Abstract
Mice engrafted with human CD34+ hematopoietic stem and progenitor cells (CD34+-HSPCs) have been used to study human infection, diabetes, sepsis and burn, suggesting that they could be highly amenable to characterizing the human inflammatory response to injury. To this end, we analyzed human leukocytes infiltrating subcutaneous implants of polytetrafluoroethylene (PVA) sponges in immunodeficient NSG mice reconstituted with CD34+-HSPCs. We report that human CD45+ (hCD45+) leukocytes were present in PVA sponges 3 and 7 days post-implantation and could be localized within the sponges by immunohistochemistry. The different CD45+ sub-types were characterized by flow cytometry and the profile of human cytokines they secreted into PVA wound fluid was assessed using a human-specific multiplex bead analyses of human IL-12p70, TNFα, IL-10, IL-6, IL1β, and IL-8. This enabled tracking the functional contributions of HLA-DR+, CD33+, CD19+, CD62L+, CD11b+ or CX3CR1+ hCD45+ infiltrating inflammatory leukocytes. PCR of cDNA prepared from these cells enabled the assessment and differentiation of human, mouse and uniquely human genes. These findings support the hypothesis that mice engrafted with CD34+-HSPCs can be deployed as precision avatars to study the human inflammatory response to injury.
Introduction
The possibility that there might exist species-specific differences in the response to injury was first raised when global leukocyte gene expression levels were compared in humans and mice after trauma, burn or infection (1). Although initially controversial (2–4), these reports renewed a long-standing scientific dialogue on whether differences in mouse and human responses can be ignored in translational models of human disease (5–10). Fortunately, several approaches exist that enable investigators to study human genes and human cell responsiveness in an experimentally tractable mouse background. For example, when the HIV/AIDS research community encountered problems modeling the disease progression in standard mouse models because of the human-specific tropism of the HIV virus, they created chimeric mice with human CD34+-HSPCs grafts to study infection (11–13). Since then, the wider availability of these models has led to their widespread use to study other human diseases including diabetes, sepsis and burn injury (14–21).
Mice engrafted with human immune cells offer several advantages to injury research. Historically, research in wound repair and regeneration has almost universally focused on genes that are highly conserved between species, with little attention to the contribution of genes that are exclusively present (i.e. uniquely human genes) or absent in one or another species. Such protein coding uniquely human genes are generated by complete and/or partial gene duplication, gene rearrangement, silencing, mutagenesis or endogenization of foreign DNA that have a defined biological activity (22). Indeed, genomic comparisons between humans and mice reveal that many genes in the human genome have no mouse ortholog, and vice versa (23). For example, some mouse genes such as the SIGLEC encoding cmah are not expressed in humans (24), which when knocked out of the mouse genome creates a “human phenotype” (25). In this paper we focused on uniquely human genes that have clearly defined biological activities (i.e. TBC1D3, ARHGAP11B, CHRFAM7A) (22, 26–29), and specific relevance to signaling and human disease (30–36). Moreover, because these genes are over-represented in inflammatory cells (22), there is a need to define their role in inflammation and injury. Together, they are thought to participate in biochemical networks of signal transduction and, growth factor and cytokine responsiveness (37) leading to the hypothesis that species-specific inflammatory responses may be implicated in human wound repair and tissue regeneration (22).
Building on an emerging interest in uniquely human genes and their possible role(s) in the human response to injury, we explored the feasibility of adapting subcutaneous implants of polyvinylalcohol (PVA) sponges as an experimental model of human inflammation (38, 39). This model is based on human studies of the inflammatory response to foreign objects and to characterize the subtypes of cells that infiltrate human wounds (40–42). Here, we compare the inflammation response of mice reconstituted with human CD34+-hematopoietic stem and progenitor cells (CD34+-HSPCs) with control mice lacking CD34+-HSPCs using a combination of flow cytometry and immunohistochemistry to define the kinetics and profile of cells recruited to the PVA sponge implant. Species-specific multiplex cytokine bead arrays were used to identify human cytokines secreted into the PVA sponge fluid, and using RT-PCR, identified uniquely human genes expressed in mice engrafted with human CD34+-HSPCs compared to control mice. Together, these studies define a novel strategy in which the human response to inflammation can be analyzed in a mouse model.
Materials and Methods
Animals
All immunodeficient NOD-SCID-IL2Rγ−− (NSG) mice were reconstituted with human CD34+-HSPCs by Jackson Labs as previously described (#715557, Jackson Labs, Bar Harbor, ME) (43, 44). Briefly, 3 week old NSG female mice were subjected to whole body sub-lethal 140 cGy irradiation and administered 105 human fetal liver CD34+ HSPCs 4 hours later by intravenous tail vein injection. Engraftment was evaluated 12 weeks post-injection by flow cytometry after red blood cell lysis by Jackson Labs to quantify levels of human circulating cells expressing the human CD45 (hCD45+) antigen (See Figure 1A). Mice were then used at 14–18 weeks of age for subsequent PVA sponge model assays to examine hCD45+ cells in peripheral sites of inflammation (45). All animal procedures were approved by the UCSD Institutional Animal Care and Use Committee.
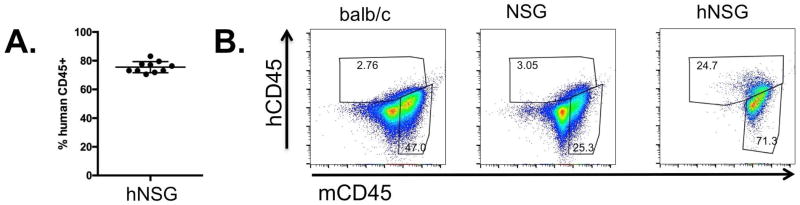
FIGURE 1. Characterization of the NSG mice reconstituted with human CD34+-HSPCs.
(A) Flow cytometric analysis of peripheral blood of NSG mice 12 weeks post-engraftment with human CD34+-HSPCs (hNSG). (B) Flow cytometric analysis of hCD45+ vs. mCD45+ cells in subcutaneous PVA sponge 7 days post-implantation from BALB/c, NSG and hNSG mice. Each of the analyses is representative of at least three separate experiments.
PVA Sponge Model
Implantation of PVA sponges (PVA Unlimited, Inc., Warsaw, IN) was performed in NSG mice 12–16 weeks post-CD34+-HSPC engraftment. Sponges (4 mm) were first hydrated in PBS, autoclaved and then aseptically implanted (N = 3) subcutaneously on the dorsum of recipient animals (38, 40, 46). At the indicated times, PVA sponges were removed and processed for immunohistochemistry, flow cytometry, cytokine analysis, or RT-PCR as described below. For immunohistochemical analyses, intact PVA sponges were fixed immediately at the time of harvest for 16 hours in 10% buffered formalin, followed by 70% ethanol and embedding in paraffin in the UCSD Histology Core. To prepare samples for cell and cytokine analyses, cells that had infiltrated the PVA sponge were flushed using a blunt forceps to compress the sponge 2–3 times into 1 mL of sterile PBS as described previously (46, 47) and detailed in the optimization assays shown in Supplementary Data Figure 1. The resulting cell suspension was then centrifuged at 500xg for 5 minutes and the supernatant immediately frozen. The cell fraction (pellet) was processed immediately for flow cytometry as described below. When gene expression was evaluated, the cell fraction (pellet) was placed in TRIzol (see below). The supernatant was analyzed for cytokine analysis (see below).
