Abstract
Metabolic engineering offers the potential to renewably produce important classes of chemicals, particularly biofuels, at an industrial scale. DNA synthesis and editing techniques can generate large pathway libraries, yet identifying the best variants is slow and cumbersome. Traditionally, analytical methods like chromatography and mass spectrometry have been used to evaluate pathway variants, but such techniques cannot be performed with high throughput. Biosensors - genetically encoded components that actuate a cellular output in response to a change in metabolite concentration - are therefore a promising tool for rapid and high-throughput evaluation of candidate pathway variants. Applying biosensors can also dynamically tune pathways in response to metabolic changes, improving balance and productivity. Here, we describe the major classes of biosensors and briefly highlight recent progress in applying them to biofuel-related metabolic pathway engineering.
Introduction
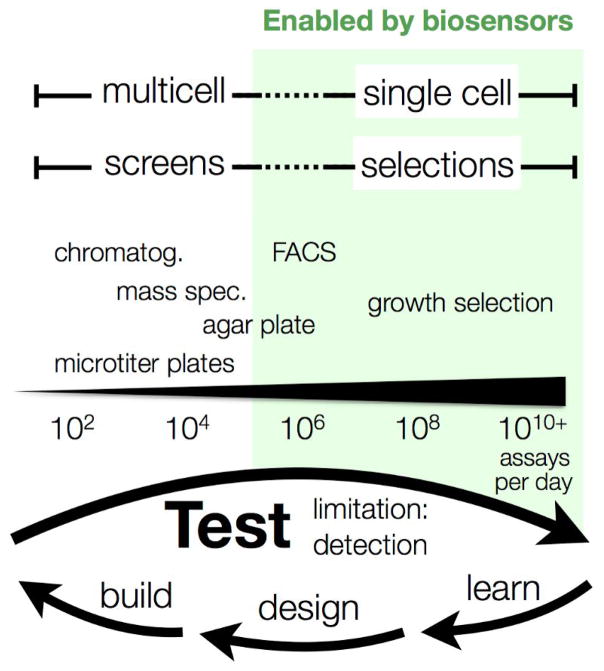
Metabolic engineering of microbes holds the promise of producing many classes of chemicals, including fuels, from renewable feedstocks [1]. However, to compete with established production methods, engineered organisms must be highly productive, efficient, and robust at industrial scales. Many factors, such as the enzymes employed, regulatory proteins and genetic regulatory elements, can affect these phenotypes, and so a fundamental aspect of pathway engineering is identifying the complex genetic alterations required to create an optimized strain. While there are numerous ways to engineer genetically diverse strain libraries - in both random and/or directed fashions [2,3] - there are few assays that scale with the bandwidth of modern genetics (Figure 1). As such, it is critical to develop novel detection technologies in order to bring the full power of genetics to bear on metabolism.
Figure 1.
Biosensors enable rapid engineering of metabolism.
Biosensors enable rapid and single-cell quantitation of metabolites allowing for high-throughput evaluation of pathway variants and improving the rate-limiting “test” step of the design-build-test-learn engineering cycle.
An effective screening tool must be specific, high throughput, and sensitive to relevant metabolite concentrations. Most metabolites, except for special cases or by the use of exogenous chemical dyes (reviewed in [4]), cannot be measured using rapid optical methods. Chromatography and mass spectrometry (MS) are thus the only analytical tools available for measuring most biofuel-related metabolites despite their low throughput. Biosensors, genetically encoded components that respond to an input signal (e.g. metabolite concentration) and transduce that signal into a detectable output (e.g. fluorescence or gene expression), are emerging as a high-throughput alternative for measuring metabolite concentrations in vivo. Often adapted from natural proteins or aptamers, biosensors can be specific, sensitive, and non-destructive.
Here, we provide an introduction to biosensors and their use in modern metabolic engineering. As a point of comparison, we start with recent advances in analytical chemistry (reviewed in greater detail in [5]) and contrast this with the commonly employed classes of biosensors. We then focus in detail on the applications of specific biosensors to biofuel-related metabolic engineering.
State-of-the-art in analytical metabolite detection
Analytical chemistry methods, including chromatography and MS, are the gold standard for measuring metabolism. These methods are label-free, sensitive, and can detect many (e.g. 100+) metabolites in a single measurement [6]. However, these methods require time- and labor-intensive metabolite extractions that result in destructive, bulk measurements that are generally low throughput (101–103 per day). Two emerging MS-based platforms that may aid in overcoming these limitations are the RapidFire high-throughput MS system from Agilent Technology, Inc. [7] and surface-based MS techniques, such as Nanostructure-Initiator MS (NIMS) [8].
RapidFire uses robotics to automate the metabolomics workflow. Samples in microtiter plates are purified by solid-phase extraction and directly injected into an MS instrument. The instrument can processes a single sample in less than 15 s, which is over 100x faster than traditional liquid-chromatography-MS measurements [7]. NIMS is a surface-assisted laser desorption/ionization technique that requires little sample preparation and uses a liquid “initiator,” instead of a co-crystallization matrix, to produce spectra with high sensitivity and lower noise in the metabolite mass region. NIMS was recently used to screen >100 glycoside hydrolases (enzymes important for biomass hydrolysis) with a wide range of substrates and reaction conditions to generate more than 10,000 data points [9]. Although surface and automated MS techniques are not yet widely used, it is likely they will continue to increase in throughput and find applications in metabolic engineering.
