliani JP, Dalotto-Moreno T, Méndez-Huergo SP, Mascanfroni ID, Dergan-Dylon S, Toscano MA, Caramelo JJ, García-Vallejo JJ, Ouyang J, et al. Glycosylation-dependent lectin-receptor interactions preserve angiogenesis in anti-VEGF refractory tumors. Cell. 2014;156:744–758. doi: 10.1016/j.cell.2014.01.043. * The authors identified the Gal1-VEGFR interaction in anti-VEGF refractory tumors, and they also demonstrated the effects of anti-Gal1 mAb in preventing compensatory angiogenesis, which has therapeutic potential.
- 21.Macor P, Tedesco F. Complement as effector system in cancer immunotherapy. Immunol. Lett. 2007;111:6–13. doi: 10.1016/j.imlet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 22.Clynes R, Takechi Y, Moroi Y, Houghton a, Ravetch JV. Fc receptors are required in passive and active immunity to melanoma. Proc. Natl. Acad. Sci. U.S.A. 1998;95:652–656. doi: 10.1073/pnas.95.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrara C, Grau S, Jager C, Sondermann P, Brunker P, Waldhauer I, Hennig M, Ruf A, Rufer AC, Stihle M, et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcγRIII and antibodies lacking core fucose. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin CW, Tsai MH, Li ST, Tsai TI, Chu KC, Liu Y, Lai MY, Wu CY, Tseng YC, Shivatare SS, et al. A common glycan structure on immunoglobulin G for enhancement of effector functions. Proc. Natl. Acad. Sci. U.S.A. 2015;112:201513456. doi: 10.1073/pnas.1513456112. ** The authors developed an effective method to modify the Fc-glycan structures to a homogeneous glycoforms, and they identified a common and optimized structure (the bi-antennary N-glycans that are terminated with two alpha-2,6-linked sialic acids) for the enhancement of effector functions.
- 25.Reusch D, Tejada ML. Fc glycans of therapeutic antibodies as critical quality attributes (CQAs) Glycobiology. 2015;25:1–45. doi: 10.1093/glycob/cwv065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayes JM, Cosgrave EFJ, Struwe WB, Wormald M, Davey GP, Jefferis R, Rudd PM. Glycosylation and Fc receptors. Curr. Top. Microbiol. Immunol. 2014;382:165–199. doi: 10.1007/978-3-319-07911-0_8. [DOI] [PubMed] [Google Scholar]
- 27.Zheng K, Yarmarkovich M, Bantog C, Bayer R, Patapoff TW. Influence of glycosylation pattern on the molecular properties of monoclonal antibodies. MAbs. 2014;6:649–658. doi: 10.4161/mabs.28588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Subedi GP, Barb AW. The Structural Role of Antibody N-Glycosylation in Receptor Interactions. Structure. 2015;23:1573–1583. doi: 10.1016/j.str.2015.06.015. * By measuring the conformation of C′E loop in several Fc variants, this study found that the structural role of N-glycan on Fc is primary to stabilize the Fc C′E loop through the intra-molecular interaction between carbohydrate and amino acids on Fc C′E loop. Moreover, this feature also confers the capability on N-glycan in tuning FcγRIIIa binding affinity.
- 29.Krapp S, Mimura Y, Jefferis R, Huber R, Sondermann P. Structural analysis of human IgG-Fc glycoforms reveals a correlation between glycosylation and structural integrity. J. Mol. Biol. 2003;325:979–989. doi: 10.1016/s0022-2836(02)01250-0. [DOI] [PubMed] [Google Scholar]
- 30. Datta-Mannan A, Huang L, Pereira J, Yaden B, Korytko A, Croy JE. Insights into the impact of heterogeneous glycosylation on the pharmacokinetic behavior of follistatin-Fc-based biotherapeutics. Drug Metab. Dispos. 2015;43:1882–1890. doi: 10.1124/dmd.115.064519. * The authors provided a LC/MS approach in characterizing the pharmacokinetic behavior of different glycans on follistatin-Fc-based antibodies. This paper highlights how glycan changes can impact the pharmacokinetic functions of the antibodies.