Antibodies and Reagents
Antibodies used for flow cytometry and for immunohistochemistry are summarized in Table I. Unless otherwise indicated, all other reagents were obtained from Sigma (St. Louis, MO).
Table 1.
Antibodies used for studies
| Antibody | Clone | Vendor |
|---|---|---|
| human CD45 | HI-30 or 2D1 | BD |
| mouse CD45 | 30-F11 | BD |
| CD11b | CBRM1/5 | eBioscience |
| CD33 | WM53 | BD |
| CD19 | SJ25C1 | BD |
| CX3CR1 | 2A9-1 | Biolegend |
| HLA-DR | G46-6 | BD |
| CD62L | DREG-56 | Biolegend |
| Fibrinogen | F0111 | DAKO |
Flow Cytometry
Cell staining of the recovered PVA exudate was first optimized using isotype-matched antibodies and/or using fluorescence minus one (FMO) analyses for analysis in FlowJo. Cells were processed for flow cytometry in PBS supplemented with 1.0% fetal bovine serum using a BD Biosciences Accuri flow cytometer and the data generated was analyzed using FlowJo software (Treestar, Ashland, OR). In all experiments, gating was set to a minimum of 10,000 viable cells, as determined by staining with 7-AAD as described by the manufacturer (BD Biosciences, San Jose, CA) and analyzed by forward and side scatter.
Immunohistochemistry
Tissue sections (10 μm) of the paraffin-embedded PVA sponges were de-waxed and incubated with citrate buffer (0.01 M Citric Acid, 0.05% Tween 20, pH 6.0) for 20 minutes at 95°C for antigen retrieval. After quenching endogenous peroxidase enzymatic activity with 3% hydrogen peroxide and blocking endogenous biotin and binding sites as described by the manufacturer (Vector Labs, Burlingame, CA), the sections were subjected to immunostaining with anti-fibrinogen antibody (DAKO, Carpenteria, CA) and detection with an anti-rabbit Vectastain kit (Vector Labs, #PK-4001). Similarly, hCD45+ cells were detected with anti-CD45 antibody (BD Biosciences) using a Vectastain kit and DAB detection. Exposure-matched micrographs of immunostained slides were captured with an FSX100 microscope (Olympus America, Center Valley, PA).
Cytometric Bead Array Analysis
The supernatants collected from flushed PVA sponges (see above) were collected in a fixed 1.0 mL volume of PBS and analyzed for levels of human IL-12p70, TNFα, IL-10, IL-6, IL1β, and IL-8 (#551811, BD Biosciences) using species-specific cytokine bead arrays (CBA). Mouse MCP-1, IL-6, TNFα, and IL-10 were also detected with CBA (#552364, BD Biosciences). Standard curves using human and mouse proteins were used to measure cytokine concentration and confirm specificity of the anti-human reagents in detecting only recombinant human cytokines, and the specificity of anti-mouse reagents in detecting only mouse cytokines. Data was analyzed with FCAP software (BD Biosciences) as described by the manufacturer.
RNA extraction and reverse transcription polymerase chain reaction
Total RNA was isolated from cells, mRNA purified over poly-T columns and 1 μg of mRNA reverse transcribed using the iScript cDNA synthesis kit (#170-889, Bio-Rad, Hercules, CA). PCR amplification of cDNA was done in a 50 μL volume containing 10 μL of 5x Buffer, 1.5 μL DMSO, 10 μM of each primer in 1 μL, 10 mM of each dNTP in 1 μL, and 0.5 μL of Phusion Taq polymerase (#M0530, New England BioLabs, Ipswich, MA). The PCR mixture was initially incubated at 98°C for 5 minutes followed by 40 cycles of denaturation at 98°C for 15 seconds, annealing at 60°C for 15 seconds and extension at 72°C for 25 seconds and a final extension at 72°C for 5 minutes. 10 μL of each PCR product was analyzed on a 2% agarose gels. The following primer pairs were used to detect gene expression:
CHRFAM7A (48):
5′-ATAGCTGCAAACTGCGATA-3′ (forward) and
5′-CAGCGTACATCGATGTAGCAG-3′ (reverse)
TBC1D3 (29):
5′-GCATCGACCGGGACGTAAG-3′ (forward) and
5′-CCTCCGGGTTGTACTCCTCAT-3′ (reverse)
ARHGAP11B (27):
5′-ATGTGGGATCAGAGGCTGGTGAA-3′ (forward) and
5′-AAGGAGGTGCAGAAGATAGGCA-3′ (reverse)
human GAPDH (48)
5′-CATGAGAAGTATGACAACAGCCT-3′ (forward) and
5′-AGTCCTTCCACGATACCAAAGT-3′ (reverse)
The sequence of mouse GAPDH primers (Qiagen Cat #QT01658692) is held by the manufacturer.
Statistical Analysis
Data analyses were performed with Graphpad Prism 6.0d (San Diego, CA), with statistical significance determined by a Mann-Whitney two-tailed t-test, and presented as standard deviation, with all data reproduced in at least two separate assays.
Results
NSG mice reconstituted with human CD34+-HSPCs support the expansion and recruitment of human leukocytes into subcutaneous implants of PVA sponge
To establish the levels of engraftment of NOD-SCID-IL2Rγ−− (NSG) mice with human CD34+-HPSCs, peripheral blood was subjected to flow cytometric analysis to detect circulating levels of human CD45+ cells (hCD45+), with 75% +/−4% of total viable circulating cells being hCD45+ (Figure 1A). This level is consistent with published data demonstrating the engraftment and expansion of human leukocytes in the murine niche of NSG mice (49–52). Next, we analyzed the profile of hCD45+ and mouse CD45+ (mCD45+) cells recruited to PVA sponges 7 days after subcutaneous implantation using species-specific anti-human and anti-mouse CD45 antibodies, to distinguish hCD45+ and mCD45+ cells, respectively. As shown in Figure 1B, we compared the recruitment of mCD45+ and hCD45+ cells in immune-competent BALB/c mice, in immune-deficient NSG mice lacking human CD34+-HSPCs, and in hNSG mice that were engrafted with human CD34+-HSPCs. While we observed a robust infiltration of mCD45+ cells to the PVA sponge in all three groups, with approximately 25–70% of cells being mCD45+, hCD45+ cells were detected only in hNSG mice. These findings demonstrate that (a) normal mouse leukocytes mobilized to subcutaneous PVA sponges within 7 days of implantation in each of the hosts, (b) human leukocytes can be readily distinguished from their murine counterpart and (c) the engraftment of CD34+-HSPCs leads to the mobilization of human leukocytes into the circulation that are recruited to PVA sponge implants in vivo, in a profile that is similar to endogenous mouse leukocytes.
