Abstract
Glutamate receptors are ligand gated tetrameric ion channels that mediate synaptic transmission in the central nervous system. They are instrumental in vertebrate cognition and their dysfunction underlies diverse diseases1,2. In both the resting and desensitized states of AMPA and kainate subtype glutamate receptors the ion channels are closed while the ligand binding domain, which is physically coupled to the channel, adopts dramatically different conformations3–6. Without an atomic model for the desensitized state, it is not possible to address a central question in receptor gating: how the resting and desensitized receptor states both display closed ion channels, even though they have major differences in quaternary structure of the ligand binding domain. By determining the cryo-EM structure of the kainate receptor GluK2 subtype in its desensitized state at 3.8 Å resolution, we show that desensitization is characterized by establishment of a ring-like structure in the ligand binding domain layer of the receptor. Formation of this “desensitization ring” is mediated by staggered helix contacts between adjacent subunits, which leads to a pseudo four-fold symmetric arrangement of the ligand binding domains, illustrating subtle changes in symmetry that are at the heart of the gating mechanism. Disruption of the desensitization ring is likely the key switch that enables restoration of the receptor to its resting state, thereby completing the gating cycle.
Ionotropic glutamate receptors (iGluRs) are tetrameric ligand gated ion channels that mediate excitatory synaptic transmission in the central nervous system7. They have been classified into large families of AMPA, kainate, and NMDA receptors that have distinct functional, pharmacological, and structural properties8,9. Understanding the structural basis of glutamate receptor function is vital to understanding their roles in learning and memory, and by extension their roles in neuropathology. Crystallographic and cryo-EM studies of AMPA and kainate receptors3–6,10–15 revealed large conformational changes between states, including dramatic changes in the arrangement of the ligand binding domain (LBD) in the desensitized state in both receptor types. While near-atomic resolution structures have been reported for the AMPA receptor GluA2 subtype in apo3, antagonist-bound6,14, and pre-activated states3,10,14, a high-resolution structure of the desensitized state of any glutamate receptor subtype has remained elusive. Thus, the central question of how a closed ion channel can be accommodated in both resting and desensitized states in the context of dramatically different LBD layer structures has remained unanswered. The structure we present here of the desensitized state of GluK2 reveals unexpected aspects of how conformational changes are coupled to symmetry mismatches across the length of the receptor and thereby allows delineation of a complete model for the glutamate receptor gating cycle.
In our experiments we used GluK2EM, a construct that binds GluK1 selective antagonists with nanomolar affinity, and trapped the desensitized state using (2S,4R)-4-methylglutamate, a high affinity agonist that promotes profound desensitization16,17. The GluK2EM desensitized state structure at 3.8 Å resolution (Extended Data Fig. 1) reveals a 2-fold symmetric amino terminal domain (ATD) layer (Figs. 1a, 1b) with an intact ATD tetramer interface (Fig. 1c), and a reorganized LBD layer resembling a pinwheel (Fig. 1d). In the transmembrane domain (TMD) layer, all three membrane-spanning helices, and the S1-M1 and M3-S2 linkers were resolved (Extended Data Figs. 2a, 2b), and the four subunits within the ion channel display an approximately 4-fold symmetric relationship (Fig. 1e). Density for complex glycans at positions Asn244, Asn347 and Asn399 is evident (Extended Data Fig. 2c), while the M2 pore helices were not resolved and the ATD-LBD linker for the B and D subunits, and the C-terminal domain appear only at low density map contours. High sequence identity (>85%) between the M3 helices of AMPA and kainate receptors suggests that the general architecture of the central pore of the GluK2 channel will be relevant to other receptor subtypes from both families. In order to validate the use of 2-fold computational symmetry in the reconstruction, the data was reprocessed entirely without symmetry (Extended Data Fig. 3). The resulting reconstruction yielded a structure with the same architecture and domain placement as when 2-fold symmetry was applied, but at slightly lowered resolution. This result thereby supports the use of 2-fold symmetry in processing the data, and shows the presence of 2-fold symmetry in the desensitized state.
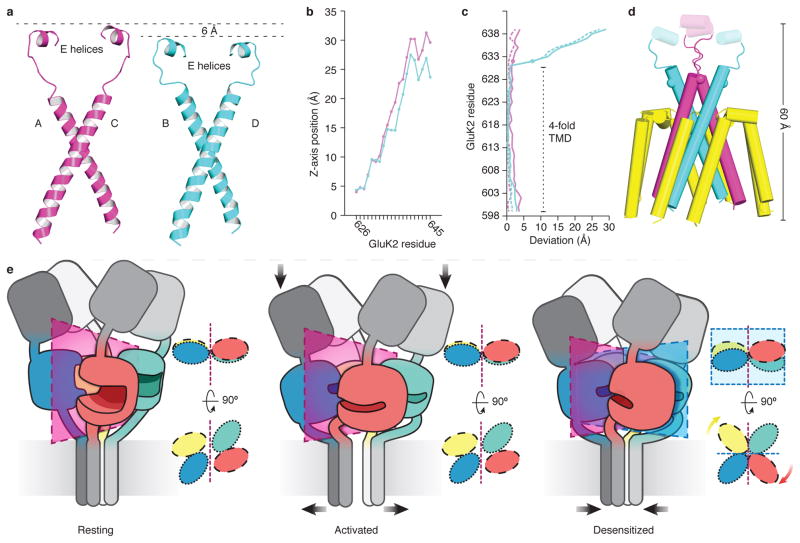
Figure 1. Desensitized kainate receptor at 3.8 Å resolution.
a, b, Cryo-EM density map (a) and atomic model (b) of 2S,4R-4-methylglutamate bound GluK2EM with each chain colored uniquely. Panel (a) has features shown from two map contours. c, Side and top view of the ATD tetramer, with cartoon highlighting 2-fold symmetry in the ATD layer. d, Top view of LBD tetramer with cartoon illustrating apparent 4-fold symmetry of the LBD layer when seen down the receptor central axis. e, Top view of the TMD with cartoon indicating domain symmetry.