Classes of genetically encoded biosensors
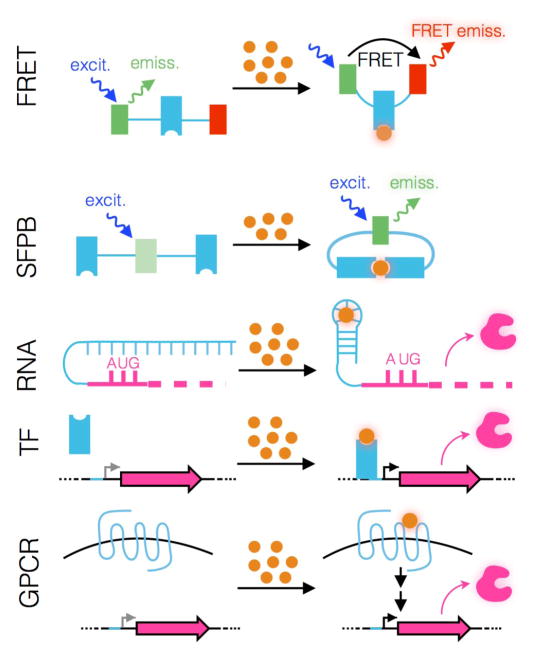
Biosensors are genetically encoded components that convert an input signal (e.g. metabolite concentration) into a measurable output like fluorescence or gene expression (Figure 2). In the following sections, we introduce common classes of biosensors constructed from fluorescent proteins, RNA, cytosolic transcription factors (TFs), G-protein-coupled receptors (GPCRs) as well as two-component systems and discuss their inherent advantages and disadvantages.
Figure 2.
Biosensor definition and types.
Biological input indicated as orange spheres. Ligand-binding component indicated in light blue. Biosensor output highlighted: Förster resonance energy transfer (FRET), energy-transfer efficiency; single fluorescent protein biosensors (SFPB), emission; RNA, translation product; transcription factor (TF), transcription product; G-protein-coupled receptor (GPCR), signaling pathway product.
Fluorescent protein biosensors
Genetically encoded biosensors based on Förster resonance energy transfer (FRET) or single fluorescent proteins are promising tools for the analysis of metabolic pathways and their products. FRET biosensors consist of a ligand-binding domain (LBD) attached to a pair of fluorescent proteins that have overlap in their emission and excitation spectra, capable of FRET (Figure 2) [10]. Binding of a metabolite to the LBD alters the distance between the two fluorophores and changes the energy-transfer efficiency, measured as a ratio of fluorescence. While FRET biosensors have been developed for many different metabolites [11,12], they typically exhibit low dynamic ranges (e.g. tens of % change in signal) that significantly impede their use in screening applications. Single fluorescent protein biosensors (SFPBs) are fluorescent proteins that either directly detect input signals or are inserted within the primary sequence of a conformationally-labile LBD such that ligand binding affects fluorescence intensity (Figure 2) [13]. While genetically encoded SFPBs have high dynamic range (e.g. 10-fold) and are used in cell biology studies [14,15], they are not widely used in metabolic engineering [16••]. There are currently few available SFPBs due to the difficulty in engineering the coupling between an LBD and a fluorescent protein partner. Methods enabling rapid SFBP engineering may therefore be useful to increase the availability of this promising class of biosensors [17].
RNA biosensors
Riboswitches are naturally evolved ligand-responsive RNA elements that possess two components: a sensor (aptamer) domain that detects metabolite binding and a regulatory domain that converts binding-induced conformational changes into changes in gene expression (Figure 2) [18]. RNA-based biosensors also benefit from known techniques (e.g. SELEX) for generating aptamers against new metabolites [19] and have been adapted as biosensors for engineered pathways [20–22]. To date, however, use in metabolic engineering has been limited, likely from the challenges of recapitulating in vitro behavior within the cellular environment.
Cytosolic transcription factor (TF) biosensors
TF-based biosensors detect environmental changes, such as metabolite levels, and alter gene expression in response (Figure 2). The most widely used are bacterial TFs, which are composed of an LBD that controls the engagement of a cognate DNA-binding domain to promoter/operator sites associated with target genes. Depending on the TF, DNA binding may lead to gene repression or activation. These biosensors can offer high sensitivity and dynamic range; small changes in ligand concentration are amplified through gene expression into large changes in protein abundance.
An early implementation of TF–based biosensors was the development of whole-cell biosensors where expressed reporter genes (e.g. luciferase or β-galactosidase) were used to detect environmental pollutants [23]. Subsequently, TF-biosensors have been used in high-throughput strain evaluation by linking metabolite levels to fluorescence [24,25] and growth advantages such as antibiotic resistance [24,26,27•]. More recently, TF-biosensors have been linked to regulatory or pathway genes to provide dynamic feedback within engineered pathways [28–31••]. This modularity of input and output domains in TF-based biosensors makes them attractive for many metabolic engineering applications.
Despite the increasingly widespread adoption of TF-based biosensors for metabolite sensing, there are potential disadvantages. First, there is a large difference in the timescales of metabolite turnover (~1 sec) and those of transcription and translation (~1–10 min [32]), which makes real-time sensing impossible when using TF-based biosensors. Additionally, TF-based biosensors are not always robust; bacterial TFs may not be portable to eukaryotes due to fundamental differences in the transcriptional process. Finally, expression of a non-native TF may have unanticipated side-effects, including non-specific binding to DNA and interfering with transcription.
G-protein coupled receptor (GPCR) and Two-component biosensors
An alternative to cytosolic TFs is GPCR-based biosensors expressed on the cell surface [33–35]. For these biosensors, the binding of an extracellular metabolite to a GPCR results in signal transduction and, ultimately, changes in gene expression (Fig. 2b). As with TF-based biosensors, the modular nature of GPCR-based biosensors and the wide variety of molecular specificities [36] make them broadly useful for metabolite sensing. However, one potential caveat is that sensing only occurs extracellularly, which may limit applications. The analog of GPCRs for prokaryotes are the two-component regulatory systems in which one component acts as a transmembrane sensor and the second component acts as an intracellular response regulator. Studies demonstrating that the extracellular sensing domain of one transmembrane sensor could be fused with the intracellular domain of another to create a hybrid biosensor, as well as studies showing that the promoter for a particular response regulator could be used to control the expression of an arbitrary output of interest, have been met with excitement in the synthetic biology community [37,38]. However, the engineering of two-component systems has met with practical difficulties [39] and more studies will be needed to determine design principles of re-engineering ligand specificity [40].