Immunolocalization of human leukocytes in PVA sponges 7 days after subcutaneous implantation
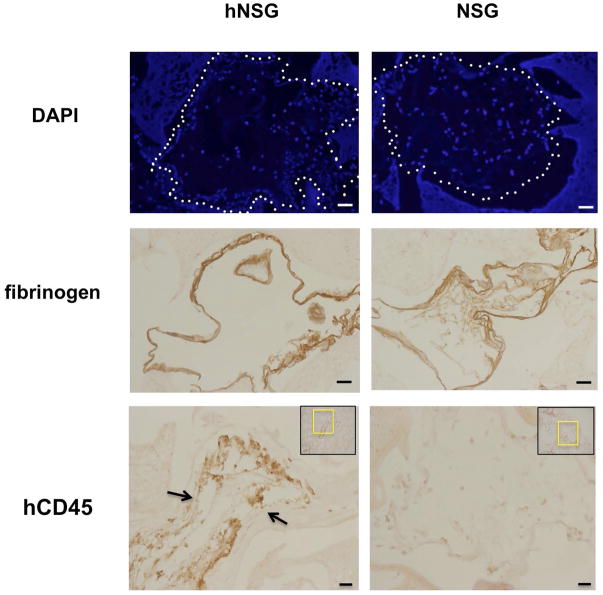
Immunohistochemistry was used to identify the distribution of hCD45+ cells in the PVA sponge implants in hNSG vs. control NSG mice (Figure 2). DAPI staining to detect nuclei was performed on paraffin sections of formalin-fixed PVA sponges to establish the localization of cells in the channels of the implant in both hNSG and NSG mice (Figure 2). Immunostaining with anti-fibrinogen antibodies (Figure 2) revealed the deposition of extracellular matrix proteins on the luminal margins of channels of the PVA sponge that were similar in PVA harvested from NSG or hNSG mice. The distribution of cells and fibrinogen staining (Figure 2) was widespread across the PVA sponge, consistent with the foreign body response to PVA. As with flow cytometry analyses (see Figure 1 above), hCD45+ cells were localized only within PVA implants harvested from hNSG mice, while no hCD45+ staining was detected in control NSG mice that had no CD34+-HSPC engraftment (Figure 2C). Immunohistochemistry data localizing hCD45+ cells in PVA sponges confirms flow cytometry analyses showing that hNSG mice support the mobilization of human leukocytes to sites of inflammation in vivo.
FIGURE 2. Immunohistochemical detection of human CD45+ cells in PVA sponge implants.
(Top Row) Cells were localized in 7 day PVA sponge implants from hNSG and NSG mice using DAPI staining of nuclei. (Middle Row) Immunohistochemical detection of fibrinogen deposition of extracellular matrix associated with inflammation. (Bottom Row) Localization of hCD45 cells in PVA sponges from hNSG vs. NSG mice using a species-specific anti-CD45 antibody. Micrographs are representative of at least three separate experiments. Size bar=32 microns.
Kinetics of human leukocyte trafficking to the implants of PVA sponge
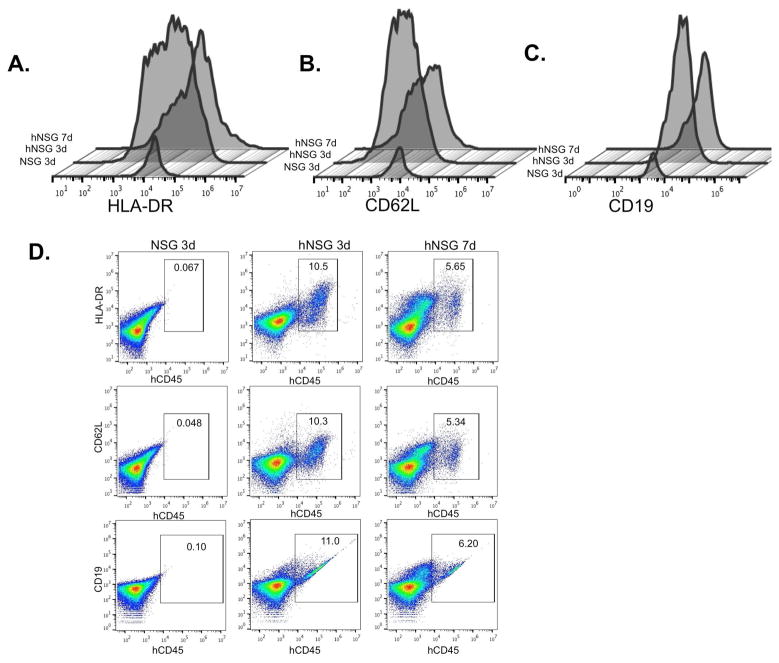
Based on the dynamic change in the profile of myeloid-derived leukocytes and similar cell types that are recruited to subcutaneous implants in a foreign body reaction (38, 40, 46, 47, 53–57), we analyzed the kinetics of human leukocyte mobilization in hNSG and NSG mice using markers for antigen presentation (HLA-DR), cell adhesion (CD62L), and B cells (i.e. CD19). We observed that there are dynamic and time-dependent changes in the recruitment of the hCD45+ cells that mobilize to PVA sponge implants in hNSG mice. As shown in Figure 3, analysis of hCD45+ cells demonstrated a peak increase in the number of MHC II+ (i.e. HLA-DR) cells at 3 vs. 7 days post-implantation (Figure 3A). Similarly, analysis of hCD45+ cells expressing L-selectin (CD62L), a protein essential for leukocyte-endothelial interactions (Figure 3B), and human CD19+, a marker for human B cells (Figure 3C), revealed peak responses at 3 days post-implantation (10–11% cells), followed by decreases by 7 days (Figure 3D). As expected, human cells were undetectable in the NSG control mice lacking human CD34+ HSPCs. These findings are consistent with the recruitment of a diverse profile of human leukocytes in hNSG mice immediately following inflammation challenge.
FIGURE 3. Kinetics of human CD45+ lineages trafficking to PVA sponge.
(A) Overlay histogram analyses of the expression levels (x-axis) on human CD45+ cells of surface markers for major histocompatibility (HLA-DR), (B) L-selectin (CD62L) and (C) B cells (CD19) 3 and 7 days post-implantation. Gating on human hCD45+ cells, representative scatterplot analyses of (D) HLA-DR, CD62L, and CD19 is shown from one of at least two separate experiments from NSG mice at 3 days (left, hNSG mice at 3 days (middle), and hNSG mice at 7 days (right).