The competitive antagonist LY466195 has nanomolar affinity for GluK2EM (Extended Data Fig. 4). We used this ligand to trap and determine the structure of the antagonist-bound resting state to ~12 Å and fit crystal structures of GluK2 ATD dimers and GluK2EM LY466195-bound LBD dimers to the cryo-EM map (Extended Data Fig. 5). The GluK2EM resting state structure is consistent with that observed previously using cryo-electron tomography5, and its structural profile matches that of the cryo-EM and crystal structures of the antagonist-bound resting state of GluA24,6. The lower resolution of the antagonist-bound resting state likely reflects greater conformational heterogeneity compared to the desensitized state, potentially arising from variability in the relative orientations of the ATD and LBD layers.
Using the cryo-EM density maps, we built a de novo atomic model of full-length desensitized state GluK2EM, and a molecular model of resting state GluK2 from crystal structures of ATD (PDB ID: 3H6G) and LBD (PDB ID: 5CMK) dimers. In the description below, we use “A”, “B”, “C” and “D” to refer to the four subunits of the tetrameric receptor, and “AC” and “BD” rather than the terms “proximal” and “distal” chains to clarify discussion of connectivity between the three (ATD, LDB and TM) layers of the receptor. Comparison of the resting and desensitized state models show that although the ATD tetrameric layers are essentially similar in both states, the LBDs splay outwards upon desensitization, tugging on the ATD assembly via the ATD-LBD linkers and pulling it downwards compared to the resting state, creating a substantial, 1,250 Å2 buried surface area interface between the ATD AC subunits and their underlying LBDs (Extended Data Fig. 6). This interface, which likely explains how the ATD contributes to stabilization of the desensitized state18, is mediated by van der Waals interactions combined with salt bridges and hydrogen bonds formed between the side chains of Lys191 and Asp476, Glu219 and Arg400, and Tyr220 and Asp480 of the ATD and LBD, respectively. Notably, linkers connecting the ATD and LBD layers adopt a helical structure that packs against the upper lobe of the LBD, a feature not seen in prior AMPA and kainate receptor structures. In the GluK2EM resting state the ATD and LBD layers do not interact and are separated by ~15 Å, similar to the GluA2 antagonist-bound resting state solved by cryo-EM4, which shows a separation of ~12 Å. This is strikingly different from GluA2 resting state crystal structures, which have 4-6 residue deletions in the ATD-LBD linkers (PDB IDs: 3KG2, 4U2P and 4U4G), and closely apposed ATD and LBD layers with a buried surface area of 400–530 Å2 per subunit, but without the helical structure found in the ATD-LBD linker of the GluK2 desensitized state3,6,14. Comparison of the GluK2EM resting state with the extended structure of the pre-activated GluA2 complex with con-ikot-ikot toxin10, for which the ATD-LBD linkers contain only 2 deletions, suggests that wild type GluA2 ATD-LBD linkers can extend to sufficient length such that the ATD and LBD are not in contact in both receptor subtypes, as also suggested by computational experiments19. We conclude that the apparent discrepancy among GluA2 and GluK2 resting states solved by cryo-EM where no buried surface is observed for the ATD-LBD interface, and the subset of GluA2 resting states solved by crystallography where these domains are in contact, is attributable to the dynamic structure of these domains19, combined with the effects of crystal packing and truncations in the ATD-LBD linkers in the GluA2 crystallization constructs. However, we note that while overall the GluA2 and GluK2 resting state structures are remarkably similar, they are both strikingly different from the compact resting state structure recently reported for GluA2/GluA3 heteromers11.
The organization of the LBD layer in the desensitized state is most strikingly characterized by ~125° rotations of AC subunit LBDs and displays ~4-fold in plane rotational symmetry (Fig. 1d) in contrast to the 2-fold symmetry of the ATD layer (Fig. 1c). Helices E and G from individual subunits abut onto the same two helices on neighboring LBDs (Figs. 2a–2c). Instead of the arrangement that would be expected if the LBD layer showed perfect 4-fold symmetry (Fig. 2d), we observe instead a circular arrangement that proceeds around the interior surface of the LBD layer with the E/G helices in an alternating, staggered pattern that we name the “desensitization ring” (Fig. 2e). This ring-like structure effectively creates a local 2-fold organization in the context of the global pseudo 4-fold arrangement of the overall LBD-layer. As a result, there are asymmetric contacts at AB and AD subunit interfaces that have buried surfaces of 860 Å2 and 447 Å2, respectively. Hydrogen bonds are formed between the Lys645 main chain and the side chain of Thr670, and salt bridges between the side chains of Asp672 and Lys667, and Asp672 and Lys673 in the AB subunit interface, with hydrogen bonds connecting the side chains of Ser639 and Arg681, and Tyr671 and Ser680 in the AD subunit interface. A structural consequence of the staggered E/G helix organization is that the BD subunit LBDs have a different pitch than the AC domains, with the AC subunits tilting away from the central receptor axis by ~10° more than their counterparts (Fig. 2e). It is this tilting which permits the E/G helices of the AC subunits to occupy the more elevated position in the staggered ring. Taken together, we observe that the LBD tetramer thus displays elements of both 4-fold and 2-fold symmetry. The apparent 4-fold symmetry manifests when considering the in-plane rotation of the LBD domains as viewed down the central axis of the receptor. The 2-fold symmetry, which ultimately defines the symmetry of the LBD layer, is apparent when accounting for the staggered helix E/G arrangement in the “desensitization ring”. This staggered ring structure demands a symmetry mismatch in AC versus BD subunit LBDs, which is accomplished by the mismatched pitches in the domains.
