Abstract
Circulating tumor cells (CTCs) in women with advanced estrogen receptor-positive/HER2-negative breast cancer acquire a HER2-positive subpopulation following multiple courses of therapy1,2. In contrast to HER2-amplified primary breast cancer, which is highly sensitive to HER2-targeted therapy, the clinical significance of acquired HER2 heterogeneity during the evolution of metastatic breast cancer is unknown. Here, we analyzed CTCs from 19 ER+/HER2− patients, 84% of whom had acquired CTCs expressing HER2. Cultured CTCs maintain discrete HER2+ and HER2− subpopulations: HER2+ CTCs are more proliferative but not addicted to HER2, consistent with activation of multiple signaling pathways. HER2− CTCs show activation of Notch and DNA damage pathways, exhibiting resistance to cytotoxic chemotherapy, but sensitivity to Notch inhibition. HER2+ and HER2− CTCs interconvert spontaneously, with cells of one phenotype producing daughters of the opposite within four cell doublings. While HER2+ and HER2− CTCs have comparable tumor initiating potential, differential proliferation favors the HER2+ state, while oxidative stress or cytotoxic chemotherapy enhances transition to the HER2− phenotype. Simultaneous treatment with paclitaxel and Notch inhibitors achieves sustained suppression of tumorigenesis in orthotopic CTC-derived tumor models. Together, these results point to distinct yet interconverting phenotypes within patient-derived CTCs, contributing to progression of breast cancer and acquisition of drug resistance.
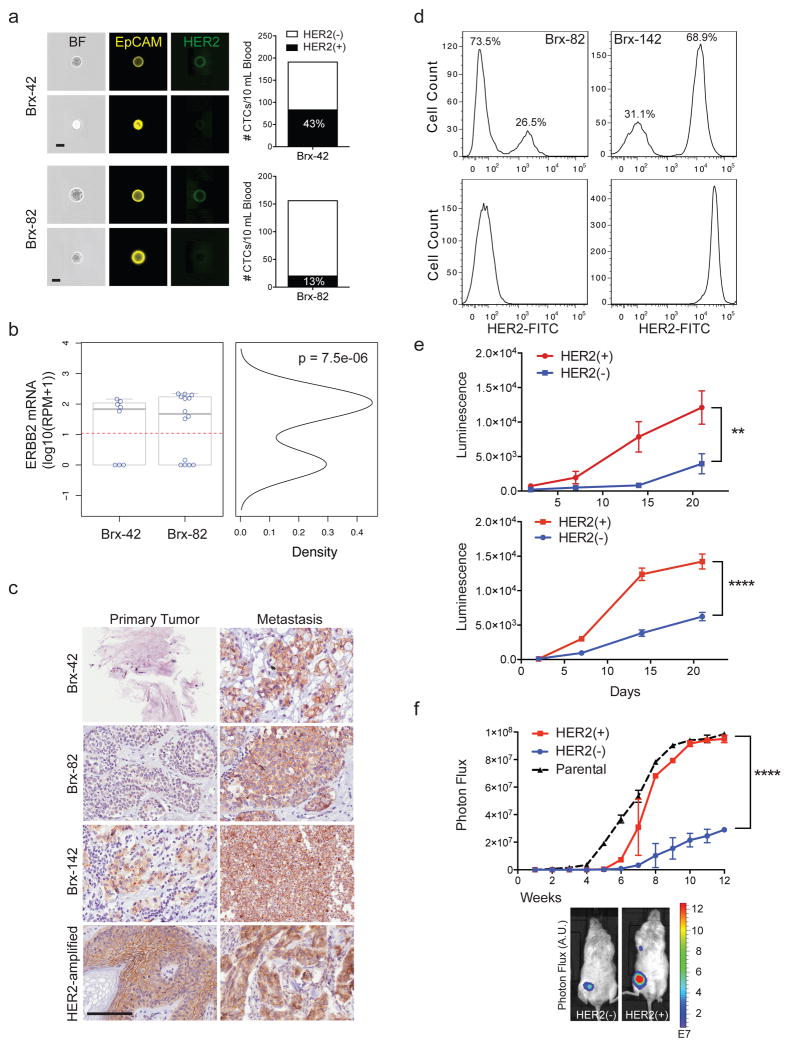
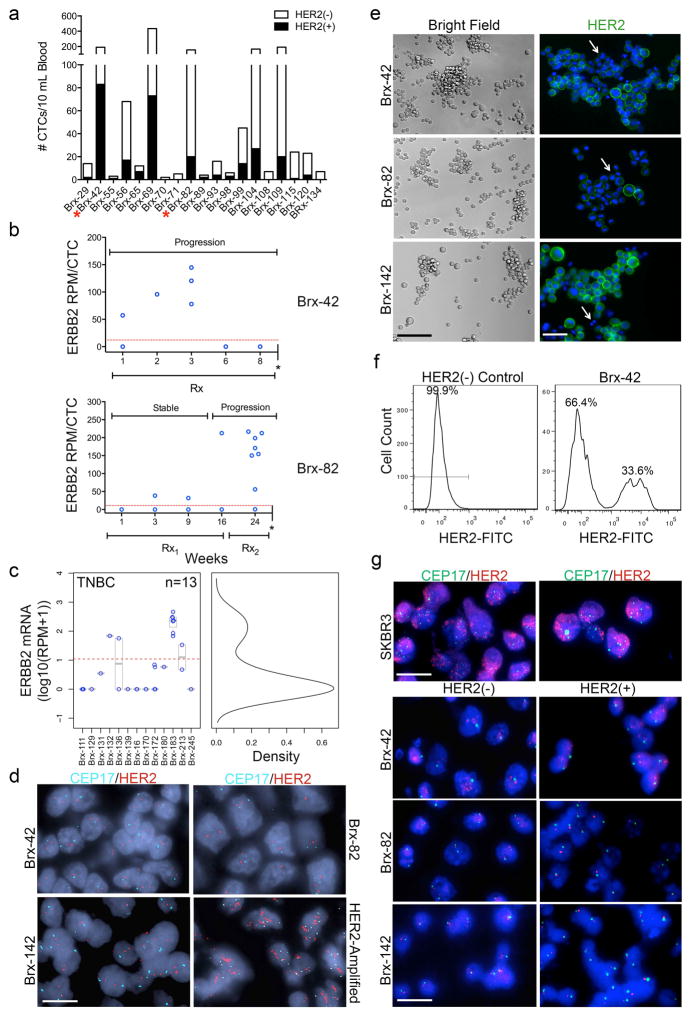
We documented the emergence of HER2+ CTCs in patients initially diagnosed with estrogen receptor-positive/HER2-negative (ER+/HER2−) breast cancer, after multiple courses of therapy for recurrent metastatic breast cancer. Using microfluidic CTC-iChip purification followed by imaging flow cytometry3, 16/19 (84%) patients had HER2+ CTCs (Fig. 1a, Extended Data Fig. 1a and Supplemental Table 1). 22 individual CTCs from two representative patients (Brx-42, Brx-82) were isolated and subjected to single-cell RNA-sequencing (scRNA-seq). HER2 expression was bimodal in distribution (≤ 1 read per million (RPM) vs median 133, range 32–217 RPM; p = 7.5e-6) (Fig. 1b), indicating the existence of discrete HER2+ and HER2− subpopulations. In these patients, the fraction of HER2+ CTCs increased with disease progression (Extended Data Fig. 1b). HER2+ CTCs were not restricted to ER+/HER2− breast cancer: 2/13 patients with ER−/PR−/HER2− breast cancer also had HER2+ and HER2− CTC subpopulations (Extended Data Fig. 1c). In ER+/HER2− breast cancers, immunohistochemical (IHC) staining of patient-matched metastatic tumor biopsies showed increased HER2+ staining, compared with primary tumors (Fig. 1c). Unlike HER2-amplified breast cancer, HER2+ tumor cells within metastatic lesions did not have evidence of gene amplification (Extended Data Fig. 1d).