Kinetics of human CD45+ myeloid cell trafficking to the implants of PVA sponge
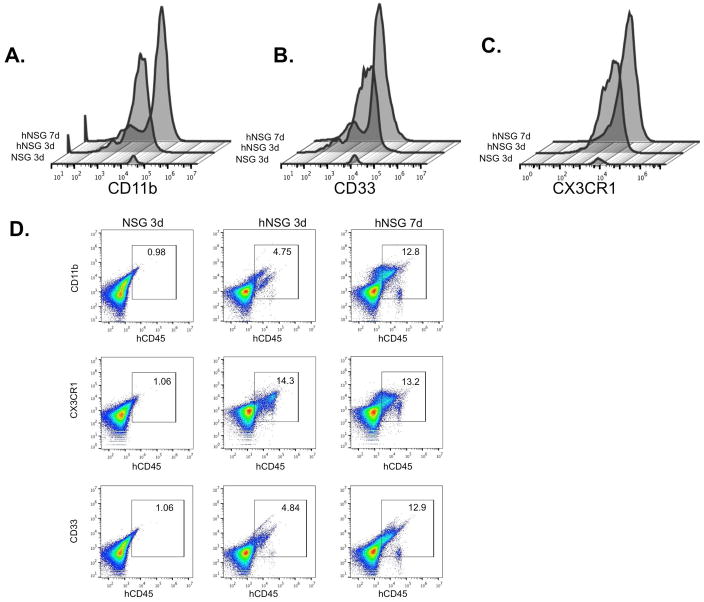
We next analyzed the profile of myeloid lineage cells that contribute to injury repair (Figure 4). Therefore, hCD45+ cells were subjected to cell surface staining with markers for monocyte integrin CD11b, the chemokine receptor CX3CR1, and myeloid cell marker CD33, to extend the profiling of hCD45+ cells in Figure 3. Analyses of CD11b+ (Figure 4A), CD33+ (Figure 4B), and CX3CR1+ (Figure 4C) markers on CD45+ cells showed a peak at 7 days for CD11b+ and CD33+ cells, and steady levels of CX3CR1+ cells at days 3 and 7 (Figure 4). These results point to changes in human myeloid cell mobilization that parallel reported profiles observed for mouse cells (46).
FIGURE 4. Analysis of myeloid cell markers in PVA sponge implants.
(A) Overlay histogram analysis of myeloid marker expression for CD11b, CX3CR1, and CD33 on human CD45+ cells 3 and 7 days post-implantation. Gating on human hCD45+ cells, representative scatterplot analyses of (B) CD11b, (C) CD33, and (D) CX3CR1 is shown from one of at least two separate experiments.
Detection of human cytokines in the wound fluid of PVA sponges is dynamic and time dependent
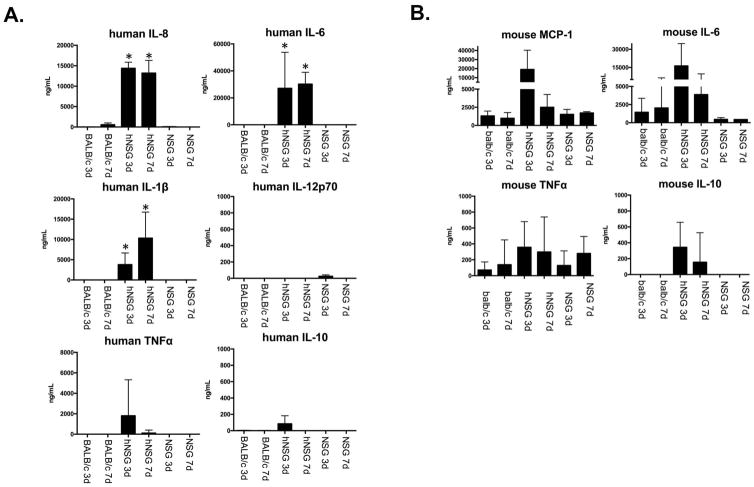
To determine whether the human cells that mobilize to PVA sponges in mice are also biologically active and contribute to the inflammatory milieu, we assessed the levels of human interleukin-8 (IL-8), interleukin-6 (IL-6), interleukin-1 beta (IL1β), interleukin-12-p70 (IL-12p70), tumor necrosis factor (TNFα), and interleukin-10 (IL-10) in the wound fluid associated with PVA sponges at 3 and 7 days post-implantation (Figure 5). The levels of human IL-6 and human IL-8 derived from human cells were elevated at both 3 and 7 days, while human IL1β levels increased from day 3 to day 7. In contrast, human TNFα peaked at day 3 compared to the lower levels at day 7. Human IL-12p70 and IL-10 were either not detected or were close to baseline at both time-points. The reagents did not cross-react with recombinant mouse proteins (data not shown), as described in more detail in the Materials and Methods. Levels of mouse cytokines were also analyzed in the PVA sponge model, focusing on the same mouse strains used for the analysis of human cytokines. These findings support our hypothesis that subcutaneous inflammation induces the dynamic recruitment of biologically active human leukocytes in hNSG mice that conditions the microenvironment of the PVA sponge with human cytokines.
FIGURE 5. Analysis of human and mouse cytokines in hNSG mice.
A) Quantification of human cytokines using human-specific cytokine bead array (CBA) detection reagents for PVA sponge supernatant 3 and 7 days post-implantation from BALB/c, NSG, and hNSG mice. Balb/c mice were used as a wildtype immune competent mouse strain for general cross-comparison. CBA detected human IL-12p70, human TNFα, human IL-10, human IL-6, human hIL1β, and human IL-8. B) Quantification of mouse cytokines using mouse-specific CBA antibodies from the same PVA sponge supernatant the human cytokine analysis. CBA detected mouse MCP-1, mouse IL-6, mouse TNFα, and mouse IL-10. Species specificity of CBA antibody reagents was tested on recombinant cytokines as shown in Supplementary Figure 1. Sample size n=2 to n=7. *, P<0.05 as compared to the day-matched NSG control.
Human genes are detected in PVA sponges when infiltrated with human leukocytes
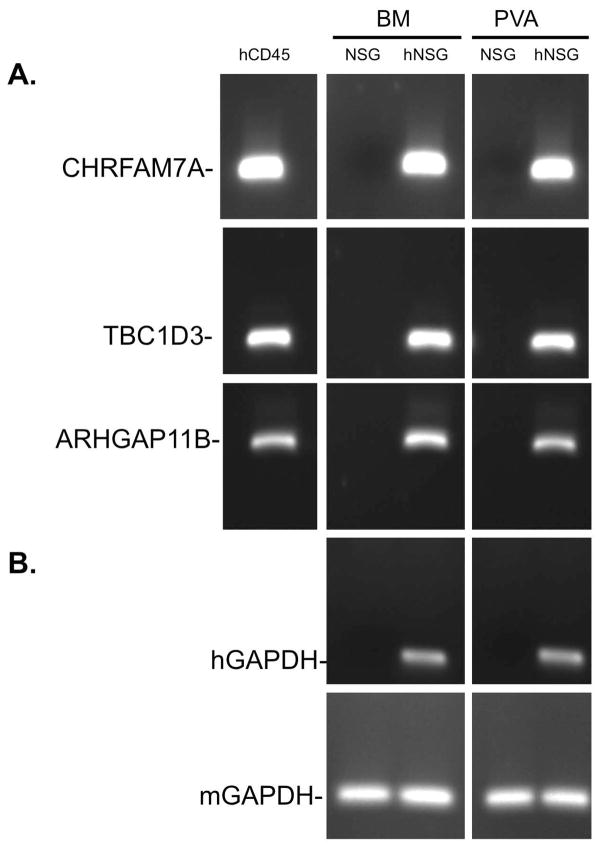
While the detection of human proteins (Figure 5) establishes that human cells produce proteins that have cognate orthologs in mouse and human, the presence of human cells in mice raises the possibility of studying human genes that have no mouse ortholog (i.e. uniquely human genes) (22). To this end, cells were recovered from PVA implants from either hNSG mice or control NSG mice and processed for RT-PCR to detect uniquely human genes. As shown in Figure 6, CHRFAM7A, a human specific gene that encodes a dominant negative subunit of the α7 nicotinic acetylcholine receptor (48, 58, 59) that is detectable in normal human leukocytes (hCD45), and in bone marrow (BM), and cells recruited to PVA sponges from hNSG mice (hNSG, top row, Figure 6A). In contrast, no gene is detected in mice when there is no human CD34+-HSPC engraftment (NSG). TBC1D3 (middle row, Figure 6B), a uniquely human gene that regulates growth factor responsiveness of human cells (28, 29, 60), and ARHGAP11B (bottom row, Figure 6C), a Rho GTPase that regulates differentiation (27), were also detected in the bone marrow and PVA sponge implants of hNSG mice. As expected, primers for both human and mouse GAPDH (Top row, Figure 6B) detected GAPDH in the appropriate cognate species, while none of the human genes were detected in NSG control samples. These findings demonstrate that hNSG mice can be used to analyze the expression of uniquely human genes in human leukocytes mobilized in an in vivo model of inflammation.