Figure 2. Desensitization ring.
a, Top view of LBD layer highlighting desensitization ring helices E and G colored magenta for AC subunits and cyan for BD subunits. Residues S669 and D672, mutated in functional experiments (g–i), are shown as sticks to highlight their positions. b, Edge-on view of desensitization ring interface with perspective indicated by eye icon in (a). c, Same perspective as in (b) but rotated 90° to show the second type of E/G interface observed in the desensitization ring. d, Illustration of hypothetical desensitization ring arrangement that would yield complete 4-fold symmetry in the LBD layer. The G helices are shown on top layer, and E helices on bottom layer. e, Illustration of the experimentally observed desensitization ring arrangement which is staggered and has 2-fold symmetry. f, The BD LBDs (left) are less inclined than the AC LBDs (right). This tilting of the AC domains elevates their E/G helices to adopt the upper position in the staggered configuration. Helix B of each LBD is highlighted in yellow as a reference to show domain tilt. g, Response to 10 mM glutamate for the D672R mutant, with the onset of desensitization fit by a single exponential of time constant 1.5 ms (red line). The upper trace shows the open tip junction current recorded at the end of the experiment. h, Recovery from desensitization, recorded from the same patch, for twin pulse applications of glutamate. The upper trace shows the command to the piezo stack. i, Recovery from desensitization for the D672R mutant (data points show mean ± SD, n=12) and wild type GluK2 (dashed line) fit with exponential functions.
To test if the desensitization ring contributes to the stability of the desensitized state, we used the structure to guide placement of arginine mutations at positions that would be expected to disrupt inter-subunit interactions. We chose S669R and D672R, both on helix G of the LBD, and measured their effects on the extent and rates of onset and recovery from desensitization. The extent of desensitization did not decrease (Fig. 2g), but both mutations speed recovery from desensitization (Figs. 2h, 2i, Extended Data Fig. 7, and Extended Data Table 1). Additionally, the nature and magnitude of this effect is consistent with previous reports identifying A676T and S679R mutations20,21, both of which, like the residues chosen in our functional experiments, reside on helix G (Extended Data Fig. 7). These results substantiate the hypothesis that the desensitization ring contributes to the stability of the state. Moreover, because these mutations do not reduce the extent of desensitization, other structural features must contribute to the deep energy well of the desensitized state. The ion channel region of the structure appears to be such a candidate, with its high relative structural stability in the cryo-EM density map (Extended Data Fig. 1e).
The desensitized state structure also reveals disruption of allosteric ion binding sites that facilitate receptor activation22–24 and suggests an attractive mechanism for the recovery to the resting state from the desensitized state. Superposition of an open cleft GluK2EM LBD crystal structure (PDB ID: 5CMK) via domain 1 coordinates on the closed cleft LBDs within the desensitized structure reveals severe steric clashes, which implies that the desensitization ring must start to reorganize before the LBD can fully open. This observation that the desensitization ring must begin reorganizing before the LBD can adopt an open cleft conformation immediately provides a structural explanation for the electrophysiological observation of channel activity during recovery from desensitization25.
Inspection of the TM region in the desensitized state shows that the overall channel architecture is remarkably similar to that reported for AMPA receptor structures in the apo and antagonist-bound resting states (Figs. 3a–3d), with residues M633, T629 and T621 forming constrictions in the pore that prevent ion permeation. High sequence similarity in the transmembrane regions of AMPA and kainate receptors, with the GluK2 desensitized structure reported here complementing previously reported GluA2 structures in apo, resting, and pre-activated states, makes possible a comparative analysis of closed channel glutamate receptor structures. This comparison makes clear that while the ion channel in these different states adopts a similar overall profile (Fig. 3e), there is a substantial difference at the entrance to the pore in the desensitized and agonist-bound pre-activated states, with a collapse to the resting state structure in the desensitized state compared to the expanded conformation observed in the pre-activated state10. That all apo, resting, and desensitized channels show similar profiles, suggests that rearrangement in the LBD layer upon desensitization is sufficient to allow the desensitized state channel to adopt a nearly identical non-conducting conformation as that present prior to activation.
Figure 3. Desensitized state ion channel.
a–c, Selected sidechain densities with atomic model for M1 (a), M3 (b), and M4 (c) helices. d, The ion channel with surface modeling of the channel. Regions with pore radius less than 1.15 Å are red, water accessible parts with radius between 1.15 Å and 2.30 Å are green, and wide areas with pore radius greater than 2.30 Å are blue. e, Plot of pore radius as a function of channel position with desensitized GluK2 (black), apo GluA2 (blue, PDB ID: 4U2P), antagonist-bound resting GluA2 (orange, PDB ID: 3KG2), and pre-activated GluA2 (magenta, PDB ID: 4U5B). Residue labels on right hand Y-axis mark the position of the respective channel forming side chains.
The desensitized state atomic model allows examination of the question of how the M3-S2 linkers connecting the ion channel and LBD layers mediate the striking differences between resting, pre-activated and desensitized state LBD structures. One aspect of these large changes in structure is that the M3-S2 linkers that couple LBD cleft closure to channel opening must somehow accommodate the height and pitch differences between AC and BD LBDs (Fig. 2f). We first considered differences between AC and BD chains of the TMD, and their connectivity to the LBD layer. Notably, while the two pairs of chains have nearly identical conformations in the TM domain, the linkers adopt different conformations as evidenced by Phi-Psi analysis, and the heights of the structures gradually diverge while still retaining overall ~4-fold in plane rotational symmetry (Fig. 4a). Specifically, beginning at M633 the AC linkers rise at a faster rate than their BD counterparts, with the difference reaching its maximum of ~6 Å at K645 (Fig. 4a, 4c). Thus, it is this height discrepancy that allows for the difference in pitch of the AC versus BD LBDs (Fig. 2f), permitting the E/G helices of the AC LBDs to occupy the elevated and tilted position in the staggered desensitization ring, whereas the compact BD subunit linkers map to the lower height of the BD LBDs. Despite the vertical asymmetry of linkers, their in-plane rotational symmetry is ~4-fold, as evidenced by orientations of helices E, the site at which linkers couple to the LBD. This mirrors the in-plane rotational symmetry of the LBD tetramer (Fig. 1d).