Figure 1. Distinct properties of HER2+ and HER2− CTC subpopulations from patients with advanced ER+/HER2− breast cancer.
(a) Quantitation by imaging flow cytometry of HER2+ and HER2− CTCs isolated from patients Brx-42, Brx-82. EpCAM (yellow) and HER2 (green). Scale bar: 10 μm. (b) Bimodal distribution of ERBB2 RNA-seq reads from single CTCs, Hartigans’ dip test p = 7.5e-6, n=22 (HER2− ≤ 1 read per million (RPM); HER2+ > 133, range 32–217). (c) IHC for HER2 (brown) in matched metastatic vs primary tumors (Brx-42, Brx-82, Brx-142) compared to HER2-amplified tumor (control). Scale bar: 100 μm; Tumor data (Supplemental Table 1). (d) FACS of cultured CTCs, showing discrete HER2+ and HER2− subpopulations. MDA-231 (TNBC) and SKBR3 (HER2-amplified) cells are shown as control. (e) Differential proliferation of FACS-purified HER2+ (red) and HER2− (blue) subpopulations from cultured CTCs; Two-way ANOVA p < 0.01 (Brx-82), p < 0.0001 (Brx-142); n=6; S.D. (error bar). (f) Increased in vivo growth of orthotopic mammary tumors derived from FACS-purified, HER2+ CTCs compared with HER2- cells; n=8; Two-way ANOVA p < 0.0001; S.D. (error bar).
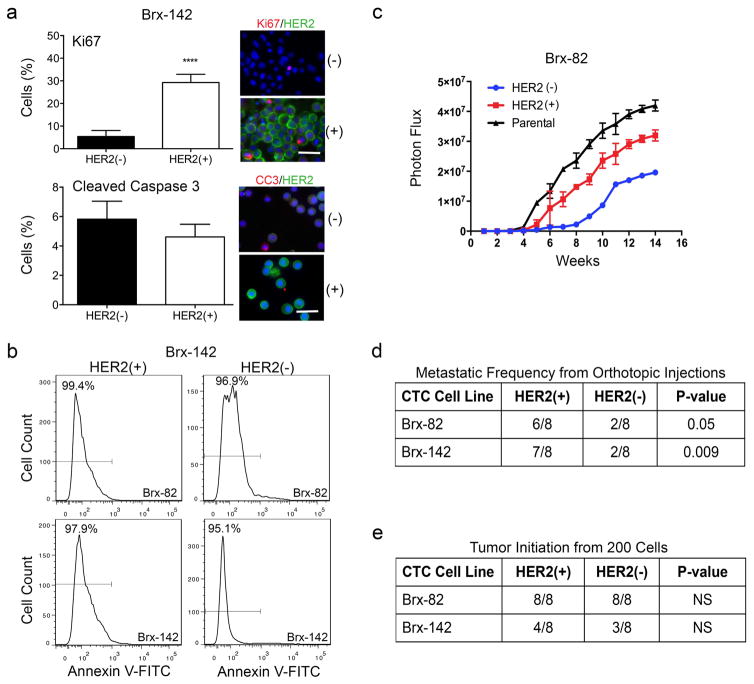
The CTC-iChip efficiently captures viable CTCs3, enabling derivation of CTC cultures4. We established CTC lines (Brx-42, Brx-82, Brx-142) with discrete HER2+/HER2− subpopulations comparable to patient-matched primary CTCs (Fig. 1a, d and Extended Data Fig. 1e, f). HER2 overexpression was not due to gene amplification, and no distinguishing mutations were identified between HER2+ and HER2− subpopulations (Extended Data Fig. 1g and Supplemental Table 2). Fluorescence-activated cell sorting (FACS) of HER2+ vs HER2− subpopulations showed distinct functional properties: HER2+ CTCs had a higher proliferation rate (Fig. 1e), with increased staining for the proliferation marker Ki67, but no change in apoptotic markers cleaved-caspase 3 or annexin 5 (Extended Data Fig. 2a, b).
We tested the relative tumorigenicity of HER2+ vs HER2− CTCs following injection into the mouse mammary fat pad. Both FACS-purified HER2+ and HER2− CTCs generated tumors, with HER2+ tumors being larger and having a higher frequency of lung metastases (Fig. 1f and Extended Data Fig. 2c, d). Despite differences in proliferation, limiting dilution studies showed that HER2+ and HER2− CTCs initiate tumors from as few as 200 cells, pointing to comparable progenitor potential (Extended Data Fig. 2e).