FIGURE 6. Detection of uniquely human genes in the PVA sponge model in hNSG mice.
RT-PCR was used for the detection of the expression of (A) uniquely human genes CHRFAM7A (Top row), TBC1D3 (middle row), and ARHGAP11B (bottom row) in hNSG mice from bone marrow (BM) and PVA sponge (PVA). Cells analyzed from NSG mice were used a negative control and primary human leukocytes cDNA (hCD45) used as a positive control. (B) Detection of human GAPDH in hNSG samples was a species specificity control, and detection of mouse GAPDH in all samples was a loading control.
Discussion
For several years now, the HIV/AIDS research community has used human bone marrow grafts in immuno-deficient mice to study HIV/AIDS because HIV tropism for human cells made it impossible to study the biology of HIV infectivity in mice (61–64). More recently, and in large part because of their success, these human-mouse chimeric models have been adapted to study human responses in diabetes, sepsis and burn injury (14–21, 65). Here we build on the development of these chimeric mouse technologies (66, 67) to show that human myeloid cells can be analyzed in an in vivo model of cutaneous inflammation (46, 68, 69). In light of the fact that both differences and similarities exist between rodents and humans in their gene expression response to severe injury (2–4), it is noteworthy that mice grafted with human CD34+-HSPCs can contribute a additional repetoire of uniquely human genes that are normally absent from the mouse genome. This allows the study of their biological activity in an experimental model that is conducive to prospective interventions like diabetes, wounding and injury.
The PVA sponge is a well-established model to study the foreign body response and the inflammatory response to a sterile implant that can be used to study inflammation responses associated with wound healing (38, 53, 57, 69). Here, we focused on the mobilization of human leukocytes to subcutaneous PVA sponge implants to characterize the human inflammation response to wound healing. We show that human leukocytes are actively mobilized to the implanted PVA sponges and identified the populations of infiltrating human leukocytes. In the past, the study of human myeloid cell differentiation at sites of human inflammation has been limited because of the focus on either blood and bone marrow-derived macrophages for in vitro analysis (66, 70) or subcutaneous synthetic implants from human volunteers in vivo (38, 40–42). In contrast, mechanisms of mouse inflammation have advanced rapidly because of the availability of gene knockout and transgenic tools for the study of myeloid cell responses (i.e. macrophage and dendritic cell biology). Albina and colleagues have previously defined the murine myeloid response to PVA implants in immune competent mice, demonstrating that monocytes are recruited by Day 1 post-implantation and develop into alternately activated macrophages by Day 7 (46). It should be noted that murine cytokine responses to the PVA sponge implants may be affected by the underlying mutations for NOD, Scid, and IL2Rγ when compared with the cytokine responses observed in a wildtype immune-competent strains such as Balb/c. In this study we are focused on characterizing human myeloid cell responses, therefore, we used NSG mice that support efficient engraftment of human cells.
Our focus on human myeloid cells in a PVA sponge model is based on NSG mice that lack NK cells, important mediators of immune cell-mediated clearance (18, 50, 51). Studies in standard and knockout mouse models have facilitated the improvement of ‘humanized’ mouse models by gene knock-in of human cytokine orthologs that enhance human-specific myeloid and lymphoid lineage levels and function (71, 72). Profiling of mouse cytokines in the hNSG model, as we have shown here, provides insight into the murine response to transplanted human cells. These observations can direct the refinement of next generation cytokine knock-in mice. While NSG mouse are immunodeficient in order to support the optimal engraftment of human CD34+-HSPCs in the bone marrow, these mice have also been well-documented to support the mobilization and circulation of human myeloid and lymphoid cells into stroma and peripheral organs (71, 73).
On a final note, while human genes lacking a mouse ortholog are not represented in current animal models, human genes can be introduced into the mouse genome by genetic engineering. However, such approaches may lack biochemical activity in mouse signaling networks. As shown here, the profile of human inflammatory cytokines that are secreted into the wound fluid in vivo can be assessed at specific endpoints. The fact that uniquely human genes like CHRFAM7A, TBC1D3, and ARHGAP11B can be analyzed in vivo shows that mice engineered to support human CD34+-HSPCs are amenable to a contextual study of uniquely human genes in human cells that are otherwise absent from the mice.
With global analyses of segmental duplications in the human genome showing an enrichment of genes associated with immune adaption and defense (74), it is interesting to speculate that they contribute to the human-specific responses to inflammation as originally suggested by Seok et al (1). One example is TBC1D3, a Rab-GAP gene that evolved through the partial duplication of the RNTRE locus late in primate speciation but that lacks Rab-GAP activity. Instead, it regulates ARF6-dependent macropinocytosis (60) presumably by regulating pools of GTP/GDP. In another, CHRFAM7A evolved as a partial recombination of a kinase gene fragment and a partial duplication of the α7 nicotinic acetylcholine anti-inflammatory receptor (α7 nAchR). It functions as a human specific dominant negative inhibitor of the anti-inflammatory α7 nAchR (26, 48, 58, 59). Finally ARHGAP11B encodes a RhoGTPase that selectively regulates intracellular signaling in human progenitor cells (27) and, as we show here, is expressed in human leukocytes. Accordingly, it will be increasingly important to study human inflammation (23, 75) in systems where the function of uniquely human genes in myeloid cells (76, 77) can be explored in intact animal models.
Supplementary Material
Acknowledgments
Funding: NIH NCI R01 CA17040
We thank Emelie Amburn, Ann-Marie Hageny and Jim Putnam for expert technical support. Studies were supported in part by an NIH R01 CA170140 grant (B.P.E.) and the Re-Investment fund of the Division of Trauma, Acute Care Surgery and Burns of the Department of Surgery, University of California San Diego.