Figure 4. LBD-TM linkers mediate channel closing and LBD reorganization.
a, Comparison of the regions encompassing the M3, M3-S2 linkers, and E helices for AC (magenta) and BD (cyan) chains of desensitized state GluK2EM. b, The vertical rise in Cα position as a function of residue number for chains AC (magenta) versus BD (cyan). c, Measurement of how Cα positions on desensitized state GluK2 chains A, C, B, and D deviate from corresponding positions on resting state GluA2cryst which was used as a reference. Line coloring is the same as in panel (b). Dotted lines indicate AB chains, with solid lines for CD chains. The magenta traces show low deviation, reflecting the similarity between these regions of the AC subunits of desensitized GluK2 and the resting state GluA2 structure used as a reference (PDB ID: 3KG2). The cyan trace begins to deviate significantly at the circular markers corresponding to R632 (R628) GluK2 (GluA2). This reflects the dramatic difference in BD linker arrangements between the resting and desensitized states. d, The desensitized GluK2 channel with the region considered in (c). M3 helices and M3-S2 linkers are colored as in other panels, with pre-M1, M1 and M4 regions colored in yellow. Spheres mark residue R632 on all four TM chains. e, Schematic view of structural rearrangements that are involved in transition from resting to activated to desensitized states of glutamate receptors. Supplementary Video 1 shows an animation of the gating cycle.
Comparison of differences between the M3-S2 regions of the AC and BD subunit enables identification of the position that serves as the hinge for the transition to the ~4-fold symmetry in the desensitized state (Fig. 4d). As a structural reference for this analysis, we used the antagonist-bound resting state of GluA26. Chains A and C have similar conformations (Fig. 4a), consistent with the observation of small movements of the corresponding LBDs between resting and desensitized states. Conversely, in the B and D chains, there are 135–180° changes in main chain Psi angles over a 3 residue stretch, from E634-P636 in the GluK2 desensitized state, the trajectory of which deviates significantly from that observed in the GluA2 resting state, indicating that this region serves as a pivot point around which channel closure drives LBD layer rearrangement during desensitization4.
The central new findings of our study are that the GluK2 desensitized state displays 2-fold, quasi 4-fold and 4-fold in-plane rotational symmetries, for the ATD, LBD, and TM regions, respectively. However, surprising mismatches between axial and in-plane symmetry are found embedded in the LBD layer. These features ultimately confer the receptor with overall 2-2-4 symmetry with the LBD layer mediating an elegant structural compromise between the 2-fold symmetric ATD layer, and the 4-fold symmetric ion channel. The 2-fold character of the LBD desensitization ring is defined by the need to accommodate the ATD layer, which effectively “rocks” the AC LBDs backwards away from the central axis. The 4-fold character of the LBD layer arises from the critical need for it to match the symmetry of the ion channel, and thereby allow closing of the ion channel to relieve receptor “tension” driven by LBD closure during activation3,4,10.
The high-resolution desensitized GluK2 structure reported here helps close an important gap in glutamate receptor biology, permitting extensive comparison between high-resolution structures of resting and pre-activated iGluRs, thus providing a more comprehensive picture of the receptor gating cycle. We conclude that the previously undocumented 2-fold symmetric desensitization ring formed by the inner face of the LBD tetramer, combined with ATD-LBD interfaces, and most likely the high stability of the ion channel in its closed conformation, contribute to the deep energy well that the receptor enters from the active state. Furthermore, the staggered helix interactions in the desensitization ring likely serve collectively as a molecular switch that, upon agonist unbinding and opening of the jaws of individual LBDs, is disrupted and triggers destabilization of the desensitized state. This event would thereby permit a structural rearrangement back to the resting state with an intact LBD dimer of dimers assembly, thus completing the gating cycle.
METHODS
Protein expression and purification
The full-length rat GluK2 subunit cDNA sequence (UniProt ID: P42260) was cloned into the pFastBac1 vector for protein expression in insect cells. GluK2EM was created by introducing four mutations in the LBD (A487T, A658S, N690S and F704L) which convert the sequence to that found in GluK1, creating a high affinity binding site for the GluK1 selective antagonist LY46619516. The construct was RNA edited at position 536 (I to V) and had two mutations (C545V (M1) and C564S (M1-M2 loop) which increased yield and tetramer stability. For fluorescence detection and affinity purification, a thrombin recognition site and linker sequence (GLVPRGSAAAA) was inserted between GluK2EM and the coding sequence for the A207K dimerization suppressed EGFP mutant, with a C-terminal SGLRHis8 affinity tag. Membranes isolated from Sf9 cells were solubilized and purified as described previously using n-dodecyl-β-D-maltopyranoside (DDM) and cholesterol hemisuccinate (CHS)5. The GluK2EM S1S2 LBD construct encoding residues S398-K513 and P636-E775 connected by a GT dipeptide linker was expressed in Escherichia coli and purified as reported previously for wild type GluK226.
Specimen preparation and cryo-electron microscopy
Vitrified samples of GluK2 solublized in DDM-CHS were prepared with protein concentrated to 4.2 mg/mL. A volume of 3 μL was added to R1.2/1.3 holey carbon grids (Quantifoil) rendered hydrophilic with self-assembled monolayer functionalization27, and grids were frozen with a Vitrobot Mk IV robot (FEI Company, Hillsboro, OR, USA) or a Leica EM GP (Leica Microsystems Inc., Buffalo Grove, IL, USA).
Data was collected using a Titan Krios, operated at 300 kV, aligned for parallel illumination, and equipped with a GIF Quantum Energy Filter (Gatan, Inc.) operated in zero-energy-loss mode with a slit width of 20 eV (Extended Data Table 2). Images were acquired manually on a K2 Summit camera (Gatan, Inc.), at 105,000 X nominal magnification corresponding to a 1.324 Å physical pixel size. Each exposure was recorded in super-resolution mode as a 38-frame movie, with dose rate and exposure time of 3 e Å−1 s−1 and 15 s, respectively.
Image processing and structure analysis
Movie frames from the K2 Summit were aligned using the UCSF drift correction software28. Data was 2 × 2 binned, yielding a pixel size of 1.324 Å. Particles were manually identified and selected using the program e2boxer within the EMAN2 program suite29. Integrated and unintegrated multi-frame images were processed in the framework of either Relion version 1.3 or 1.430. The integrated images were used for CTF estimation with CTFFIND331 as implemented in the Relion workflow. Single particles were subjected to 2D (T = 2) and 3D (T = 4) classifications with C2 symmetry imposed (or C1 symmetry where specified) and low-population or poorly-defined classes were discarded at both stages. Data was subjected to particle polishing as implemented in Relion32, and resulting “shiny” particles refined to give final structures that were B-factor corrected in Relion. “Gold standard” FSC resolution plots were calculated using the EMAN2 program e2proc3d29 with a soft shape mask applied to independent unfiltered half maps from Relion. Three-dimensional classification was bootstrapped using previous GluK2 cryo-EM maps filtered to 60 Å as initial models. Extended Data Table 2 contains numbers of micrographs and particles used at all stages of image processing. Graphics for figures were prepared using UCSF Chimera33 and PyMOL34. The ion channel profiles in Figs. 3d and 3e were determined using the HOLE program35. Local resolution visualizations in Extended Data Figs. 1e and 3e were generated using the blocres utility in the Bsoft package36.