Biosensor Applications
In the following sections we highlight recent applications of biosensors to the i) isolation of improved mutants and ii) dynamic control of metabolic pathways (Figure 3a). TF-based biosensors predominate in these examples as they have been the most widely adopted, to date, in biofuel-related metabolic engineering.
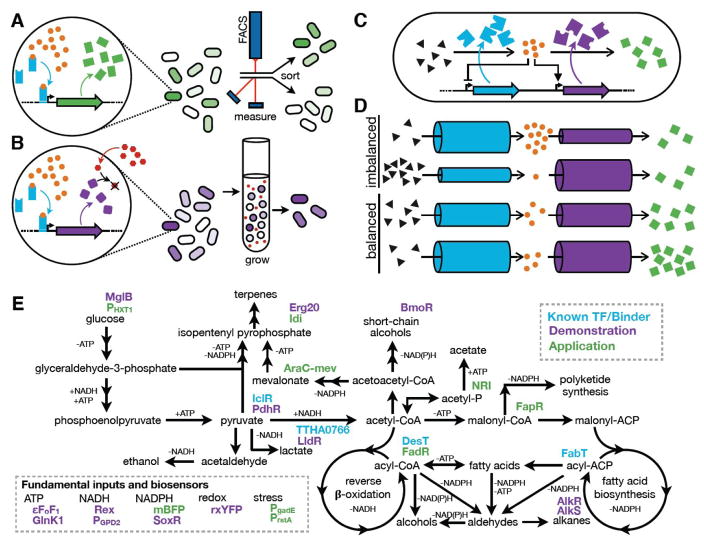
Figure 3.
Applications of biosensors.
(a) A biosensor with an output, such as fluorescence, can be used in a screen. The metabolite of interest (MOI, orange) is detected by a biosensor, which drives the expression of an output signal (green) in proportion to the MOI concentration. The output signal, often fluorescence, is used to isolate high-producing variants through screening (e.g. FACS or plate reader assays). (b) A biosensor with a selectable output. The MOI drives the expression of a protein (purple) that provides a growth advantage to the cell. As depicted, an enzyme that neutralizes an antibiotic (red) is expressed in a ligand-dependent manner. Growing variants under selective pressure (e.g. in the presence of antibiotic) enriches the population with high-producing variants. (c) Biosensors for dynamic regulation of pathways. The MOI (orange) is detected by a biosensor (not depicted – biosensor actions are represented by feedback symbols) which alters expression of enzymes in the MOI pathway. The cartoon illustrates balancing of a pathway intermediate (MOI, orange) by repressing the preceding enzyme and activating the subsequent enzyme in the pathway (similar to [31]). (d) Visualization of pathway balancing. Enzyme activity is represented by tubes (i.e. maximal flux that can be carried). Regulation of enzyme concentration (by TFs or degradation) or activity (allostery) alters the flux capacity between pathway intermediates. When enzyme flux is imbalanced (top two examples), starting materials or intermediates accumulate and product formation is limited. When flux is balanced (bottom two examples), accumulation does not occur at any step. (e) Potential biofuel pathways and biosensors. Binding proteins and transcription factors (or regulated promoters) are shown with colors designating: known binder with no biosensor use published; demonstrated use as a biosensor; or biosensor applied to screening, selection, or pathway-balancing. AlkR [61]; AlkS [62] AraC-mev [63]; BmoR [24]; DesT [64]; εFoF1 [65]; Erg20 [27]; FabT [64]; FadR [30]; FapR [31]; GlnK1 [14]; Idi [27]; IclR (ecocyc.org); LldR [66], mBFP [16••]; MglB [67]; NRI [28]; PdhR [68]; PgadE [69]; PGPD2 [70]; PHXT1 [71]; PrstA [69]; Rex [15]; rxYFP [72]; SoxR [44]; TTHA0766 [73].
Biosensors with phenotypic output
Biosensors are often used to generate a phenotypic output that can be screened or selected (Figure 3a–b). The sensed ligand is generally an intermediate or the product of a desired pathway and the biosensor is used to isolate genotypes with higher titers and improved pathway flux. For example, malonyl-CoA production, the first committed step of fatty acid biosynthesis (Figure 3e), was targeted for improvement in S. cerevisiae using a TF-based biosensor [25]. FapR, a TF that represses expression in the absence of malonyl-CoA, was used in a FACS screen to isolate genes from a cDNA library of ~106 variants that improved malonyl-CoA production (Figure 3a). Similarly, an SFPB for the fundamental co-factor NADPH was used to isolate production strains with more biosynthetic potential. This biosensor allowed rapid micro-well plate quantification of 624 computationally designed synthetic-pathway variants for high NADPH titer in E. coli [16]. When combined with a terpenoid biosynthetic pathway, the improved NADPH pool increased production by nearly 2-fold (Figure 3e). The rapid screening achieved with these examples illustrates the advances that can be made when production strains are evaluated in high throughput.
Biosensors can also generate an output that imparts a growth advantage to cells, allowing for growth selection and increasing the potential number of testable designs by orders of magnitude (Figures 1 and 3b). Raman and coworkers optimized a TolC antibiotic-resistance output linked to a TF biosensor for the commodity chemical glucaric acid [26]. Selection of a 107 library after multiplex genome engineering led to a strain with 22-fold greater glucaric acid titer. Alternatively, an innovative biosensor design developed by Chou and Keasling to increase isopentenyl pyrophosphate (IPP, a terpene building block, Figure 3e) production had mutation rate as the output [27•]. Starting with a high mutation rate, an artificial TF decreased the expression of the mutD5 polymerase (mutator) as IPP concentration increased. This stabilized high IPP-producing genotypes, resulting in a ~17-fold improvement in lycopene (terpene) production over strains with no link between IPP concentration and mutD5. Coupling the appropriate biosensor with the right selection can therefore enrich immense libraries (>106 variants) to isolate improved strains.