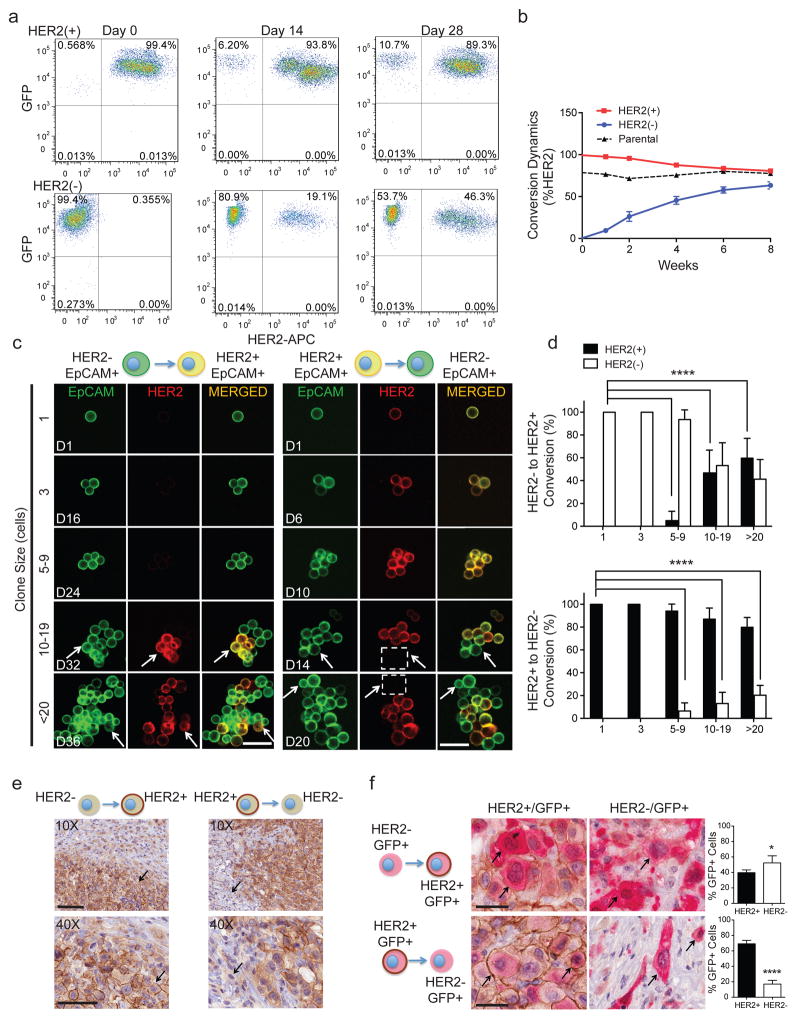
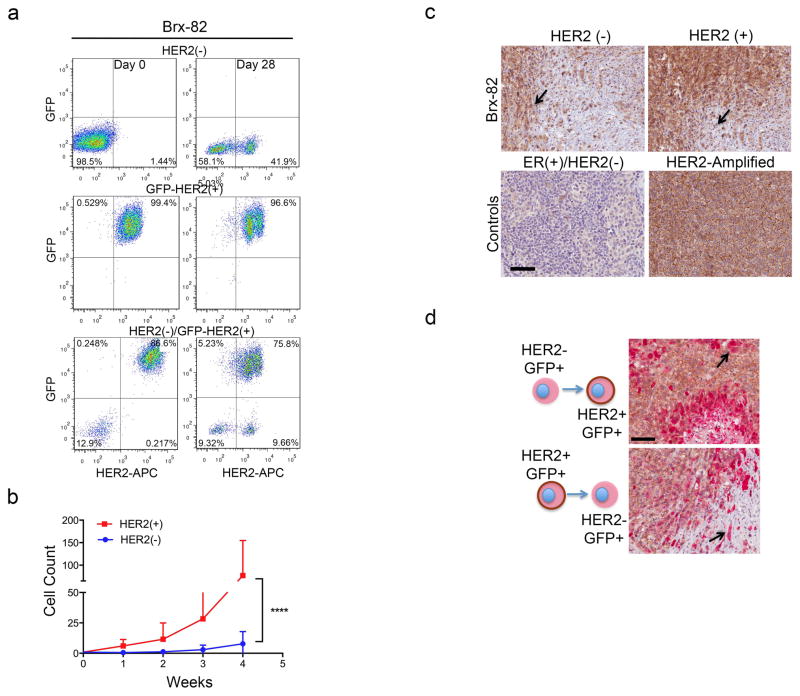
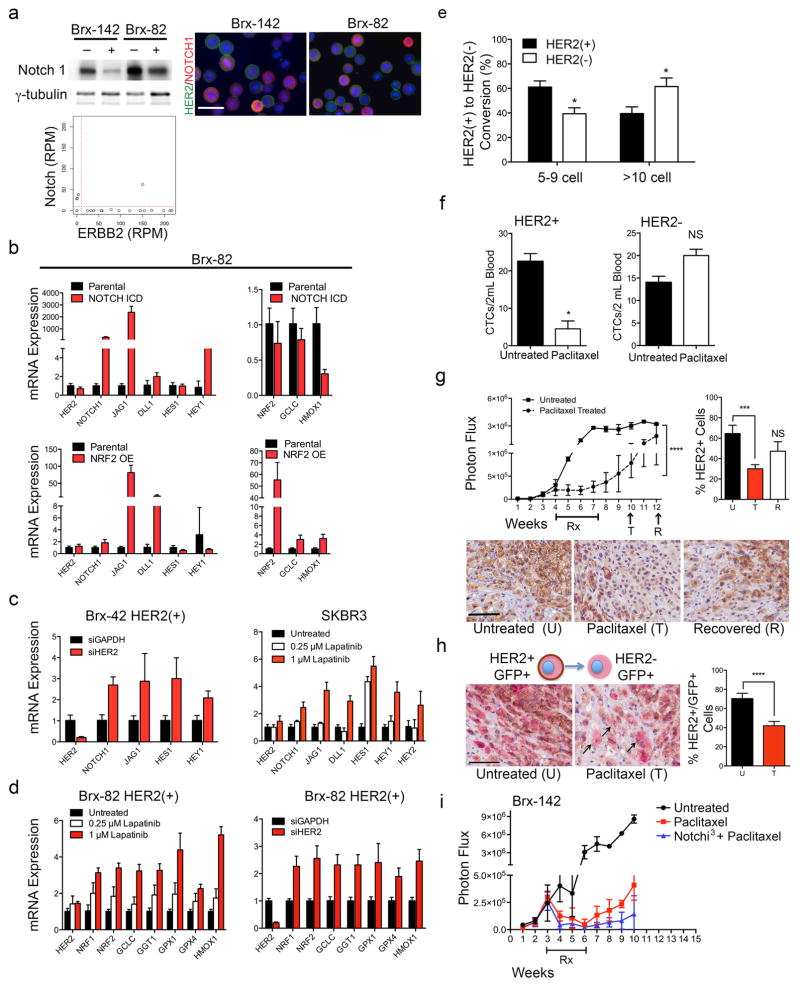
The coexistence of HER2+ and HER2− CTCs, despite differing proliferation rates, led us to test whether these subpopulations are capable of interconversion. After 4 weeks in culture, FACS-purified GFP-tagged HER2− CTCs acquired HER2+ cells (Brx-82: 42%; Brx-142: 46%), while HER2+ CTCs generated HER2− cells at lower efficiency (Brx-82: 5%; Brx-142: 11%) (Fig. 2a, b and Extended Data Fig. 3a). By 8 weeks, the parental HER2+/HER2− composition was nearly reestablished (Fig. 2b). This interconversion was also evident by mixing equal proportions of GFP+/HER2+ and GFP−/HER2− CTCs, with the emergence of GFP+/HER2− and GFP−/HER2+ cells, respectively (Extended Data Fig. 3a).
Figure 2. Interconversion of HER2+ and HER2− phenotypes.
(a) FACS-purified GFP-tagged HER2+ and HER2− CTCs generate HER2− (upper panels) and HER2+ cells (lower panels), respectively. (b) Time-course of HER2+/HER2− interconversion following FACS-isolation of HER2+ (red) and HER2− (blue) cells; n=3; S.D. (error bar). Parental cultured CTCs (black dotted) are shown as control. (c) Representative confocal microscopic images depicting HER2+/HER2− interconversion within single cell-derived clones at indicated timepoints (D=days) and colony sizes. EpCAM (green), HER2 (red) and MERGED (gold). Scale bar: 20 μm; n=20. Arrows and dashed boxes indicate interconverting cells, with loss/gain of HER2. (d) Quantitation of HER2+/HER2− interconversion from single cell-derived colonies at each colony size; T-test p-value < 0.0001; n=20; S.D. (error bar). (e) Gain/loss of HER2+ cells (brown, arrow) in tumor xenografts derived from purified HER2− (left)/HER2+ (right) CTCs. Scale bar: 100 μm (upper panels); 50 μm (lower panels); n=8. (f) IHC imaging and quantitation of GFP+/HER2+ cells within tumors generated from GFP-tagged/HER2− and untagged HER2+ CTCs (upper panels), and the converse (lower panels). GFP: cytoplasmic red, HER2: membrane brown. Scale bar: 20 μm; T-test p-value <0.05 (*), p-value < 0.0001 (****); n=8; S.D. (error bar).