LBD crystallization and atomic model building of full-length receptor
Crystals for the GluK2EM LBD complex with (2S,4R)-4-methylglutamate were grown using a reservoir containing 18% PEG 8K, with a buffer solution of 20 mM NaCl, 1 mM EDTA, 5 mM 2S,4R-4-methylglutamate and 2 mM Tris pH 8.0. Crystals for the GluK2EM LBD complex with LY466195 were grown using a reservoir containing 2M Li2SO4, 3% PEG 4K, 0.1 M MgSO4, 5 mM LY466195 and 0.1 M Na Acetate pH 5.5. The structures were solved by molecular replacement, using the upper and lower lobes of the wild type GluK2 LBD (PDB ID: 3G3F) glutamate complex as search probes, and refined to good statistics using PHENIX37 and COOT38 (Extended Data Table 3).
The GluK2EM LBD complex with 2S,4R-4-methylglutamate crystallized with one protomer in the asymmetric unit. Due in large part to the N690S and F704L mutations the solvent accessible volume of the ligand binding pocket increased from 270 Å3 for wild type GluK2 to 310 Å3 for GluK2EM, comparable to the value of 305 Å3 for GluK126. Identical to the structure of GluK1 an additional trapped water molecule entered the binding pocket, forming a hydrogen bond network made by the Asn690 side chain carbonyl oxygen atom in the wild type protein. A dimer assembly essentially identical to that for wild type GluK2 (PDB ID: 3G3F) was created by least squares superposition using domain one coordinates (RMSD 0.23 Å) to position two GluK2EM protomers. The GluK2EM LBD complex with LY466195 crystallized as a dimer, but surprisingly one protomer contained a bound glutamate molecule while the second protomer bound LY466195. The glutamate bound protomer adopted a closed cleft conformation, while the LY466196 bound protomer adopted an open cleft conformation produced by a 27° rotation of domain 2. The structure revealed hydrogen bonds with the bound LY466195 ligand formed by the hydroxyl groups of the A487T and A658S mutant side chains, while the N690S mutation relieved a bad steric clash that prevents binding of LY466195 to wild type GluK2. A dimer with two open cleft protomers was created by least squares superposition using domain one coordinates (RMSD 0.31 Å) to position a copy of the LY466195 bound protomer in place of the glutamate bound protomer.
The GluK2EM desensitized state atomic model was built using rigid body fitting in UCSF Chimera33 of two copies of a GluK2 ATD dimer crystal structure (PDB ID: 3H6G) and four copies of a 2S,4R-4-methylglutamate-bound GluK2EM LBD protomer crystal structure (PDB ID: 5CMM) to the cryo-EM density map, followed by rebuilding in COOT. The ion channel and linkers to the LBD were also built in COOT using the GluA2 resting state crystal structure (PDB ID: 3KG2) as a guide, followed by real space refinement of the complete model using PHENIX. The ATD-LBD linker regions spanning residues 385-398 were then modeled using RosettaCM39 with C2 symmetry and the real space refined GluK2EM model as input. The antagonist-bound resting state model of GluK2EM was built from rigid body fits of two copies of a GluK2 ATD dimer crystal structure (PDB ID: 3H6G) and two copies of the manually generated dimer corresponding to the LY466195-bound GluK2EM LBD (PDB ID: 5CMK).
Electrophysiological experiments
Outside-out patch recordings from HEK cells transfected with wild type and mutant GluK2 constructs, with fast solution exchange achieved using four-bore glass tubing mounted on a P245.30 piezoelectric stack driven by a P-270 HVA amplifier (Physik Instrumente), were performed at room temperature using a Axopatch 200A amplifier as described previously40. The external solution contained (in mM) 145 NaCl, 2.5 KCl, 1.8 CaCl2, 1 MgCl2, 5 HEPES (pH 7.3), and 10 glucose, with 10 mM L-glutamate used for activation. The internal solution contained (in mM) 105 NaCl, 20 NaF, 5 Na4BAPTA, 0.5 CaCl2, 5 HEPES (pH 7.3), and 10 mM Na2ATP. Two electrode voltage clamp recordings at a holding potential of −60 mV, with 3M KCl agarose tipped electrodes of resistance 0.1 – 0.8 MΩ, were performed using stage 5–6 Xenopus oocytes, 2–3 days after injection of cRNA for either GluK2EM or wild type GluK2. The bath solution contained 100 mM NaCl, 1 mM KCl, 5 mM HEPES pH 7.5, 0.8 mM BaCl2 and 1 mM MgCl2, with concanavalin A (Sigma Type IV) 0.6 mg/ml used to block desensitization. The initial response to 100 μM glutamate following preincubation with 1.5 – 300 nM LY466195 was recorded as a step response before the slow increase in current due to dissociation of antagonist and fit with the Hill equation.