Biosensors for dynamic pathway regulation
Metabolic pathways are complex and tightly regulated in their native context. Maximizing pathway yields requires careful balancing of pathway flux to prevent bottlenecks and/or the accumulation of toxic intermediates. Dynamic pathway regulation, using TF-based biosensors, allows for flux to be altered in response to changing cellular and environmental conditions (Figure 3c). A groundbreaking study in 2000 by Farmer and Liao demonstrated the utility of this approach by improving lycopene yield in E. coli using a biosensor for acetyl-phosphate [28]. The concentration of acetyl-phosphate is a proxy for glucose availability and reduced growth (Figure 3e). Lycopene pathway genes, which normally inhibit growth, were gradually activated by the TF-biosensor as cells exited exponential phase. This strategy improved lycopene titer by almost 20-fold over static expression. Pathway feedback can be especially beneficial when intermediates are toxic, preventing dangerous buildups. For example, a prominent study engineered a fatty acid ethyl ester (FAEE) pathway using a fatty acyl-CoA TF biosensor (FadR, [30]). The introduced feedback both reduced toxic ethanol accumulation and prevented depletion of cellular fatty acids, thereby creating a strain with 3-fold greater FAEE yield.
As mentioned in the previous section, malonyl-CoA is a crucial intermediate for the production of biofuel-related molecules (Figure 3e). Because of this, it has been targeted in multiple efforts to implement biosensor-based regulation. Xu and Koffas used a single TF biosensor (FapR) to repress the acc operon (catalyzing acetyl-CoA carboxylation to malonyl-CoA) and, in the presence of malonyl-CoA, induce a fatty acid biosynthesis operon (malonyl-CoA conversion to fatty acid) [31••] (Figure 3d). This combined regulation led to a >3-fold increase in fatty acid yield and improved cell growth as compared to the un-regulated strain. Interestingly, while FapR naturally represses fatty acid biosynthesis in B. subtilis, this work serendipitously identified FapR activation behavior in the promoter region of gapA (glyceraldehyde-3-phosphate dehydrogenase A) within the E. coli host, providing both types of control in the engineered strain. To achieve the same positive control with FapR in E. coli, another group used a genetic invertor to regulate acc expression, thus avoiding the need to re-engineer the TF or operator [41,42]. The same bacterial TF was also recently employed as a biosensor in S. cerevisiae to provide negative control of mcr (malonyl-CoA conversion to 3-hydroxypropionic acid) [43]. Fatty acids and their intermediates are desirable end products and these successful biosensor engineering efforts point to exciting future possibilities. We expect further integration of new biosensors, such as those for NADPH [44], to improve yields.
Outlook
In addition to the biosensor designs discussed above, there have been several recent innovative approaches to biosensor construction. For example, Feng and colleagues developed eukaryotic biosensors based on protein lifetime that could be applied to biofuel-related molecules [45]. In this approach, a library of mutagenized LBDs fused to a reporter are screened for high signal in the presence of ligand, due to conditional protein stabilization, and low signal in the absence of ligand, due to degradation by the proteasome. Enzyme-based biosensors are another alternative. Such biosensors enzymatically convert a metabolite of interest to a molecule that is directly detectable, or for which there is a pre-existing genetically encoded biosensor [46, 47•,48]. SensiPath, a recently reported web-based tool (http://sensipath.micalis.fr) that searches for enzymatic pathways that convert metabolites to detectable molecules, may be especially useful in trying to achieve these goals [49•].
Despite these promising approaches, we believe that there is significant potential for single-component allosteric biosensors. Single protein molecules that link input and output domains via allostery, such as SFPBs, are powerful tools due to their genetic portability and fast response rates. We have already highlighted the benefit of biosensor-mediated feedback for pathway balancing and improved strain performance. However, to date, only multi-component, TF-based biosensors have been used for this purpose. These biosensors respond on a timescale that is too slow for many metabolic applications, as metabolic fluctuations are 10–100 times faster than transcription and translation [32]. In addition, delayed negative feedback can lead to oscillations in the concentration of regulated species [31••,50]. Using a pathway enzyme as an output in a single-component biosensor, as is common in natural metabolism [51], would dramatically improve the speed and robustness of feedback control. In this section, we highlight some of the future challenges and opportunities in developing this type of biosensor by focusing on techniques for the identification of novel input domains, the engineering of allostery and the exploration of new functional outputs.
Input domains
A significant barrier to the development of biosensors is identifying novel LBDs. One approach to overcoming this problem is to mine the large numbers of naturally occurring proteins that are being identified in genome and metagenome sequencing projects (Figure 4a). Vetting and coworkers recently highlighted this strategy by using high-throughput protein expression and differential scanning fluorimetry to screen 158 candidate LBDs against 189 ligands to identify 40 new ligand-LBD interactions [52••]. Alternatively, techniques such as substrate-induced gene expression screening (SIGEX), which involves inserting restriction enzyme-digested (meta)genome fragments upstream of a reporter gene, may also be adapted to identify previously uncharacterized LBDs [53].
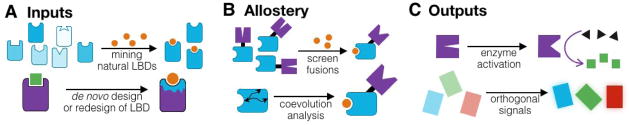
Figure 4.
Future challenges and opportunities. (a) If no known Ligand-Binding Domain (LBD) is known for a molecule of interest (orange), techniques are needed to discover or create the necessary biosensor input. Uncharacterized potential LBDs from (meta)genome databases can be mined for the desired binding property. Alternatively, in silico approaches of creating new binding proteins or redesigning characterized LBDs can lead to a desired input domain. (b) Engineering allosteric communication between input and output domains is challenging. Methods that create and test many fusion variants have led to successful biosensor function. Computational analysis of coevolving residues in a protein can predict surface sites for functional allosteric fusions. (c) Many new output functions are available for exploration in biosensors. Enzyme output domains provide opportunities for growth selection as well as direct metabolic pathway feedback control. Detectable outputs that are orthogonal can be used in unique biosensors for multiplex measurements of different molecules of interest.