To better define the timing of HER2+/HER2− interconversion, we established, single cell-derived CTC colonies using HER2-based FACS, followed by sequential confocal microscopy. Colonies were scored for HER2 and EpCAM expression at 1, 3, 5–9, 10–19 and >20 cell stages. Single HER2− CTCs initially proliferated slowly (Extended Data Fig. 3b), and first acquired HER2+ daughter cells at the 5–9 cell stage (6.5%), with rapid interconversion thereafter (10–19 cells: 47%, >20 cells: 59%; Fig. 2c, d). The more rapidly proliferating single HER2+ CTCs also generated HER2− progeny at the 5–9 cell stage (5%), but the proportion of HER2− CTCs rose more slowly (10–19 cells: 17%, >20 cells: 22%; Fig. 2c, d). Thus, interconversion between HER2+/HER2− phenotypes occurs spontaneously as early as four cell doublings.
Interconversion between HER2+ and HER2− phenotypes was also tested in vivo by orthotopic inoculation of FACS-purified CTCs. Tumors established from HER2− CTCs displayed HER2+ subpopulations, and vice versa (Fig. 2e and Extended Data Fig. 3c). In vivo interconversion was confirmed by injecting a 1:1 mixture of GFP+/HER2+ and GFP−/HER2− CTCs (or the converse), followed by dual GFP and HER2 IHC. Within mixed tumors, GFP-tagged HER2− CTCs produced GFP+/HER2+ cells (44%), and in separate tumors, GFP-tagged HER2+ CTCs generated HER2− cells (21%) (Fig. 2f and Extended Data Fig. 3d).
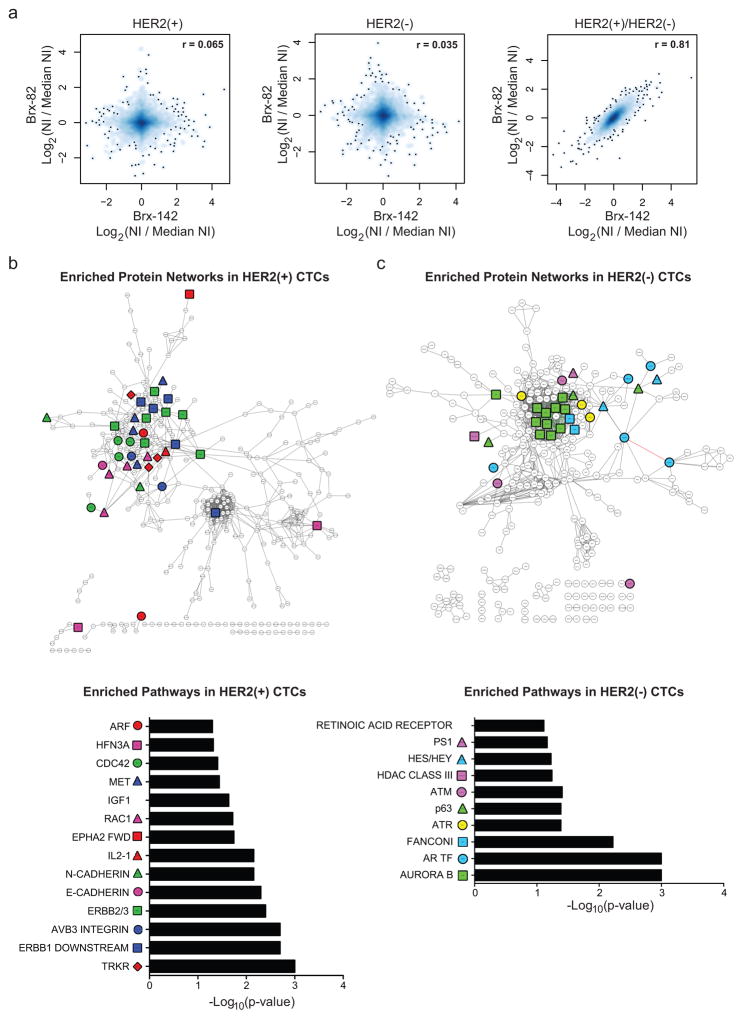
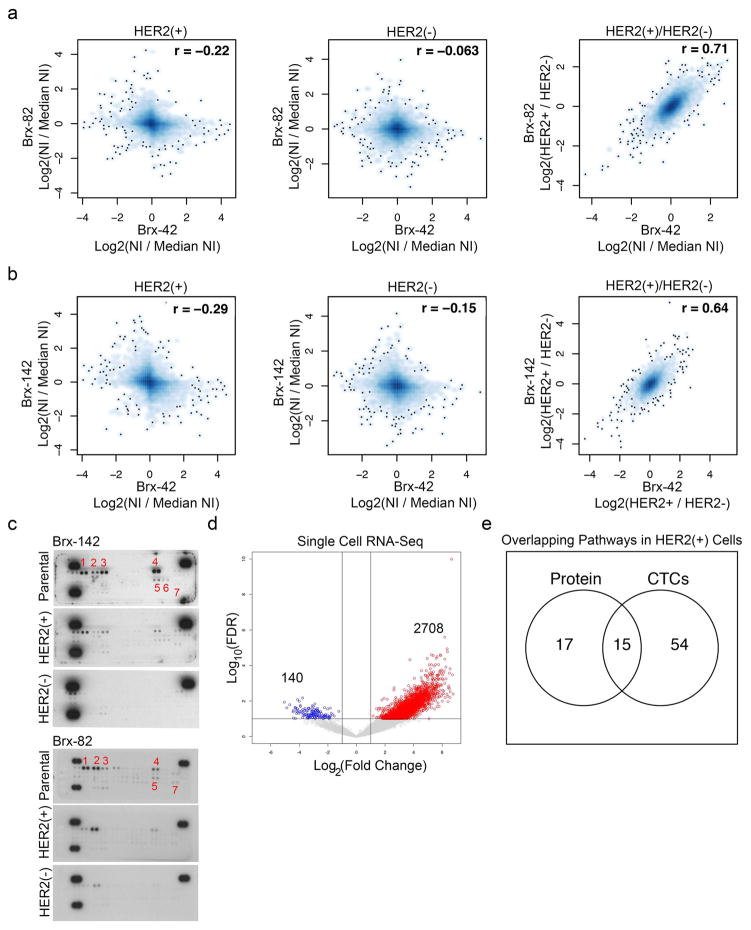
To define the molecular characteristics of HER2+ vs HER2− CTCs, we quantitatively mapped the global proteomes (>6300 proteins) of FACS-purified subpopulations (Brx-42, Brx-82, Brx-142) using multiplexed mass spectrometry (MS) with isobaric tandem mass tags (TMT)5 (Supplemental Table 3). While proteome profiles of individual cell lines were distinct, they shared, differences between HER2+ and HER2− subpopulations (Pearson correlation coefficients: Brx-82 vs. Brx-142=0.81; Brx-82 vs. Brx-42=0.71; Brx-42 vs. Brx-142=0.64) (Fig. 3a and Extended Data Fig. 4a, b). HER2+ CTCs showed enrichment (Pathway Interaction Database; PID) of receptor tyrosine kinase (RTK) and pro-growth signaling (GSEA, FDR ≤ 0.25) (Fig. 3b and Supplemental Tables 3, 4). Phosphotyrosine blots from the HER2+ subpopulations confirmed RTK phosphorylation (HER2, HER3, HER4, Insulin Receptor (INSR), AXL/DTK, EPHA1 and EPHA2), which was absent from matched HER2− CTCs (Extended Data Fig. 4c). scRNA-seq analysis of 15 primary HER2+ CTCs compared with 7 HER2− CTCs from matched patient blood samples showed enrichment for 15 of 32 shared pathways (ERBB1, ERBB2/ERBB3, IGF1, EPHA2, MET) identified by MS analysis of CTC lines (Fig. 3b, Extended Data Fig. 4d, e and Supplemental Tables 4, 5). In contrast to HER2+ CTCs, MS analysis of cultured HER2− CTCs showed increased expression of proteins enriched in Notch (HES/HEY, Presenilin 1 (PS1)) and DNA damage pathways (AuroraB, ATM, ATR, Fanconi) (GSEA, FDR ≤ 0.25) (Fig. 3c and Supplemental Tables 3, 4).