Extended Data
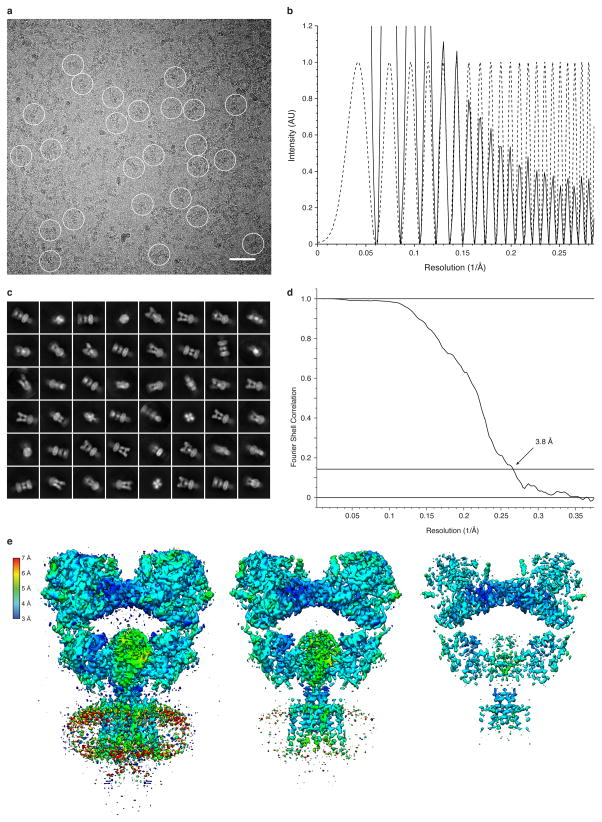
Extended Data Figure 1. Desensitized GluK2 imaging and structure determination.
a, b, Representative cryo-EM image of GluK2EM solubilized in DDM-CHS and bound by 2S,4R-4-methylglutamate (a), with the corresponding image power spectrum and CTF estimate showing signal beyond 3 Å resolution (b, solid and dotted lines, respectively). The defocus value for the image is 1.5 μm. In panel (a) particles are highlighted with circles, and image binning at 4x and uniform level adjustments were used to make particles apparent. Scale bar is 500 Å. c, Subset of selected two-dimensional class averages. d, FSC curve with reported resolution of 3.8 Å at the 0.143 crossing. e, Structure of agonist bound GluK2EM colored according to local resolution, shown at three progressively increasing contours.
Extended Data Figure 2. Desensitized GluK2 transmembrane and glycosylation features.
a, b, Cryo-EM density for resolved S1-M1 (a) and M3-S2 (b) linkers and TM helices, displayed with Cα trace of the atomic model. c, Representative sites with densities for complex glycans at Asn244, Asn347, and Asn399.
Extended Data Figure 3. Reconstruction of desensitized GluK2 without computational symmetry.
a, Cryo-EM density map for the reconstruction of agonist bound GluK2 without imposition of computational symmetry, and colored according to local resolution. b, FSC curve with reported resolution of 4.4 Å at the 0.143 crossing. c, Segmentation of individual GluK2 chains of the asymmetric reconstruction. The density map segmentation is shown fitted with a trace representation of the model displayed in Fig. 1b.
Extended Data Figure 4. Inhibition of GluK2EM by LY466195 and LBD crystal structures for agonist and antagonist complexes.
a, Crystal structure for the GluK2EM isolated LBD dimer assembly complex with 2S,4R-4-methylglutamate; the upper/lower lobes for the two subunits are colored orange/pale yellow and teal/pale cyan respectively; the dashed line indicates the separation of the lower lobes measured as the distance between the Cα positions of Ile637. b, Crystal structure for the GluK2EM isolated LBD dimer assembly complex with LY466195 illustrating the large decrease in separation of the lower lobes compared to the agonist complex. Coloring is the same as in (a). c, Responses to 100 μM glutamate recorded under two electrode voltage clamp for GluK2EM (top) and wild type GluK2 (bottom); the initial response to glutamate recorded after prior application of 300 nM LY466195 showed nearly complete block for GluK2EM with no change in amplitude for wild type. d, Concentration dependence for inhibition of GluK2EM by LY466195 yielded an IC50 of 30 nM.
Extended Data Figure 5. Imaging and structure of GluK2EM bound by antagonist LY466195.
a, b, Representative cryo-EM image of GluK2EM solubilized in DDM-CHS and bound by LY466195 (a), with the corresponding image power spectrum and CTF estimate showing signal beyond 6 Å resolution (b, solid and dotted lines, respectively). The defocus value for the image is 2.7 μm. In panel (a) particles are highlighted with circles, and image binning at 4x and uniform level adjustments were used to make particles apparent. Scale bar is 500 Å. c, Subset of selected two-dimensional class averages. d, Cryo-EM density map for GluA2 bound to ZK2007754 (left), density map for GluK2EM bound to LY466195 (middle) and its corresponding molecular model built from ATD and LBD dimers (right). e, FSC curve with reported resolution of 11.6 Å at 0.143 crossing.
Extended Data Figure 6. ATD-LBD interface of desensitized GluK2.
a, Desensitized GluK2 shown in surface representation with ATD-LBD interfaces highlighted. b, Top down view of LBD layer shown with perspective indicated by eye icon in (a). c, Underside of the ATD layer as viewed after peeling away from LBD layer. Dashed lines in (a) highlight where the layers are separated. In all panels, interfaces on chain A and C are in green and blue, respectively. d, Table with residues that mediate ATD-LBD interaction. e, f, Cartoon representation of LBD and ATD layers from same views as in (b) and (c), with interface residues colored to correspond with table in (d).
Extended Data Figure 7. Desensitization ring residues that influence recovery kinetics.
a–d, Rate of recovery from desensitization measured using twin pulse applications of 10 mM glutamate (data points show mean ± SD; fits are shown in red). a, Wild type GluK2 fit with a single exponential. b, D672R fit with the sum of two exponentials with the response for wild type shown as a black line; c, S669R D672R double mutant fit with the sum of two exponentials with the response for wild type shown as a black line. d, Time to 50% recovery in seconds. e–h, Top views of the GluK2 desensitization ring with residues found to influence recovery kinetics when mutated. Each panel shows the wild-type residue alpha carbon position as a sphere. Positions for the S669R and D672R mutations from the present study are shown in (e) and (f), respectively. The A676T and S679R positions found in previous studies20,21, are in (g) and (h), respectively.
Extended Data Table 1. Electrophysiological analysis.