Abbreviations
- PVA
polyvinyl alcohol
- NOD
Non-obese diabetic
- SCID
Severe combined immunodeficiency
- SSC
side scatter
- FSC
Forward scatter
- DAPI
4′,6-diamidino-2-phenylindole
- NSG
NOD-Scid-IL2Rγ−−
- hNSG
humanized NSG
- CD34+-HSPC
CD34+ hematopoietic stem and progenitor cells
Footnotes
Authorship: B.P.E. and A.B. conceived of the experiments and worked with B.E.T., and T.C. in the specific design and interpretation. C.D., M.H.E., C.T., X.D. and F.H. performed specific experiments. B.P.E. and A.B. wrote the manuscript with editing and revisions by B.E.T., R.C. and T.C. and corrections by C.D., M.H.E., C.T., X.D. and F.H.
Conflict of Interest Disclosure: The authors report having no conflicts of interest with this study.
References
- 1.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2013;110(9):3507–12. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Correction for Takao and Miyakawa. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2015;112(10):E1163–7. doi: 10.1073/pnas.1502188112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takao K, Miyakawa T. Genomic responses in mouse models greatly mimic human inflammatory diseases. Proc Natl Acad Sci U S A. 2015;112(4):1167–72. doi: 10.1073/pnas.1401965111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warren HS, Tompkins RG, Moldawer LL, Seok J, Xu W, Mindrinos MN, et al. Mice are not men. Proc Natl Acad Sci U S A. 2015;112(4):E345. doi: 10.1073/pnas.1414857111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osuchowski MF, Remick DG, Lederer JA, Lang CH, Aasen AO, Aibiki M, et al. Abandon the mouse research ship? Not just yet! Shock. 2014;41(6):463–75. doi: 10.1097/SHK.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobezda T, Ghassemi-Nejad S, Mikecz K, Glant TT, Szekanecz Z. Of mice and men: how animal models advance our understanding of T-cell function in RA. Nature reviews Rheumatology. 2014;10(3):160–70. doi: 10.1038/nrrheum.2013.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bryant CE, Monie TP. Mice, men and the relatives: cross-species studies underpin innate immunity. Open biology. 2012;2(4):120015. doi: 10.1098/rsob.120015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romieu-Mourez R, Coutu DL, Galipeau J. The immune plasticity of mesenchymal stromal cells from mice and men: concordances and discrepancies. Frontiers in bioscience. 2012;4:824–37. doi: 10.2741/E422. [DOI] [PubMed] [Google Scholar]
- 9.Zschaler J, Schlorke D, Arnhold J. Differences in innate immune response between man and mouse. Critical reviews in immunology. 2014;34(5):433–54. [PubMed] [Google Scholar]
- 10.Smithey MJ, Uhrlaub JL, Li G, Vukmanovic-Stejic M, Akbar AN, Nikolich-Zugich J. Lost in translation: mice, men and cutaneous immunity in old age. Biogerontology. 2015;16(2):203–8. doi: 10.1007/s10522-014-9517-0. [DOI] [PubMed] [Google Scholar]
- 11.Dudek TE, No DC, Seung E, Vrbanac VD, Fadda L, Bhoumik P, et al. Rapid evolution of HIV-1 to functional CD8(+) T cell responses in humanized BLT mice. Sci Transl Med. 2012;4(143):143ra98. doi: 10.1126/scitranslmed.3003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olesen R, Wahl A, Denton PW, Garcia JV. Immune reconstitution of the female reproductive tract of humanized BLT mice and their susceptibility to human immunodeficiency virus infection. J Reprod Immunol. 2011;88(2):195–203. doi: 10.1016/j.jri.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun Z, Denton PW, Estes JD, Othieno FA, Wei BL, Wege AK, et al. Intrarectal transmission, systemic infection, and CD4+ T cell depletion in humanized mice infected with HIV-1. J Exp Med. 2007;204(4):705–14. doi: 10.1084/jem.20062411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ernst W, Zimara N, Hanses F, Mannel DN, Seelbach-Gobel B, Wege AK. Humanized mice, a new model to study the influence of drug treatment on neonatal sepsis. Infection and immunity. 2013;81(5):1520–31. doi: 10.1128/IAI.01235-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harding MJ, Lepus CM, Gibson TF, Shepherd BR, Gerber SA, Graham M, et al. An implantable vascularized protein gel construct that supports human fetal hepatoblast survival and infection by hepatitis C virus in mice. PloS one. 2010;5(4):e9987. doi: 10.1371/journal.pone.0009987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Misharin AV, Haines GK, 3rd, Rose S, Gierut AK, Hotchkiss RS, Perlman H. Development of a new humanized mouse model to study acute inflammatory arthritis. Journal of translational medicine. 2012;10:190. doi: 10.1186/1479-5876-10-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramer PC, Chijioke O, Meixlsperger S, Leung CS, Munz C. Mice with human immune system components as in vivo models for infections with human pathogens. Immunology and cell biology. 2011;89(3):408–16. doi: 10.1038/icb.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Unsinger J, McDonough JS, Shultz LD, Ferguson TA, Hotchkiss RS. Sepsis-induced human lymphocyte apoptosis and cytokine production in “humanized” mice. Journal of leukocyte biology. 2009;86(2):219–27. doi: 10.1189/jlb.1008615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Washburn ML, Bility MT, Zhang L, Kovalev GI, Buntzman A, Frelinger JA, et al. A humanized mouse model to study hepatitis C virus infection, immune response, and liver disease. Gastroenterology. 2011;140(4):1334–44. doi: 10.1053/j.gastro.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye C, Choi JG, Abraham S, Wu H, Diaz D, Terreros D, et al. Human macrophage and dendritic cell-specific silencing of high-mobility group protein B1 ameliorates sepsis in a humanized mouse model. Proc Natl Acad Sci U S A. 2012;109(51):21052–7. doi: 10.1073/pnas.1216195109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skirecki T, Kawiak J, Machaj E, Pojda Z, Wasilewska D, Czubak J, et al. Early severe impairment of hematopoietic stem and progenitor cells from the bone marrow caused by CLP sepsis and endotoxemia in a humanized mice model. Stem Cell Res Ther. 2015;6:142. doi: 10.1186/s13287-015-0135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baird A, Costantini T, Coimbra R, Eliceiri BP. Injury, Inflammation and the Emergence of Human-Specific Genes. Wound repair and regeneration: official publication of the Wound Healing Society [and] the European Tissue Repair Society; 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Bleness M, Searles VB, Varki A, Gagneux P, Sikela JM. Evolution of genetic and genomic features unique to the human lineage. Nature reviews Genetics. 2012;13(12):853–66. doi: 10.1038/nrg3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deng L, Song J, Gao X, Wang J, Yu H, Chen X, et al. Host adaptation of a bacterial toxin from the human pathogen Salmonella Typhi. Cell. 2014;159(6):1290–9. doi: 10.1016/j.cell.2014.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hedlund M, Tangvoranuntakul P, Takematsu H, Long JM, Housley GD, Kozutsumi Y, et al. N-glycolylneuraminic acid deficiency in mice: implications for human biology and evolution. Molecular and cellular biology. 2007;27(12):4340–6. doi: 10.1128/MCB.00379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Araud T, Graw S, Berger R, Lee M, Neveu E, Bertrand D, et al. The chimeric gene CHRFAM7A, a partial duplication of the CHRNA7 gene, is a dominant negative regulator of alpha7*nAChR function. Biochemical pharmacology. 2011;82(8):904–14. doi: 10.1016/j.bcp.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347(6229):1465–70. doi: 10.1126/science.aaa1975. [DOI] [PubMed] [Google Scholar]