Figure 3. Molecular pathways differentially activated in HER2− versus HER2+ cultured CTCs.
(a) Comparison of quantitative MS proteomes (6349 proteins) showing distinct profiles for individual cultured CTCs (Brx-82, Brx-142), but linear correlation between proteins differentially expressed in HER2+ and HER2− subpopulations; NI = Normalized Intensity; n=2 biological replicates per CTC line Brx-42, Brx-82, Brx-142 (Supplemental Table 3). (b–c) Cytoscape network maps (upper panels) and GSEA pathway analysis (lower panels) depicting proteins enriched by greater than log2(0.5) by quantitative MS in (b) HER2+ and (c) HER2− CTCs (GSEA FDR ≤ 0.25; Nominal p-value cut-off < 0.05; Supplemental Table 4). Colored shapes represent proteins within denoted pathways. Red asterisks highlight RTK pathways in (b) and Notch pathways in (c).
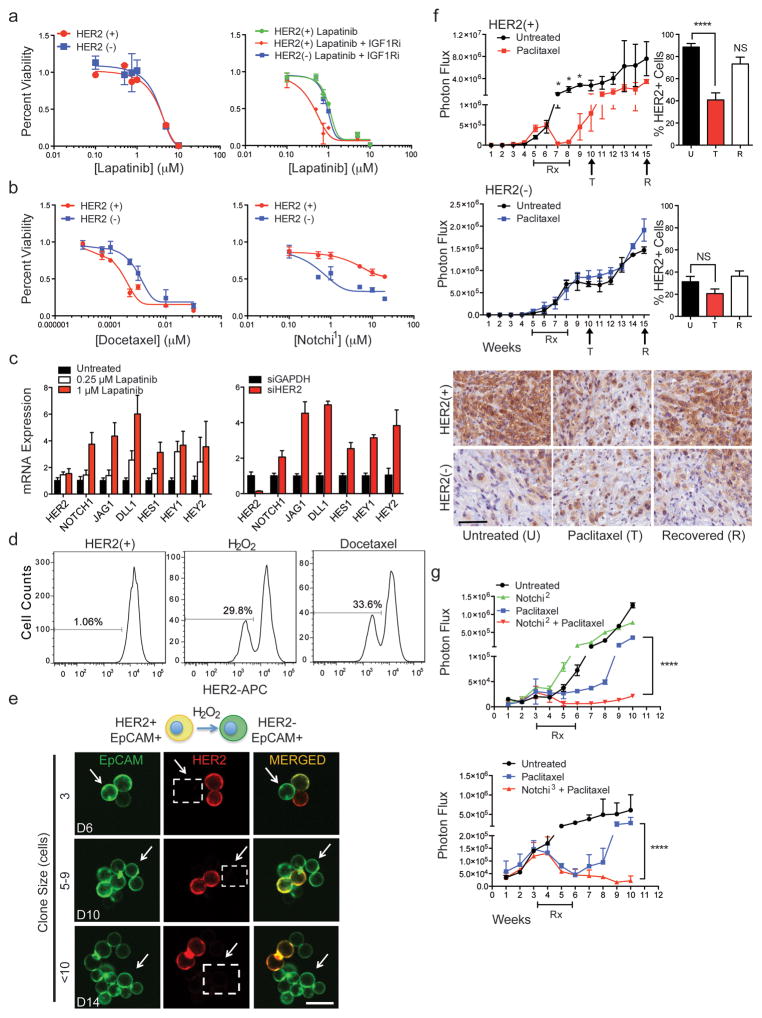
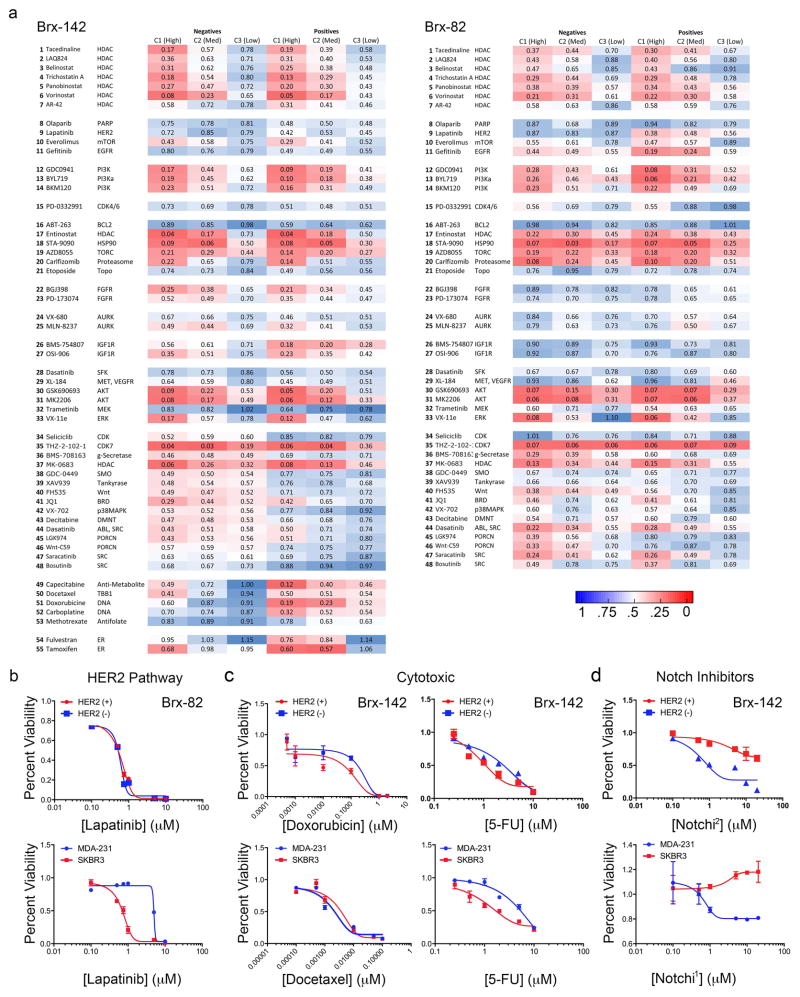
To explore the potential therapeutic significance of pathways differentially activated in HER2+ vs HER2− CTC subpopulations, we screened a panel of 55 drugs selected both for clinical relevance and ability to target MS-identified pathways (Supplemental Table 6). HER2+ CTCs were no more sensitive to the HER2 inhibitor lapatinib than HER2− CTCs (IC50 = 1μM), indicating they are not “oncogene addicted” to HER2, unlike the HER2-amplified SKBR3 cells (IC50=5 nM) (Fig. 4a, Extended Data Fig. 5a, b). However, dual inhibition of HER2 and IGF1R, another RTK activated in HER2+ CTCs, was cytotoxic to HER2+ but not HER2− CTCs (Fig. 4a), suggesting inhibition of multiple kinases may be effective in treating HER2+ CTCs. Compared with HER2+ CTCs, HER2− CTCs showed reduced sensitivity to chemotherapeutic agents docetaxel, doxorubicin and 5-fluorouracil (5-FU) (Fig. 4b and Extended Data Fig. 5a, c), but increased sensitivity to γ-secretase inhibitors, which suppress Notch activity (Fig. 4b and Extended Data Fig. 5a, d). Despite proteomic enrichment for Aurora B signaling, HER2− CTCs were not differentially sensitive to Aurora family inhibitors (Fig. 3c, Extended Data Fig. 5a and Supplemental Tables 3, 4).