- 31.Solá RJ, Griebenow K. Glycosylation of therapeutic proteins: An effective strategy to optimize efficacy. BioDrugs. 2010;24(1):9–21. doi: 10.2165/11530550-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng K, Bantog C, Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. MAbs. 2011;3 doi: 10.4161/mabs.3.6.17922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raju TS, Scallon B. Fc glycans terminated with N-acetylglucosamine residues increase antibody resistance to papain. Biotechnol. Prog. 2007;23:964–971. doi: 10.1021/bp070118k. [DOI] [PubMed] [Google Scholar]
- 34.Beck A, Reichert JM. Marketing approval of mogamulizumab: A triumph for glyco-engineering. MAbs. 2012;4:419–425. doi: 10.4161/mabs.20996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ishii T, Ishida T, Utsunomiya A, Inagaki A, Yano H, Komatsu H, Iida S, Imada K, Uchiyama T, Akinaga S, et al. Defucosylated humanized aanti-CCR4 monoclonal antibody KW-0761 as a novel immunotherapeutic agent for adult T-cell leukemia/lymphoma. Clin. Cancer Res. 2010;16:1520–1531. doi: 10.1158/1078-0432.CCR-09-2697. [DOI] [PubMed] [Google Scholar]
- 36. Illidge T, Klein C, Sehn LH, Davies A, Salles G, Cartron G. Obinutuzumab in hematologic malignancies: Lessons learned to date. Cancer Treat. Rev. 2015;41:784–792. doi: 10.1016/j.ctrv.2015.07.003. * A detailed review of the glycoengineered mAb obinutuzumab in the treatment of chronic lymphocytic leukemia (CLL) with emphasis on discussing preclinical data and the potential for improving drug efficacy in a number of ongoing phase III clinical trials therapeutics for CLL.
- 37. Liu L. Antibody glycosylation and its impact on the pharmacokinetics and pharmacodynamics of monoclonal antibodies and Fc-fusion proteins. J. Pharm. Sci. 2015;104:1866–1884. doi: 10.1002/jps.24444. ** An up-to-date and comprehensive review of the impacts of antibody glycosylation on the pharmacokinetics and pharmacodynamics.
- 38.Carter PJ. Potent antibody therapeutics by design. Nat. Rev. Immunol. 2006;6:343–357. doi: 10.1038/nri1837. [DOI] [PubMed] [Google Scholar]
- 39.Zhong X, Wright JF. Biological insights into therapeutic protein modifications throughout trafficking and their biopharmaceutical applications. Int. J. Cell Biol. 2013;2013:273086. doi: 10.1155/2013/273086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tekoah Y, Ko K, Koprowski H, Harvey DJ, Wormald MR, Dwek RA, Rudd PM. Controlled glycosylation of therapeutic antibodies in plants. Arch. Biochem. Biophys. 2004;426:266–278. doi: 10.1016/j.abb.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 41.Kenter M, Cohen A. Establishing risk of human experimentation with drugs: lessons from TGN1412. Lancet. 2006;368:1387–1391. doi: 10.1016/S0140-6736(06)69562-7. [DOI] [PubMed] [Google Scholar]
- 42.Gramer MJ, Eckblad JJ, Donahue R, Brown J, Shultz C, Vickerman K, Priem P, van den Bremer ETJ, Gerritsen J, van Berkel PHC. Modulation of antibody galactosylation through feeding of uridine, manganese chloride, and galactose. Biotechnol. Bioeng. 2011;108:1591–1602. doi: 10.1002/bit.23075. [DOI] [PubMed] [Google Scholar]
- 43.Boyd PN, Lines AC, Patel AK. The effect of the removal of sialic acid, galactose and total carbohydrate on the functional activity of Campath-1H. Mol. Immunol. 1995;32:1311–1318. doi: 10.1016/0161-5890(95)00118-2. [DOI] [PubMed] [Google Scholar]
- 44.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 45.Fan Y, Jimenez Del Val I, Müller C, Wagtberg Sen J, Rasmussen SK, Kontoravdi C, Weilguny D, Andersen MR. Amino acid and glucose metabolism in fed-batch CHO cell culture affects antibody production and glycosylation. Biotechnol. Bioeng. 2015;112:521–535. doi: 10.1002/bit.25450. [DOI] [PubMed] [Google Scholar]
- 46. Kaas CS, Fan Y, Weilguny D, Kristensen C, Kildegaard HF, Andersen MR. Toward genome-scale models of the Chinese hamster ovary cells: incentives, status and perspectives. Pharm. Bioprocess. 2014;2:437–448. * A review of current progress of genome-scale modeling across several species and discuss their potential application in CHO cell metabolic engineering and genome annotation.