- 28.Wainszelbaum MJ, Charron AJ, Kong C, Kirkpatrick DS, Srikanth P, Barbieri MA, et al. The hominoid-specific oncogene TBC1D3 activates Ras and modulates epidermal growth factor receptor signaling and trafficking. The Journal of biological chemistry. 2008;283(19):13233–42. doi: 10.1074/jbc.M800234200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wainszelbaum MJ, Liu J, Kong C, Srikanth P, Samovski D, Su X, et al. TBC1D3, a hominoid-specific gene, delays IRS-1 degradation and promotes insulin signaling by modulating p70 S6 kinase activity. PloS one. 2012;7(2):e31225. doi: 10.1371/journal.pone.0031225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper DN, Kehrer-Sawatzki H. Exploring the potential relevance of human-specific genes to complex disease. Hum Genomics. 2011;5(2):99–107. doi: 10.1186/1479-7364-5-2-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. Journal of immunology. 2004;172(5):2731–8. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- 32.Garralda E, Paz K, Lopez-Casas PP, Jones S, Katz A, Kann LM, et al. Integrated next-generation sequencing and avatar mouse models for personalized cancer treatment. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20(9):2476–84. doi: 10.1158/1078-0432.CCR-13-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gosain A, DiPietro LA. Aging and wound healing. World journal of surgery. 2004;28(3):321–6. doi: 10.1007/s00268-003-7397-6. [DOI] [PubMed] [Google Scholar]
- 34.Lin-Tsai O, Taylor JA, 3rd, Clark PE, Adam RM, Wu XR, DeGraff DJ. Progress made in the use of animal models for the study of high-risk, nonmuscle invasive bladder cancer. Current opinion in urology. 2014;24(5):512–6. doi: 10.1097/MOU.0000000000000087. [DOI] [PubMed] [Google Scholar]
- 35.Malaney P, Nicosia SV, Dave V. One mouse, one patient paradigm: New avatars of personalized cancer therapy. Cancer letters. 2014;344(1):1–12. doi: 10.1016/j.canlet.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sen CK, Gordillo GM, Roy S, Kirsner R, Lambert L, Hunt TK, et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound repair and regeneration: official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2009;17(6):763–71. doi: 10.1111/j.1524-475X.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stahl PD, Wainszelbaum MJ. Human-specific genes may offer a unique window into human cell signaling. Science signaling. 2009;2(89):pe59. doi: 10.1126/scisignal.289pe59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deskins DL, Ardestani S, Young PP. The polyvinyl alcohol sponge model implantation. Journal of visualized experiments: JoVE. 2012;(62) doi: 10.3791/3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diegelmann RF, Kim JC, Lindblad WJ, Smith TC, Harris TM, Cohen IK. Collection of leukocytes, fibroblasts, and collagen within an implantable reservoir tube during tissue repair. Journal of leukocyte biology. 1987;42(6):667–72. doi: 10.1002/jlb.42.6.667. [DOI] [PubMed] [Google Scholar]
- 40.Diegelmann RF, Lindblad WJ, Cohen IK. A subcutaneous implant for wound healing studies in humans. The Journal of surgical research. 1986;40(3):229–37. doi: 10.1016/0022-4804(86)90156-3. [DOI] [PubMed] [Google Scholar]
- 41.Alaish SM, Bettinger DA, Olutoye OO, Gould LJ, Yager DR, Davis A, et al. Comparison of the polyvinyl alcohol sponge and expanded polytetrafluoroethylene subcutaneous implants as models to evaluate wound healing potential in human beings. Wound repair and regeneration: official publication of the Wound Healing Society [and] the European Tissue Repair Society. 1995;3(3):292–8. doi: 10.1046/j.1524-475X.1995.30309.x. [DOI] [PubMed] [Google Scholar]
- 42.Jorgensen LN, Olsen L, Kallehave F, Karlsmark T, Diegelmann RF, Cohen IK, et al. The wound healing process in surgical patients evaluated by the expanded polytetrafluoroethylene and the polyvinyl alcohol sponge: a comparison with special reference to intrapatient variability. Wound repair and regeneration: official publication of the Wound Healing Society [and] the European Tissue Repair Society. 1995;3(4):527–32. doi: 10.1046/j.1524-475X.1995.30419.x. [DOI] [PubMed] [Google Scholar]
- 43.Rongvaux A, Takizawa H, Strowig T, Willinger T, Eynon EE, Flavell RA, et al. Human hemato-lymphoid system mice: current use and future potential for medicine. Annu Rev Immunol. 2013;31:635–74. doi: 10.1146/annurev-immunol-032712-095921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lepus CM, Gibson TF, Gerber SA, Kawikova I, Szczepanik M, Hossain J, et al. Comparison of human fetal liver, umbilical cord blood, and adult blood hematopoietic stem cell engraftment in NOD-scid/gammac−/−, Balb/c-Rag1−/−gammac−/−, and C. B-17-scid/bg immunodeficient mice. Hum Immunol. 2009;70(10):790–802. doi: 10.1016/j.humimm.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.King M, Pearson T, Shultz LD, Leif J, Bottino R, Trucco M, et al. A new Hu-PBL model for the study of human islet alloreactivity based on NOD-scid mice bearing a targeted mutation in the IL-2 receptor gamma chain gene. Clin Immunol. 2008;126(3):303–14. doi: 10.1016/j.clim.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Daley JM, Brancato SK, Thomay AA, Reichner JS, Albina JE. The phenotype of murine wound macrophages. Journal of leukocyte biology. 2010;87(1):59–67. doi: 10.1189/jlb.0409236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Daley JM, Reichner JS, Mahoney EJ, Manfield L, Henry WL, Jr, Mastrofrancesco B, et al. Modulation of macrophage phenotype by soluble product(s) released from neutrophils. Journal of immunology. 2005;174(4):2265–72. doi: 10.4049/jimmunol.174.4.2265. [DOI] [PubMed] [Google Scholar]