Figure 4. Cooperative targeting of HER2+ and HER2− CTC subpopulations suppresses tumor growth.
(a) HER2+ CTCs show no change in sensitivity to lapatinib alone, compared with matched HER2− CTCs (Brx-142), but have increased sensitivity to combined HER2 and IGF1R (BMS-754807) inhibitors; n=6; S.D (error bar). (b) HER2− CTCs demonstrate reduced chemosensitivity (docetaxel) but have enhanced sensitivity to Notch inhibition (BMS-708163, Notchi1), compared with HER2+ CTCs; n=6; S.D. (error bar). (c) Inhibition of HER2 with lapatinib or siRNA-mediated knockdown in HER2+ CTCs (Brx-82) results in dose-dependent increase of Notch-related genes: NOTCH1, JAG1, DLL1, HES1, HEY1, HEY2; p <0.05; n=6; S.E.M (error bar). (d) Rapid emergence (96 hrs) of HER2− CTCs following treatment of HER2+ CTCs with H2O2 (10 mM) or docetaxel (1 nM). (e) Confocal microscopy showing rapid appearance of HER2− progeny (3 cell) from single-CTC derived HER2+ colonies treated with H2O2. EpCAM (green), HER2 (red) and MERGED (gold). Scale bar: 20 μm; n=10. Arrows and dashed boxes indicate cells with loss of HER2 at indicated timepoints (D=days). (f) Paclitaxel treatment (4 weeks) of mice with CTC-derived (Brx-142) orthotopic mammary tumors. Upper panel: HER2+/HER2− tumor growth curves with paclitaxel treatment; Lower panels: Representative IHC for HER2 (brown) in HER2+ and HER2− derived tumors at the U (untreated), T (2-weeks post-treatment) and R (7-weeks post-treatment) timepoints. Scale bar: 100 μm; p-value T-test < 0.05 n=8; p-value < 0.05. (g) Simultaneous treatment (4 weeks) of mammary xenografts (Brx-82) with paclitaxel and either Notch inhibitor RO4929097 (Notchi2) or LY-411575 (Notchi3), showing sustained responses for the combination, compared with paclitaxel alone. Rx denotes treatment duration; Two-way ANOVA p-value < 0.0001 n=8.
The increased NOTCH1 in HER2− CTCs observed by quantitative MS and confirmed by western blot (Extended Data Fig. 6a and Supplemental Tables 3, 4) was inversely correlated with HER2 expression within primary CTCs and CTC lines, shown by scRNA-seq and immunostaining (Extended Data Fig. 6a). We therefore tested the consequences of suppressing HER2 or activating Notch signaling in HER2+ CTCs.
Manipulation of NOTCH1 or its downstream effector NFE2L2/NRF2 in cultured HER2+ CTCs did not reduce HER2 expression (Extended Data Fig. 6b). However, inhibition of HER2 using lapatinib or siRNA led to increased expression of NOTCH1, its ligands JAG1 and DLL1, and Notch-regulated genes HES1, HEY1 and HEY2 (Fig. 4c and Extended Data Fig. 6c), confirming previous reports from HER2-amplified breast cancer cells6,7 (Extended Data Fig. 6c). Suppression of HER2 also resulted in increased expression of genes (GCLC, GGT1, GPX1, GPX4, HMOX1) downstream of Notch-regulated NRF2, a transcriptional regulator of anti-oxidant/glutathione metabolism pathways8,9 (Extended Data Fig. 6d). Thus, expression of HER2 in CTCs appears to mediate downregulation of the NOTCH1/NRF2 axis, potentially switching between proliferative and survival-prone phenotypes.
In addition to suppressing HER2 directly, we tested additional stimuli capable of modulating the HER2+/HER2− interconversion. Treatment of HER2+ CTCs with low doses of docetaxel (1 nM) or induction of oxidative stress with hydrogen peroxide (H2O2; 10 mM) induced rapid shifts from HER2+ to HER2− (30% conversion, >70% survival) (Fig. 4d). To exclude differential cell death, we demonstrated acceleration in the appearance of HER2− progeny from FACS-purified single HER2+ CTCs (5–9 cells: 45%, >10 cells: 62%) (Fig. 4e and Extended Data Fig. 6e). Thus, exposure to cytotoxic/oxidative stress mediates a switch to a less proliferative but more drug resistant phenotype.
To model the potential significance of HER2+/HER2− interconversion in vivo, we generated orthotopic mammary xenografts from FACS-purified subpopulations and analyzed tumors before and after treatment with paclitaxel. Purified HER2+ CTCs generated mixed tumors (88% HER2+, 12% HER2−) and showed dramatic tumor shrinkage following paclitaxel treatment. The recurrent tumor showed a transient reduction in HER2+ with a corresponding increase in HER2− composition following chemotherapy (2-weeks: 39% HER2+; 7-weeks: 74% HER2+; Figure 4f). Purified HER2− CTCs also gave rise to a mixed tumor (35% HER2+, 65% HER2−), but paclitaxel induced only a limited delay in tumor growth with a minimal effect on HER2 content. Shedding of CTCs was also suppressed by paclitaxel in HER2+ but not HER2− tumors (Extended Data Fig. 6f). The chemotherapy-induced shift in HER2 composition was also evident following inoculation of parental CTC cultures (Untreated 65% HER2+; Post-therapy 30% HER2+; Extended Data Fig. 6g). Finally, we generated tumors from a 1:1 mixture of GFP-tagged HER2+ and untagged HER2− cells, demonstrating a shift from GFP+/HER2+ to GFP+/HER2− cells following paclitaxel treatment (Untreated: 70% GFP+/HER2+, Post-therapy: 42% GFP+/HER2+; Extended Data Fig. 6h). The potent effect of chemotherapy on HER2+/HER2− phenotypes in vivo may reflect both reduced drug-sensitivity of HER2− cells, as well as stress-induced HER2+ to HER2− switching.
Given the demonstrated susceptibility of HER2− CTCs to Notch inhibitors, we combined paclitaxel with either of two γ-secretase inhibitors (LY-411575; RO4929097) in treating mice with tumors initiated from parental CTC lines. Compared with paclitaxel alone, the combination therapy significantly delayed onset of tumor recurrence, while Notch inhibition alone had no effect on tumor growth (Fig. 4g and Extended Data Fig. 6f).