- 47. Spahn PN, Hansen AH, Hansen HG, Arnsdorf J, Kildegaard HF, Lewis NE. A Markov chain model for N-linked protein glycosylation - towards a low-parameter tool for model-driven glycoengineering. Metab. Eng. 2016;33:52–66. doi: 10.1016/j.ymben.2015.10.007. * A Markov chain model of N-linked glycosylation, in which the model learns glycosyl-transferase activities for a given protein by fitting experimentally measured levels of each glycan to the model. Then, the model predicts glycosylation after glycosyltransferase knock-downs or overexpression.
- 48.Seth G, Hossler P, Yee JC, Hu WS. Engineering cells for cell culture bioprocessing--physiological fundamentals. Adv. Biochem. Eng. Biotechnol. 2006;101:119–164. doi: 10.1007/10_017. [DOI] [PubMed] [Google Scholar]
- 49.Nocon J, Steiger MG, Pfeffer M, Sohn SB, Kim TY, Maurer M, Rußmayer H, Pflügl S, Ask M, Haberhauer-Troyer C, et al. Model based engineering of Pichia pastoris central metabolism enhances recombinant protein production. Metab. Eng. 2014;24:129–138. doi: 10.1016/j.ymben.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutierrez JM, Lewis NE. Optimizing eukaryotic cell hosts for protein production through systems biotechnology and genome-scale modeling. Biotechnol. J. 2015;10:939–949. doi: 10.1002/biot.201400647. [DOI] [PubMed] [Google Scholar]
- 51. Sha S, Agarabi C, Brorson K, Lee DY, Yoon S. N-Glycosylation Design and Control of Therapeutic Monoclonal Antibodies. Trends Biotechnol. 2016 doi: 10.1016/j.tibtech.2016.02.013. pii:S0167-7799(16)00047-0. ** A comprehensive review of current progress of systems biology on the N-glycosylation design and control of therapeutic monoclonal antibodies.
- 52.Castilho A. Glyco-Engineering. Methods Mol. Biol. 2015;1321:v–vii. doi: 10.1007/978-1-4939-2760-9. [DOI] [PubMed] [Google Scholar]
- 53.Gentzsch M, Tanner W. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 1996;15:5752–5759. [PMC free article] [PubMed] [Google Scholar]
- 54.Könitzer JD, Müller MM, Leparc G, Pauers M, Bechmann J, Schulz P, Schaub J, Enenkel B, Hildebrandt T, Hampel M, et al. A global RNA-seq-driven analysis of CHO host and production cell lines reveals distinct differential expression patterns of genes contributing to recombinant antibody glycosylation. Biotechnol. J. 2015;10:1412–1423. doi: 10.1002/biot.201400652. [DOI] [PubMed] [Google Scholar]
- 55.Jamnikar U, Nikolic P, Belic A, Blas M, Gaser D, Francky A, Laux H, Blejec A, Baebler S, Gruden K. Transcriptome study and identification of potential marker genes related to the stable expression of recombinant proteins in CHO clones. BMC Biotechnol. 2015;15:98. doi: 10.1186/s12896-015-0218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ranzinger R, Herget S, von der Lieth CW, Frank M. GlycomeDB-A unified database for carbohydrate structures. Nucleic Acids Res. 2011;39:D373–D376. doi: 10.1093/nar/gkq1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meuris L, Santens F, Elson G, Festjens N, Boone M, Dos Santos A, Devos S, Rousseau F, Plets E, Houthuys E, et al. GlycoDelete engineering of mammalian cells simplifies N-glycosylation of recombinant proteins. Nat. Biotechnol. 2014;32:485–489. doi: 10.1038/nbt.2885. ** The authors introduced a strategy to reduce N-glycosylation heterogeneity in mammalian cell-based glycoprotein production by truncating glycans.
- 58.Bennun SV, Yarema KJ, Betenbaugh MJ, Krambeck FJ. Integration of the Transcriptome and Glycome for Identification of Glycan Cell Signatures. PLoS Comput. Biol. 2013;9(1):e1002813. doi: 10.1371/journal.pcbi.1002813. [DOI] [PMC free article] [PubMed] [Google Scholar]