An alternative method for expanding the number of LBDs is to employ computational design approaches (Figure 4a). Tinberg and colleagues reported the design of a ligand-binding protein for the steroid digoxigenin (DIG) by computationally designing the protein-ligand interface and intramolecular interactions using a protein scaffold of unknown function that did not originally bind the target molecule. Of 17 designs that were experimentally characterized, two were functional for DIG-binding, and optimization of one of these constructs using site-saturation mutagenesis coupled with selections resulted in a 75-fold improvement in binding affinity [54]. Similarly, Taylor and coworkers have reported the redesign of the lac repressor transcription factor for a number of novel inducers using a combination of computational protein design, mutagenesis and gene shuffling [55]. As computational design of binding improves, a potential application of such work would be in tuning the affinity of a biosensor, and thus the operational range, for various applications. The utility of such biosensor tuning was demonstrated by the Frommer lab by developing a family of glucose biosensors with a range of affinities to visualize the different responses to glucose perfusion in various areas of the plant [56]. While still very challenging, continued advances in this area using de novo protein design, the redesign of existing LBD scaffolds and directed evolution will expand the number of LBDs and consequently the number of metabolite biosensors.
Allostery
Once an LBD is identified for a small molecule of interest, the greatest challenge in creating a single-component biosensor is often engineering the allosteric connection to the desired output domain. One proven method for doing so has been via domain insertion whereby one protein domain is inserted into another such that the functions of the two independent domains are coupled (Figure 4b). However, while this strategy has proven successful, for example, as demonstrated by Guntas and coworkers in the development of a maltose-dependent β–lactamase biosensor [57], it is plagued by its low-throughput nature due to the difficulties in reliably predicting the insertion sites for linking the associated domains. To overcome this, our group recently reported a strategy for the rapid construction of biosensors termed domain-insertion profiling with sequencing (DIP-seq) [17]. In this approach, we created diverse libraries of potential SFPBs using modified transposons and then used high-throughput activity assays to identify functional biosensors. While we have applied this tool to the rapid construction of SFPBs, it may also be applied to the generation of allosteric biosensors of any type and function [58].
Engineering allostery through the redesign of protein surfaces to include ligand-binding sites may be another route (Figure 4b). Work from the Ranganathan group has shown that there are networks of physically connected and coevolving amino acids that link protein active sites to spatially distinct surface sites [59]. With continued computational advances, it may one day be possible to reliably predict these surface sites and further engineer them to include ligand-binding sites for the development of allosterically regulated biosensors.
Output domains
The majority of reported biosensors employ fluorescence as an output, which is effective for visualization and screening. However, given the availability of alternative functional output domains, there are likely many alternative novel biosensor applications (Figure 4c). As mentioned above, the use of selection over screening can enable testing of orders of magnitude more designs by linking metabolite binding to host fitness. For example, by allosterically coupling E. coli MBP to TEM1 β-lactamase, Guntas and coworkers developed maltose-dependent switches that conferred growth selection phenotypes in the presence of β–lactam antibiotics [57]. Similar strategies may also be adopted for biofuels by linking the detection of the desired molecule to the growth of the host.
Compared with most available biosensors that have only single detection channels, multiplexed biosensors could provide more information by increasing the number of available outputs that can be detected at the same time (Figure 4c). For biosensors that provide fluorescence readout, the most common form of multiplexing involves combining biosensors with different wavelengths but this approach is limited due to spectral overlap. However, combining intensity measurements with time-resolved measurements may provide a means to multiplex in both the wavelength and time domains. Employing such time-resolved fluorescence lifetime biosensors are advantageous because they are quantitative and independent of biosensor concentration. Recently, Mongeon and coworkers demonstrated that Peredox, a previously reported SFPB for NADH:NAD+ ratio, could also be used as a fluorescence lifetime biosensor because it showed a large change in fluorescence lifetime over its sensing range [60].
Along with identifying and designing new ligand-binding domains, engineering methods for allosteric enzyme design are long-term goals that will benefit the field. We expect that future improvements in the identification of ligand-binding domains and in the predictable engineering of allostery in fluorescent proteins and enzymes will provide a biotechnological backbone that vastly improves our capacity to design, screen, and select pathway variants for the biological production of fuels. Ultimately, these improvements may enable biocatalysis to compete with fossil fuels in reliability and economic terms.
Highlights.
Biosensors and their use in biofuel-related metabolic engineering are introduced.
The pros and cons of major biosensor classes are reviewed.
Biosensors enable rapid screening and growth selection of diverse libraries.
Dynamic pathway regulation with biosensors can improve productivity.
Continued biosensor development will further accelerate bioenergy engineering.