Taken together, we have used primary and cultured CTCs from ER+/HER2− breast cancer patients who developed metastatic multidrug resistant disease to show that coexisting distinct HER2+ and HER2− tumor cell subpopulations may interconvert, with striking consequences for disease progression and drug response. The comparable tumor initiating potential and similar expression of stem cell marker ALDH1 in HER2+ and HER2− CTCs suggests underlying tumor cell plasticity in these advanced patient-derived breast CTC lines, rather than a hierarchical cancer stem cell model as described in drug resistant subpopulations within established breast cancer cell lines7,10–15. While expression of NOTCH1 and other embryonic markers has been reported in rare, quiescent cells within primary breast tumors7,16,17,18, the NOTCH1+ CTCs reported here constitute a major cell population, exhibiting both persistent cell proliferation in vitro and tumorigenesis in vivo. Thus, we propose a dynamic model, in which the equilibrium between HER2+ and HER2− cells within a heterogeneous tumor population is driven by spontaneous interconversion between these phenotypes, with the more rapidly proliferating HER2+ cells prevalent under baseline conditions, and environmental or therapy-induced stress enhancing conversion to the more resistant HER2− phenotype. Neither molecular profiling nor functional studies have revealed secreted factors that affect the mutual survival of HER2+ and HER2− CTCs, but we cannot exclude such additional factors.
Finally, the properties of patient-derived CTC lines established after multiple courses of therapy provide relevant insight to the treatment of drug-refractory, advanced breast cancer. While clinical trials are evaluating the efficacy of HER2-targeted therapy in HER2− breast cancer with acquired HER2+ CTCs1,19–21, our observations indicate that acquisition of HER2 does not indicate HER2 oncogene dependence and drug susceptibility; instead it constitutes a marker of a proliferative, multi-RTK state. Furthermore, the interconversion of chemotherapy-sensitive HER2+/NOTCH1− and NOTCH inhibitor-sensitive HER2−/NOTCH1+ CTCs suggests that dual treatment, as modeled here, may be required for effective treatment. Clinical trials to date have had limited success sequentially administering embryonic pathway inhibitors targeting Hedgehog, Wnt or Notch to inhibit cancer stem cells following initial chemotherapy16,22–25. The rapid interconversion between proliferative and drug resistant CTC subpopulations raises the possibility that simultaneous combination therapy may provide a novel strategy for clinical validation.
Methods
Patient Selection and CTC isolation
Patients with a diagnosis of metastatic breast cancer provided informed consent for de-identified blood collection, as per Institutional Review Board (IRB) approved protocol (DF/HCC 05-300). Enrolled patients had received multiple courses of therapy, which is typical in advanced ER+ breast cancer, and we did not have sufficient power in this pilot study to enable a statistically significant correlation between the number of therapeutic interventions and the frequency of HER2+ CTCs. Patient matched primary and metastatic tumor specimens were collected per IRB approved protocol (2002-P-002059), and relevant tumor source data is provided in Supplementary Table 1.
Single CTCs were isolated from fresh whole blood by depleting leukocytes using the microfluidic CTC-iChip as previously described3. Briefly, whole blood samples were incubated with biotinylated antibodies against CD45 (R&D Systems, clone 2D1), CD66b (AbD Serotec, clone 80H3), and CD16 (BD, clone 3G8) followed by incubation with Dynabeads MyOne Streptavidin T1 (Invitrogen) to achieve magnetic labeling of white blood cells. This mixture was processed through the CTC-iChip, and the CTCs were stained in solution with Alexa 488-conjugated antibodies against EpCAM (Cell Signaling Technology, clone VU1D9) and HER2 (Cell Signaling Technology, clone 29D8 or Janssen R&D) and identified by imaging flow cytometry (Amnis). Individual CTCs were picked after staining as described above, and PE-CF594-conjugated antibody against CD45 (BD Biosciences, clone HI30) was included to stain contaminating leukocytes. CTCs were individually micromanipulated using a 10 μm transfer tip on an Eppendorf TransferMan NK 2 micromanipulator, transferred into PCR tubes containing RNA protective lysis buffer, and flash frozen in liquid nitrogen as previously described26. Standard CTC enumeration of fixed samples is performed on the BioView high content imaging system following Megafunnel fixation and staining with the combination of wide spectrum cytokeratin (Abcam, ab9377), EpCAM (Cell Signaling Technology, clone VU1D9), EGFR (Cell Signaling Technology, clone D38B1) and HER2 (Cell Signaling Technology, clone 29D8) antibodies.
For mouse xenograft studies, blood was collected via cardiac puncture and ~1mL of blood was processed through the microfluidic CTC iChip. CTCs were enumerated on the BioView imaging system after staining with Alexa 488-conjugated antibodies against EpCAM (Cell Signaling Technology, clone VU1D9), HER2 (Janssen R&D or Cell Signaling Technology, clone 29D8) and GFP (ab13970) followed by secondary antibodies conjugated with Alexa-488 (Invitrogen).
Immunohistochemistry
Tissues were sectioned, and slides were incubated in 0.3% hydrogen peroxide in methanol for 20 minutes to block endogenous peroxidase activity. Tissues were permeabilized, and antigen retrieval was performed in 1X citrate buffer (pH 6) for 15 minutes. Slides were washed and blocked for 30 minutes with 5% Goat serum. Primary HER2 (Cell Signaling, 29D8) or GFP (Living Colors AV 632381) antibodies were diluted 1:75 or 1:250 in DAKO antibody diluent and samples were incubated for one hour at room temperature. Slides were incubated with HRP anti-rabbit antibody (EnVision + DAKO) for 30 minutes. After washing with PBS, the peroxidase reaction was performed with 3,3′-diaminobenzidine (DAB) from Vector lab kits for 10 minutes. Cells were counterstained with Gill’s #2 hematoxylin for 10–15 seconds, dehydrated with ethanol and cleared with xylene before mounting. Images represent at least 5 independent fields from 8 tumors per condition.
Fluorescence-in-situ Hybridization (FISH)
FISH was performed as described previously27,28. Briefly, 5-μm sections of formalin-fixed, paraffin-embedded tumor samples were deparaffinized, hydrated, and pretreated with 0.1% pepsin for 1–2 hours. Slides were then washed in 2x saline-sodium citrate buffer (SSC), dehydrated, air dried, and co-denatured at 80°C for 5 minutes with a mixture of CEP17 and HER2 probes and hybridized at 40°C overnight using the Hybrite Hybridization System (Abbott). Two minute post-hybridization washes were performed in 2x SSC/0.3%NP40 at 72°C followed by a 1 min wash in 2x SSC at room temperature. Slides were mounted with Vectashield containing 4′,6-diamidino-2-phenylindole (Vector, Burlingame, CA, USA). Entire sections were observed with an Olympus BX61 fluorescent microscope equipped with a charge-coupled device camera and analyzed with Cytovision software (Applied Imaging, Santa Clara, CA).
The HER2 and CEP17 signals were quantified in 50 randomly selected, non-overlapping nuclei, and mean numbers of HER2 and CEP17 copies per nucleus were calculated. HER2 was considered amplified when the HER2:CEP17 ratio was ≥2.0 or HER2 signals per nuclei was >6 following the guidelines of the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP)29. The probes used in this study consisted of centromeric CEP: 17p11.1-q11.1, spectrum aqua (Abbott Molecular, Des Plaines, IL) and locus-specific identifier (LSI) probes derived from bacterial artificial chromosome RP11-94L15 (17q12-17q21.1, spectrum orange probe (CHORI, Oakland, CA).