- 48.Costantini TW, Dang X, Yurchyshyna MV, Coimbra R, Eliceiri BP, Baird A. A Human-Specific alpha7-Nicotinic Acetylcholine Receptor Gene in Human Leukocytes: Identification, Regulation and the Consequences of CHRFAM7A Expression. Molecular medicine. 2015;21:323–36. doi: 10.2119/molmed.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito M, Hiramatsu H, Kobayashi K, Suzue K, Kawahata M, Hioki K, et al. NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood. 2002;100(9):3175–82. doi: 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- 50.Shultz LD, Ishikawa F, Greiner DL. Humanized mice in translational biomedical research. Nature reviews Immunology. 2007;7(2):118–30. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- 51.Pearson T, Greiner DL, Shultz LD. Creation of “humanized” mice to study human immunity. In: Coligan John E, et al., editors. Current protocols in immunology. Unit 15. Chapter 15. 2008. p. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brehm MA, Cuthbert A, Yang C, Miller DM, DiIorio P, Laning J, et al. Parameters for establishing humanized mouse models to study human immunity: analysis of human hematopoietic stem cell engraftment in three immunodeficient strains of mice bearing the IL2rgamma(null) mutation. Clin Immunol. 2010;135(1):84–98. doi: 10.1016/j.clim.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brancato SK, Thomay AA, Daley JM, Crane MJ, Reichner JS, Sabo E, et al. Toll-like receptor 4 signaling regulates the acute local inflammatory response to injury and the fibrosis/neovascularization of sterile wounds. Wound repair and regeneration: official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2013;21(4):624–33. doi: 10.1111/wrr.12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brubaker AL, Schneider DF, Palmer JL, Faunce DE, Kovacs EJ. An improved cell isolation method for flow cytometric and functional analyses of cutaneous wound leukocytes. Journal of immunological methods. 2011;373(1–2):161–6. doi: 10.1016/j.jim.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khanna S, Biswas S, Shang Y, Collard E, Azad A, Kauh C, et al. Macrophage dysfunction impairs resolution of inflammation in the wounds of diabetic mice. PloS one. 2010;5(3):e9539. doi: 10.1371/journal.pone.0009539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schwacha MG, Thobe BM, Daniel T, Hubbard WJ. Impact of thermal injury on wound infiltration and the dermal inflammatory response. The Journal of surgical research. 2010;158(1):112–20. doi: 10.1016/j.jss.2008.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Major MR, Wong VW, Nelson ER, Longaker MT, Gurtner GC. The foreign body response: at the interface of surgery and bioengineering. Plast Reconstr Surg. 2015;135(5):1489–98. doi: 10.1097/PRS.0000000000001193. [DOI] [PubMed] [Google Scholar]
- 58.Costantini TW, Dang X, Coimbra R, Eliceiri BP, Baird A. CHRFAM7A, a human-specific and partially duplicated alpha7-nicotinic acetylcholine receptor gene with the potential to specify a human-specific inflammatory response to injury. Journal of leukocyte biology. 2015;97(2):247–57. doi: 10.1189/jlb.4RU0814-381R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinkus ML, Graw S, Freedman R, Ross RG, Lester HA, Leonard S. The human CHRNA7 and CHRFAM7A genes: A review of the genetics, regulation, and function. Neuropharmacology. 2015;96(Pt B):274–88. doi: 10.1016/j.neuropharm.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frittoli E, Palamidessi A, Pizzigoni A, Lanzetti L, Garre M, Troglio F, et al. The primate-specific protein TBC1D3 is required for optimal macropinocytosis in a novel ARF6-dependent pathway. Molecular biology of the cell. 2008;19(4):1304–16. doi: 10.1091/mbc.E07-06-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arainga M, Su H, Poluektova LY, Gorantla S, Gendelman HE. HIV-1 cellular and tissue replication patterns in infected humanized mice. Sci Rep. 2016;6:23513. doi: 10.1038/srep23513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Denton PW, Garcia JV. Humanized mouse models of HIV infection. AIDS Rev. 2011;13(3):135–48. [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L, Su L. HIV-1 immunopathogenesis in humanized mouse models. Cell Mol Immunol. 2012;9(3):237–44. doi: 10.1038/cmi.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tager AM, Pensiero M, Allen TM. Recent advances in humanized mice: accelerating the development of an HIV vaccine. J Infect Dis. 2013;208(Suppl 2):S121–4. doi: 10.1093/infdis/jit451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Serr I, Furst RW, Achenbach P, Scherm MG, Gokmen F, Haupt F, et al. Type 1 diabetes vaccine candidates promote human Foxp3(+)Treg induction in humanized mice. Nat Commun. 2016;7:10991. doi: 10.1038/ncomms10991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reynolds G, Haniffa M. Human and Mouse Mononuclear Phagocyte Networks: A Tale of Two Species? Frontiers in immunology. 2015;6:330. doi: 10.3389/fimmu.2015.00330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iwasaki A. The use of bone marrow-chimeric mice in elucidating immune mechanisms. Methods in molecular medicine. 2006;127:281–92. doi: 10.1385/1-59745-168-1:281. [DOI] [PubMed] [Google Scholar]
- 68.Tanaka S, Saito Y, Kunisawa J, Kurashima Y, Wake T, Suzuki N, et al. Development of mature and functional human myeloid subsets in hematopoietic stem cell-engrafted NOD/SCID/IL2rgammaKO mice. Journal of immunology. 2012;188(12):6145–55. doi: 10.4049/jimmunol.1103660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crane MJ, Daley JM, van Houtte O, Brancato SK, Henry WL, Jr, Albina JE. The monocyte to macrophage transition in the murine sterile wound. PloS one. 2014;9(1):e86660. doi: 10.1371/journal.pone.0086660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hume DA. The Many Alternative Faces of Macrophage Activation. Frontiers in immunology. 2015;6:370. doi: 10.3389/fimmu.2015.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rathinam C, Poueymirou WT, Rojas J, Murphy AJ, Valenzuela DM, Yancopoulos GD, et al. Efficient differentiation and function of human macrophages in humanized CSF-1 mice. Blood. 2011;118(11):3119–28. doi: 10.1182/blood-2010-12-326926. [DOI] [PubMed] [Google Scholar]
- 72.Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, et al. Development and function of human innate immune cells in a humanized mouse model. Nature biotechnology. 2014;32(4):364–72. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sato K, Misawa N, Nie C, Satou Y, Iwakiri D, Matsuoka M, et al. A novel animal model of Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis in humanized mice. Blood. 2011;117(21):5663–73. doi: 10.1182/blood-2010-09-305979. [DOI] [PubMed] [Google Scholar]
- 74.Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome research. 2001;11(6):1005–17. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sikela JM. The jewels of our genome: the search for the genomic changes underlying the evolutionarily unique capacities of the human brain. PLoS genetics. 2006;2(5):e80. doi: 10.1371/journal.pgen.0020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Uchida N, Hsieh MM, Hayakawa J, Madison C, Washington KN, Tisdale JF. Optimal conditions for lentiviral transduction of engrafting human CD34+ cells. Gene therapy. 2011;18(11):1078–86. doi: 10.1038/gt.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang CX, Sather BD, Wang X, Adair J, Khan I, Singh S, et al. Rapamycin relieves lentiviral vector transduction resistance in human and mouse hematopoietic stem cells. Blood. 2014;124(6):913–23. doi: 10.1182/blood-2013-12-546218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.