Acknowledgments
We thank Avi Flamholz and Sean Higgins for help in preparing the manuscript. This work was supported by the Energy Biosciences Institute (OO1J25), a NIH New Innovator Award (1DP2EB018658-01), and a Basil O’Connor Starter Scholar Research Award from the March of Dimes to D.F.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keasling JD. Manufacturing Molecules Through Metabolic Engineering. Science. 2010;330:1355–1358. doi: 10.1126/science.1193990. [DOI] [PubMed] [Google Scholar]
- 2.Haimovich AD, Muir P, Isaacs FJ. Genomes by design. Nat Rev Genet. 2015;16:501–516. doi: 10.1038/nrg3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu R, Bassalo MC, Zeitoun RI, Gill RT. Genome scale engineering techniques for metabolic engineering. Metab Eng. 2015;32:143–154. doi: 10.1016/j.ymben.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 4.Hounslow E, Noirel J, Gilmour DJ, Wright PC. Lipid quantification techniques for screening oleaginous species of microalgae for biofuel production. Eur J Lipid Sci Technol. 2016 doi: 10.1002/ejlt.201500469. [DOI] [Google Scholar]
- 5.Petzold CJ, Chan LJG, Nhan M, Adams PD. Analytics for Metabolic Engineering. Front Bioeng Biotech. 2015;3:135. doi: 10.3389/fbioe.2015.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett BD, Yuan J, Kimball EH, Rabinowitz JD. Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc. 2008;3:1299–1311. doi: 10.1038/nprot.2008.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanderporten E, Frick L, Turincio R, Thana P, Lamarr W, Liu Y. Label-free high-throughput assays to screen and characterize novel lactate dehydrogenase inhibitors. Anal Biochem. 2013;441:115–122. doi: 10.1016/j.ab.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, Golledge SL, Nordström A, Siuzdak G. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 9.Heins RA, Cheng X, Nath S, Deng K, Bowen BP, Chivian DC, Datta S, Friedland GD, D’Haeseleer P, Wu D, et al. Phylogenomically guided identification of industrially relevant GH1 β-glucosidases through DNA synthesis and nanostructure-initiator mass spectrometry. ACS Chem Biol. 2014;9:2082–2091. doi: 10.1021/cb500244v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 11.Miyawaki A, Llopis J, Heim R, McCaffery JM, Adams JA, Ikura M, Tsien RY. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 1997;388:882–887. doi: 10.1038/42264. [DOI] [PubMed] [Google Scholar]
- 12.Deuschle K, Okumoto S, Fehr M, Looger LL, Kozhukh L, Frommer WB. Construction and optimization of a family of genetically encoded metabolite sensors by semirational protein engineering. Protein Sci. 2005;14:2304–2314. doi: 10.1110/ps.051508105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci USA. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat Meth. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hung YP, Albeck JG, Tantama M, Yellen G. Imaging Cytosolic NADH-NAD(+) Redox State with a Genetically Encoded Fluorescent Biosensor. Cell Metab. 2011;14:545–554. doi: 10.1016/j.cmet.2011.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••16.Ng CY, Farasat I, Maranas CD, Salis HM. Rational design of a synthetic Entner-Doudoroff pathway for improved and controllable NADPH regeneration. Metab Eng. 2015;29:86–96. doi: 10.1016/j.ymben.2015.03.001. The authors used MAGE strain engineering to create computationally designed RBS variants of a synthetic Entner-Doudoroff pathway within E. coli and screened for NADPH production in microwell plates with a SFPB. This is the first published example of high-throughput screening using an SFPB. [DOI] [PubMed] [Google Scholar]
- 17.Nadler DC, Morgan S-A, Flamholz A, Kortright KE, Savage DF. Rapid construction of metabolite biosensors using domain-insertion profiling. Nat Comm. 2016;7:12266. doi: 10.1038/ncomms12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKeague M, Derosa MC. Challenges and opportunities for small molecule aptamer development. J Nucleic Acids. 2012;2012:748913. doi: 10.1155/2012/748913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michener JK, Smolke CD. High-throughput enzyme evolution in Saccharomyces cerevisiae using a synthetic RNA switch. Metab Eng. 2012;14:306–316. doi: 10.1016/j.ymben.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 21.Meyer A, Pellaux R, Potot S, Becker K, Hohmann H-P, Panke S, Held M. Optimization of a whole-cell biocatalyst by employing genetically encoded product sensors inside nanolitre reactors. Nat Chem. 2015;7:673–678. doi: 10.1038/nchem.2301. [DOI] [PubMed] [Google Scholar]
- 22.Lee S-W, Oh M-K. A synthetic suicide riboswitch for the high-throughput screening of metabolite production in Saccharomyces cerevisiae. Metab Eng. 2015;28:143–150. doi: 10.1016/j.ymben.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Gu MB, Mitchell RJ, Kim BC. Whole-cell-based biosensors for environmental biomonitoring and application. Adv Biochem Eng Biotechnol. 2004;87:269–305. doi: 10.1007/b13533. [DOI] [PubMed] [Google Scholar]
- 24.Dietrich JA, Shis DL, Alikhani A, Keasling JD. Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis. ACS Synth Biol. 2013;2:47–58. doi: 10.1021/sb300091d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li S, Si T, Wang M, Zhao H. Development of a Synthetic Malonyl-CoA Sensor in Saccharomyces cerevisiae for Intracellular Metabolite Monitoring and Genetic Screening. ACS Synth Biol. 2015;4:1308–1315. doi: 10.1021/acssynbio.5b00069. [DOI] [PubMed] [Google Scholar]
- 26.Raman S, Rogers JK, Taylor ND, Church GM. Evolution-guided optimization of biosynthetic pathways. Proc Natl Acad Sci USA. 2014;111:17803–17808. doi: 10.1073/pnas.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •27.Chou HH, Keasling JD. Programming adaptive control to evolve increased metabolite production. Nat Comm. 2013;4:2595. doi: 10.1038/ncomms3595. This paper used a synthetic-TF biosensor to repress the expression of a mutator (broken proof-reading exonuclease). This system increases mutations and explores genetic diversity while ligand levels were low, then ‘locks in’ the genotype as metabolite levels rise. [DOI] [PubMed] [Google Scholar]