For wild type GluK2, the D672R mutant and the S669R/D672R double mutant values (mean ± SEM) are reported for the peak amplitude of the response to 10 mM glutamate (Amp); the percent desensitization measured 50 ms after the start of the application of glutamate; the time constant of onset of desensitization determined using a single exponential fit (Tau des); the amplitude of the fast component of recovery from desensitization (Amp fast); and the rate constants of the fast (kfast) and slow (kslow) components of recovery from desensitization determined from double exponential fits. Values for S679R are from Carbone and Plested20; values for A676T are from Fleck et al21.
| Amp (nA) | % Desens | Tau des | Amp fast % | kfast s−1 | kslow s−1 | |
|---|---|---|---|---|---|---|
| WT | 1.41 ± 0.27 (n=13) | 99.3 ± 0.19 | 3.56 ± 0.40 | 100 | 0.79 ± 0.09 | |
| D672R | 0.56 ± 0.10 (n=12) | 99.0 ± 0.21 | 2.06 ± 0.11 | 85.3 ± 3.4 | 2.48 ± 0.27 | 0.14 ± 0.04 |
| S669R/D672R | 0.21 ± 0.06 (n=10) | 99.4 ± 0.17 | 2.08 ± 0.15 | 74.8 ± 3.7 | 3.25 ± 0.31 | 0.20 ± 0.06 |
| S679R | NR | 99.5 | 4.8 | 100 | 1.3 | |
| A676T | NR | NR | 3.0 | 100 | 1.4 |
Extended Data Table 2. Cryo-EM data collection and structural analysis.
Imaging conditions and data processing. The table shows the robot or robots used to make samples; the number of micrographs used for image processing; the number of particles manually designated in the micrographs; the number of particles retained after 2D classification; the number of particles retained after 3D classification and used for structure refinement; and the resolution and symmetry.
| Sample | Robot(s) | Micrographs | Particles prior to depletion | Particles retained after 2D classification | Particles in 3D reconstruction | Resolution (Å) |
|---|---|---|---|---|---|---|
| GluK2-SYM | Leica EM GP Vitrobot Mk IV |
2,454 | 166,311 | 166,284 | 62,244 (C2 symmetry) 62,552 (C1 symmetry) |
3.76 (C2 symmetry) 4.36 (C1 symmetry) |
| GluK2-LY466195 | Vitrobot Mk IV | 1,774 | 93,583 | 92,945 | 31,000 | 11.60 (C2 symmetry) |
Extended Data Table 3. Crystallographic data collection and refinement statistics.
Crystallographic data collection and refinement statistics for LBD structures.
| DATA COLLECTION | (2S,4R)-4-MethylGlu | LY466195 - Glu |
| Space group | P61 | P6122 |
| Unit cell a, b, c (Å) | 52.3, 52.3, 170.9 | 102.8, 102.8, 282.0 |
| a, β, g | 90, 90, 120 | 90, 90, 120 |
| Number per a.u. | 1 | 2 |
| Wavelength (Å) | 1.0000 | 1.0000 |
| Resolution (Å) a | 30 – 1.27 (1.29) | 30 – 1.8 (1.83) |
| Unique observations | 68757 | 82330 |
| Mean redundancy b | 7.0 (4.7) | 14.5 (14.2) |
| Completeness (%) b | 99.4 (95.2) | 100 (100) |
| Rmergebc | 0.043 (0.335) | 0.058 (> 1) |
| Rpimbd | 0.017 (0.166) | 0.017 (0.326) |
| I/s(I)b | 45.6 (3.8) | 48.4 (2.8) |
| REFINEMENT | ||
| Resolution (Å) | 27.29 – 1.27 | 29.86 – 1.80 |
| Protein atoms (AC) e | 2127 (80) | 4125 (95) |
| Ligand atoms | 11 | 34 |
| Li/Cl/SO4 ions | -/-/- | 2/10/2 |
| Water atoms | 410 | 533 |
| Rwork/Rfree (%) f | 14.3/16.3 | 16.4/18.7 |
| rms deviations | ||
| Bond lengths (Å) | 0.009 | 0.014 |
| Bond angles ° | 1.24 | 1.40 |
| Mean B-Values (Å2) | ||
| Protein overall | 20.9 | 27.5 |
| MC/SC g | 18.6/23.2 | 23.4/31.7 |
| Ligand | 13.2 | 19.8 |
| Li/Cl/SO4 ions | -/-/- | 23.8/57.1/34.8 |
| Water | 34.9 | 34.1 |
| Ramachandran % h | 97.8/0 | 97.9/0 |
Superscripts a–h denote the following:
Values in parenthesis indicate the low resolution limit for the highest-resolution shell of data.
Values in parenthesis indicate statistics for the highest-resolution shell of data.
Rmerge = (Σ|II − <II>|)/ΣI |II|, where <II> is the mean II over symmetry–equivalent reflections.
, where <II> is the mean II over symmetry–equivalent reflections.
Alternate conformations.
Rwork = (Σ ||Fo| − |Fc||)/Σ |Fo|, where Fo and Fc denote observed and calculated structure factors, respectively; 5% of the reflections were set aside for the calculation of the Rfree value.
Main chain/Side chain.
Preferred/Disallowed conformations.
Supplementary Material
Acknowledgments
This research was supported by the intramural programs of the NCI, and NICHD, NIH, the IATAP program at NIH, and the NIH-FEI Living Lab for Structural Biology. Synchrotron diffraction data was collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory. Use of APS was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. We thank Dr. David Bleakman for the gift of LY466195, Carla Glasser for preparing DNA constructs, and Prof. Frank DiMaio and colleagues for assistance with use of the program Rosetta. We also thank Veronica Falconieri for preparing the schematic illustration panel. This study utilized the capabilities of the HPC Biowulf cluster at NIH, Bethesda, MD (http://hpc.nih.gov).
Footnotes
AUTHOR CONTRIBUTIONS
J.R.M., M.L.M. and S.S. were involved in all stages of design of experiments and interpretation of results; S.C. carried out protein expression and purification; S.C. and M.L.M. carried out X-ray crystallography; T.H., E.S and M.L.M. performed electrophysiological experiments; J.R.M., A.M., P.R. and S.C. carried out cryo-EM data collection; J.R.M carried out cryo-EM image processing; J.R.M. and M.L.M. carried out structural analysis; J.R.M., M.L.M. and S.S. integrated all of the data, analysis of the implications and mechanism, and wrote the manuscript, with help from S.C.