- 28.Farmer WR, Liao JC. Improving lycopene production in Escherichia coli by engineering metabolic control. Nat Biotechnol. 2000;18:533–537. doi: 10.1038/75398. [DOI] [PubMed] [Google Scholar]
- 29.Scalcinati G, Partow S, Siewers V, Schalk M, Daviet L, Nielsen J. Combined metabolic engineering of precursor and co-factor supply to increase α-santalene production by Saccharomyces cerevisiae. Microb Cell Fact. 2012;11:117. doi: 10.1186/1475-2859-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F, Carothers JM, Keasling JD. Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nature Biotechnol. 2012;30:354–359. doi: 10.1038/nbt.2149. [DOI] [PubMed] [Google Scholar]
- ••31.Xu P, Li L, Zhang F, Stephanopoulos G, Koffas M. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc Natl Acad Sci USA. 2014;111:11299–11304. doi: 10.1073/pnas.1406401111. Using a TF, the authors engineered both up- and down-regulation of genes within a fatty acid pathway in response to malonyl-CoA concentration. This unique example of one TF providing both types of pathway control improved fatty acid production greater than threefold. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alon U. An Introduction to Systems Biology: Design Principles of Biological Circuits. 2007. [Google Scholar]
- 33.Erickson JP, Wu JJ, Goddard JG, Tigyi G, Kawanishi K, Tomei LD, Kiefer MC. Edg-2/Vzg-1 couples to the yeast pheromone response pathway selectively in response to lysophosphatidic acid. J Biol Chem. 1998;273:1506–1510. doi: 10.1074/jbc.273.3.1506. [DOI] [PubMed] [Google Scholar]
- 34.Radhika V, Proikas-Cezanne T, Jayaraman M, Onesime D, Ha JH, Dhanasekaran DN. Chemical sensing of DNT by engineered olfactory yeast strain. Nat Chem Bio. 2007;3:325–330. doi: 10.1038/nchembio882. [DOI] [PubMed] [Google Scholar]
- 35.Mukherjee K, Bhattacharyya S, Peralta-Yahya P. GPCR-Based Chemical Biosensors for Medium-Chain Fatty Acids. ACS Synth Bio. 2015;4:1261–1269. doi: 10.1021/sb500365m. [DOI] [PubMed] [Google Scholar]
- 36.Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- 37.Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM, et al. Synthetic biology: engineering Escherichia coli to see light. Nat Cell Biol. 2005;438:441–442. doi: 10.1038/nature04405. [DOI] [PubMed] [Google Scholar]
- 38.Baumgartner JW, Kim C, Brissette RE, Inouye M, Park C, Hazelbauer GL. Transmembrane signalling by a hybrid protein: communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J Bacteriol. 1994;176:1157–1163. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ninfa AJ. Use of two-component signal transduction systems in the construction of synthetic genetic networks. Curr Opin Microbiol. 2010;13:240–245. doi: 10.1016/j.mib.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganesh I, Ravikumar S, Yoo I-K, Hong SH. Construction of malate-sensing Escherichia coli by introduction of a novel chimeric two-component system. Bioprocess Biosyst Eng. 2015;38:797–804. doi: 10.1007/s00449-014-1321-3. [DOI] [PubMed] [Google Scholar]
- 41.Liu D, Xiao Y, Evans BS, Zhang F. Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor-actuator. ACS Synth Biol. 2015;4:132–140. doi: 10.1021/sb400158w. [DOI] [PubMed] [Google Scholar]
- 42.Voigt CA. Genetic parts to program bacteria. Curr Opin Biotechnol. 2006;17:548–557. doi: 10.1016/j.copbio.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 43.David F, Nielsen J, Siewers V. Flux Control at the Malonyl-CoA Node through Hierarchical Dynamic Pathway Regulation in Saccharomyces cerevisiae. ACS Synth Biol. 2016;5:224–233. doi: 10.1021/acssynbio.5b00161. [DOI] [PubMed] [Google Scholar]
- 44.Siedler S, Schendzielorz G, Binder S, Eggeling L, Bringer S, Bott M. SoxR as a single-cell biosensor for NADPH-consuming enzymes in Escherichia coli. ACS Synth Biol. 2014;3:41–47. doi: 10.1021/sb400110j. [DOI] [PubMed] [Google Scholar]
- 45.Feng J, Jester BW, Tinberg CE, Mandell DJ, Antunes MS, Chari R, Morey KJ, Rios X, Medford JI, Church GM, et al. A general strategy to construct small molecule biosensors in eukaryotes. eLife. 2015:4. doi: 10.7554/eLife.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos CNS, Stephanopoulos G. Melanin-Based High-Throughput Screen for L-Tyrosine Production in Escherichia coli. Appl Environ Microbiol. 2008;74:1190–1197. doi: 10.1128/AEM.02448-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •47.DeLoache WC, Russ ZN, Narcross L, Gonzales AM, Martin VJJ, Dueber JE. An enzyme-coupled biosensor enables (S)-reticuline production in yeast from glucose. Nat Chem Biol. 2015;11:465–471. doi: 10.1038/nchembio.1816. This paper developed an enzyme-based biosensor for L-DOPA, transforming it into a fluorescent pigment for efficient screening of tyrosine to L-DOPA conversion. This approach shows future promise for detecting biofuel-related molecules as pathways toward other fluorescent products, such as polyenes, are elucidated. [DOI] [PubMed] [Google Scholar]
- 48.Rogers JK, Church GM. Genetically encoded sensors enable real-time observation of metabolite production. Proc Natl Acad Sci USA. 2016;113:2388–2393. doi: 10.1073/pnas.1600375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •49.Delépine B, Libis V, Carbonell P, Faulon J-L. SensiPath: computer-aided design of sensing-enabling metabolic pathways. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw305. This work reports SensiPath ( http://sensipath.micalis.fr), an informative, web-based tool for the computer-aided design of metabolic pathways that can convert a non-detectable target metabolite to one that can be detected. [DOI] [PMC free article] [PubMed]
- 50.Ferrell JE, Tsai TY-C, Yang Q. Modeling the cell cycle: why do certain circuits oscillate? Cell. 2011;144:874–885. doi: 10.1016/j.cell.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 51.Lindsley JE, Rutter J. Whence cometh the allosterome? Proc Natl Acad Sci USA. 2006;103:10533–10535. doi: 10.1073/pnas.0604452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••52.Vetting MW, Al-Obaidi N, Zhao S, San Francisco B, Kim J, Wichelecki DJ, Bouvier JT, Solbiati JO, Vu H, Zhang X, et al. Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes. Biochem. 2015;54:909–931. doi: 10.1021/bi501388y. This paper used computational and high-throughput experimental approaches to determine ligands and functional annotations for members of a family of solute binding proteins. Such approaches may be useful in mining novel LBDs from putative protein sequences found in genome and metagenome sequencing projects. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uchiyama T, Abe T, Ikemura T, Watanabe K. Substrate-induced gene-expression screening of environmental metagenome libraries for isolation of catabolic genes. Nat Biotechnol. 2005;23:88–93. doi: 10.1038/nbt1048. [DOI] [PubMed] [Google Scholar]
- 54.Tinberg CE, Khare SD, Dou J, Doyle L, Nelson JW, Schena A, Jankowski W, Kalodimos CG, Johnsson K, Stoddard BL, et al. Computational design of ligand-binding proteins with high affinity and selectivity. Nature. 2013;501:212–216. doi: 10.1038/nature12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor ND, Garruss AS, Moretti R, Chan S, Arbing MA, Cascio D, Rogers JK, Isaacs FJ, Kosuri S, Baker D, et al. Engineering an allosteric transcription factor to respond to new ligands. Nat Meth. 2015;13:177–183. doi: 10.1038/nmeth.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deuschle K, Chaudhuri B, Okumoto S, Lager I, Lalonde S, Frommer WB. Rapid metabolism of glucose detected with FRET glucose nanosensors in epidermal cells and intact roots of Arabidopsis RNA-silencing mutants. Plant Cell. 2006;18:2314–2325. doi: 10.1105/tpc.106.044073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guntas G, Mansell TJ, Kim JR, Ostermeier M. Directed evolution of protein switches and their application to the creation of ligand-binding proteins. Proc Natl Acad Sci. 2005;102:11224–11229. doi: 10.1073/pnas.0502673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Oakes BL, Nadler DC, Flamholz A, Fellmann C, Staahl BT, Doudna JA, Savage DF. Profiling of engineering hotspots identifies an allosteric CRISPR-Cas9 switch. Nat Biotechnol. 2016;34:646–651. doi: 10.1038/nbt.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reynolds KA, McLaughlin RN, Ranganathan R. Hot spots for allosteric regulation on protein surfaces. Cell. 2011;147:1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mongeon R, Venkatachalam V, Yellen G. Cytosolic NADH-NAD(+) Redox Visualized in Brain Slices by Two-Photon Fluorescence Lifetime Biosensor Imaging. Antioxid Redox Signal. 2016 doi: 10.1089/ars.2015.6593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang D, He Y, Wang Y, Wang H, Wu L, Aries E, Huang WE. Whole-cell bacterial bioreporter for actively searching and sensing of alkanes and oil spills. Microbial Biotech. 2012;5:87–97. doi: 10.1111/j.1751-7915.2011.00301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sticher P, Jaspers MC, Stemmler K, Harms H, Zehnder AJ, van der Meer JR. Development and characterization of a whole-cell bioluminescent sensor for bioavailable middle-chain alkanes in contaminated groundwater samples. Appl Environ Microbiol. 1997;63:4053–4060. doi: 10.1128/aem.63.10.4053-4060.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang S-Y, Cirino PC. Design and application of a mevalonate-responsive regulatory protein. Angew Chem Int Ed Engl. 2011;50:1084–1086. doi: 10.1002/anie.201006083. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y-M, Rock CO. A rainbow coalition of lipid transcriptional regulators. Mol Microbiol. 2010;78:5–8. [PMC free article] [PubMed] [Google Scholar]
- 65.Imamura H, Nhat KPH, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci USA. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.San Martín A, Ceballo S, Ruminot I, Lerchundi R, Frommer WB, Barros LF. A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells. PLoS ONE. 2013;8:e57712. doi: 10.1371/journal.pone.0057712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fehr M, Lalonde S, Lager I, Wolff MW, Frommer WB. In vivo imaging of the dynamics of glucose uptake in the cytosol of COS-7 cells by fluorescent nanosensors. J Biol Chem. 2003;278:19127–19133. doi: 10.1074/jbc.M301333200. [DOI] [PubMed] [Google Scholar]
- 68.San Martín A, Ceballo S, Baeza-Lehnert F, Lerchundi R, Valdebenito R, Contreras-Baeza Y, Alegría K, Barros LF. Imaging mitochondrial flux in single cells with a FRET sensor for pyruvate. PLoS ONE. 2014;9:e85780. doi: 10.1371/journal.pone.0085780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dahl RH, Zhang F, Alonso-Gutierrez J, Baidoo E, Batth TS, Redding-Johanson AM, Petzold CJ, Mukhopadhyay A, Lee TS, Adams PD, et al. Engineering dynamic pathway regulation using stress-response promoters. Nat Biotechnol. 2013;31:1039–1046. doi: 10.1038/nbt.2689. [DOI] [PubMed] [Google Scholar]
- 70.Knudsen JD, Carlquist M, Gorwa-Grauslund M. NADH-dependent biosensor in Saccharomyces cerevisiae: principle and validation at the single cell level. AMB Express. 2014;4:81. doi: 10.1186/s13568-014-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scalcinati G, Knuf C, Partow S, Chen Y, Maury J, Schalk M, Daviet L, Nielsen J, Siewers V. Dynamic control of gene expression in Saccharomyces cerevisiae engineered for the production of plant sesquitepene α-santalene in a fed-batch mode. Metab Eng. 2012;14:91–103. doi: 10.1016/j.ymben.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 72.Østergaard H, Henriksen A, Hansen FG, Winther JR. Shedding light on disulfide bond formation: engineering a redox switch in green fluorescent protein. EMBO J. 2001;20:5853–5862. doi: 10.1093/emboj/20.21.5853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Akiyama N, Takeda K, Miki K. Crystal Structure of a Periplasmic Substrate-Binding Protein in Complex with Calcium Lactate. J Mol Biol. 2009;392:559–565. doi: 10.1016/j.jmb.2009.07.043. [DOI] [PubMed] [Google Scholar]