AUTHOR INFORMATION
Cryo-EM density maps for GluK2EM with 2S,4R-4-methylglutamate, and GluK2EM with LY466195, have been deposited in the EM Data Bank under accession codes EMD-8289, and EMD-8290, respectively. Atomic coordinates for GluK2EM with 2S,4R-4-methylglutamate, GluK2EM with LY466195, GluK2EM LBD with 2S,4R-4-methylglutamate, and GluK2EM LBD with LY466195, have been deposited in the Protein Data Bank under accession codes, 5KUF, 5KUH, 5CMM and 5CMK, respectively.
The authors declare no competing financial interests.
References
- 1.Burnashev N, Szepetowski P. NMDA receptor subunit mutations in neurodevelopmental disorders. Curr Opin Pharmacol. 2015;20:73–82. doi: 10.1016/j.coph.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Lesca G, et al. GRIN2A mutations in acquired epileptic aphasia and related childhood focal epilepsies and encephalopathies with speech and language dysfunction. Nat Genet. 2013;45:1061–1066. doi: 10.1038/ng.2726. [DOI] [PubMed] [Google Scholar]
- 3.Durr KL, et al. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell. 2014;158:778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyerson JR, et al. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schauder DM, et al. Glutamate receptor desensitization is mediated by changes in quaternary structure of the ligand binding domain. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5921–5926. doi: 10.1073/pnas.1217549110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacological reviews. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer ML. Glutamate receptors at atomic resolution. Nature. 2006;440:456–462. doi: 10.1038/nature04709. [DOI] [PubMed] [Google Scholar]
- 9.Watkins JC, Evans RH. Excitatory amino acid transmitters. Annual review of pharmacology and toxicology. 1981;21:165–204. doi: 10.1146/annurev.pa.21.040181.001121. [DOI] [PubMed] [Google Scholar]
- 10.Chen L, Durr KL, Gouaux E. X-ray structures of AMPA receptor-cone snail toxin complexes illuminate activation mechanism. Science. 2014;345:1021–1026. doi: 10.1126/science.1258409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herguedas B, et al. Structure and organization of heteromeric AMPA-type glutamate receptors. Science. 2016;352:aad3873. doi: 10.1126/science.aad3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- 13.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI. Elucidation of AMPA receptor-stargazin complexes by cryo-electron microscopy. Science. 2016;353:83–86. doi: 10.1126/science.aaf8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yelshanskaya MV, Li M, Sobolevsky AI. Structure of an agonist-bound ionotropic glutamate receptor. Science. 2014;345:1070–1074. doi: 10.1126/science.1256508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Chen S, Yoshioka C, Baconguis I, Gouaux E. Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM. Nature. 2016 doi: 10.1038/nature18961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alushin GM, Jane D, Mayer ML. Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists. Neuropharmacology. 2011;60:126–134. doi: 10.1016/j.neuropharm.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones KA, Wilding TJ, Huettner JE, Costa AM. Desensitization of kainate receptors by kainate, glutamate and diastereomers of 4-methylglutamate. Neuropharmacology. 1997;36:853–863. doi: 10.1016/s0028-3908(97)00066-x. [DOI] [PubMed] [Google Scholar]
- 18.Moykkynen T, Coleman SK, Semenov A, Keinanen K. The N-terminal domain modulates alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptor desensitization. J Biol Chem. 2014;289:13197–13205. doi: 10.1074/jbc.M113.526301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutta A, et al. Cooperative Dynamics of Intact AMPA and NMDA Glutamate Receptors: Similarities and Subfamily-Specific Differences. Structure. 2015;23:1692–1704. doi: 10.1016/j.str.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carbone AL, Plested AJ. Coupled control of desensitization and gating by the ligand binding domain of glutamate receptors. Neuron. 2012;74:845–857. doi: 10.1016/j.neuron.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 21.Fleck MW, Cornell E, Mah SJ. Amino-acid residues involved in glutamate receptor 6 kainate receptor gating and desensitization. J Neurosci. 2003;23:1219–1227. doi: 10.1523/JNEUROSCI.23-04-01219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaudhry C, Plested AJ, Schuck P, Mayer ML. Energetics of glutamate receptor ligand binding domain dimer assembly are modulated by allosteric ions. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:12329–12334. doi: 10.1073/pnas.0904175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dawe GB, et al. Distinct Structural Pathways Coordinate the Activation of AMPA Receptor-Auxiliary Subunit Complexes. Neuron. 2016;89:1264–1276. doi: 10.1016/j.neuron.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plested AJ, Vijayan R, Biggin PC, Mayer ML. Molecular basis of kainate receptor modulation by sodium. Neuron. 2008;58:720–735. doi: 10.1016/j.neuron.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patneau DK, Mayer ML, Jane DE, Watkins JC. Activation and desensitization of AMPA/kainate receptors by novel derivatives of willardiine. J Neurosci. 1992;12:595–606. doi: 10.1523/JNEUROSCI.12-02-00595.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 27.Meyerson JR, et al. Self-assembled monolayers improve protein distribution on holey carbon cryo-EM supports. Scientific reports. 2014;4:7084. doi: 10.1038/srep07084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nature methods. 2013;10:584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. Journal of structural biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 30.Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of structural biology. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. Journal of structural biology. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 32.Scheres SH. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife. 2014;3:e03665. doi: 10.7554/eLife.03665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC. DeLano Scientific; 2002. [Google Scholar]
- 35.Smart OS, Goodfellow JM, Wallace BA. The pore dimensions of gramicidin A. Biophysical journal. 1993;65:2455–2460. doi: 10.1016/S0006-3495(93)81293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heymann JB. Bsoft: image and molecular processing in electron microscopy. Journal of structural biology. 2001;133:156–169. doi: 10.1006/jsbi.2001.4339. [DOI] [PubMed] [Google Scholar]
- 37.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang RY, et al. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nature methods. 2015;12:335–338. doi: 10.1038/nmeth.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Horning MS, Mayer ML. Regulation of AMPA receptor gating by ligand binding core dimers. Neuron. 2004;41:379–388. doi: 10.1016/s0896-6273(04)00018-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.