CTC Cell Culture
CTC cultures were grown in suspension in ultra-low attachment plates (Corning) in tumor sphere medium [(RPMI-1640, EGF (20ng/mL), bFGF (20ng/mL), 1X B27, 1X antibiotic/antimycotic (Life Technologies)] under hypoxic (4% O2) conditions. The breast CTC cell lines, Brx-42, Brx-82 and Brx-142, were derived from CTCs isolated using the CTC-iChip as previously described4. CTC cell lines were routinely checked for mycoplasma, using the mycoplasma detection kit (MycoAlert, Lonza), and were authenticated by RNA-seq, MS and DNA-seq (1000 gene mutation panel).
Fluorescence-activated cell sorting (FACS)
Cells were trypsinized into single-cell suspensions, resuspended in Hanks’ balanced salt solution (HBSS), and incubated with Anti-HER2/NEU APC (BD, clone 42 c-erbB-2), Anti-HER2 FITC (Janssen R & D) or Annexin V FITC (BD, clone RUO) antibodies for 20 min at 4°C. Unbound antibodies were washed from cells using HBSS. For analytical flow, cells were fixed with 3% paraformaldehyde and analyzed using a Laser BD Fortessa instrument. For sterile live-cell flow cytometry, cells were sorted using a Laser BD FACS Aria Fusion Cell Sorter, BSL2+. FACS plots are representative of at least 2 independent experiments performed within 6-months of culture initiation (Fig. 1d, 2a and Extended Data Fig. 1f, 3a).
Sequencing Analysis of Genomic DNA
Genomic DNA extracted from CTC-derived cell lines was sequenced using a multiplex polymerase chain reaction (PCR) technology called Anchored Multiplex PCR (AMP) for single nucleotide variant (SNV) and insertion/deletion (indel) detection using next generation sequencing (NGS) as previously described30. Briefly, genomic DNA was isolated from cell lines and then sheared with the Covaris M220 instrument, followed by end-repair, adenylation, and ligation with an adapter. A sequencing library targeting hotspots and exons in 39 commonly mutated, cancer-associated genes was generated using two hemi-nested PCR reactions. Illumina MiSeq 2 × 151 base paired-end sequencing results were aligned to the hg19 human genome reference using BWA-MEM31. MuTect32 and a laboratory-developed insertion/deletion analysis algorithm were used for SNV and indel variant detection, respectively. This assay has been validated to detect SNV and indel variants at 5% allelic frequency or higher in target regions with sufficient read coverage.
Lentivirus Production, Infection and siRNA knockdown of CTC Cell Lines
To produce replication-incompetent lentivirus, 293T cells were cotransfected with either Lenti-Luc-GFP or Notch intracellular domain-pcw107 (Addgene #64621) constructs in combination with REV, VSVG, PDML or pMD2.G and psPAX2 (Addgene) using Lipofectamine Plus reagent (Invitrogen). Twenty-four hours later, growth medium was replenished. Viral supernatants were harvested 48 hours post-transfection, concentrated with Lenti-X Concentrator (Clontech), and viral pellets were resuspended in 400 μl base medium. CTC cultures were infected overnight with 100 μl lentivirus in 6 μg/ml Polybrene. Puromycin (3 μg/ml) was used to select transduced cells over a period of 7 days. Puromycin (3 μg/ml) was used to select transduced cells over a
For the RNAi knockdown, CTC lines Brx-42, Brx-82 and Brx-142 were reverse transfected in ultra-low attachment 6-well plates (Corning) with 25 nM siRNA smart pools (Dhamacon) containing the combination of 4 different siRNA oligonucleotides for ERBB2/HER2 (GGACGAAUUCUGCACAAUG; GACGAAUUCUGCACAAUGG; CUACAACACAGACACGUUU; AGACGAAGCAUACGUGAUG), NOTCH1 (GCGACAAGGUGUUGACGUU; GAUGCGAGAUCGACGUCAA; GAACGGGGCUAACAAAGAU; GCAAGGACCACUUCAGCGA), NRE2L2 (GAGAAAGAAUUGCCUGUAA, CCAAAGAGCAGUUCAAUGA, UAAAGUGGCUGCUCAGAAU; UGACAGAAGUUGACAAUUA) or the negative control gene GAPDH. siRNA pools for target genes were deconvolved to demonstrate targeted knockdown efficiency (> 2 siRNA siRNAs per gene).
Immunofluorescence
CTC lines were spun onto poly-L-lysine-functionalized glass slides with Spintrap, fixed with 3% paraformaldehyde, permeabilized with 0.1% Triton X, and stained with nuclear 4,6-diamidino-2-phenylindole (DAPI) stain, HER2 (Cell Signaling Technologies, clone 29D8), Ki67 (Zymed), Cleaved Caspase-3 (Cell Signaling Technologies, clone D3E9) and/or NOTCH1 (Cell Signaling Technologies, clone D1E11) antibodies. Secondary antibodies were conjugated to either Alexa Fluor 488 or Alexa Fluor 647 (Life Technologies) and fluorescence was measured using the Nikon 90-I fluorescent microscope. Images are representative of at least 3 independent images per sample.
Single Cell Lineage Tracing and Confocal Microscopy
Single HER2+ or HER2− CTCs were flow sorted in 96-well white-walled plates (Corning) using Laser BD FACS Aria Fusion Cell Sorter, BSL2+. Single cell, 1, 3, 5–9, 10–20 and <20-cell clones were analyzed for heterogeneity in HER2 expression via staining with antibodies against EpCAM (FITC labeled; Cell Signaling, clone VU1D9) and HER2 (APC labeled, BD, clone 42 c-erbB-2). Imaging and image processing was performed sequentially with the confocal microscope (Zeiss 710 Laser Scanning Confocal) followed by FIJI (Image J). Images are representative of at least 20 independent images per colony size.
Bioinformatic Analysis
Bioinformatic analysis of single cell RNA-Seq data
Determination of reads-per-million (RPM)
Trimmomatic was used to crop reads lengths to 50 nucleotides, and to remove the TruSeq3-PE-2 Illumina adapters. The paired-end reads were then aligned using tophat2 and bowtie1 with the no-novel-juncs argument set with human genome version hg19 and transcriptome defined by the hg19 genes.gtf table from http://genome.ucsc.edu. Reads that did not align or aligned to multiple locations were discarded. The number of reads aligning to each gene was then determined using htseq-count. Samples that had less than 105 reads were discarded. The read count for each gene was divided by the total counts assigned to all genes and multiplied by one million to form the reads-per-million (RPM). Samples for which the expression of the white blood cell marker PTPRC (CD45) was greater than 10 RPM were discarded. Single-cell RNA-seq data has been deposited in the Gene Expression Omnibus under accession number GSE75367.
Bimodality
To establish that the distribution of HER2 expression in CTCs is multi-modal, we applied the Hartigans’ dip test as implemented in the diptest R-package to the log10(RPM + 1) values with 10 RPM as the threshold to define HER2- versus HER2+ CTCs. To establish that the distribution has two modes and not more, we applied the density function of R with default values to the log10(RPM + 1) values.
Gene Set Enrichment Analysis of RNA-seq and Quantitative Proteomics Data
Based on the analysis of bimodality above, we defined HER2+ samples to be those for which the expression of HER2 exceeded 10 RPM and defined the rest to be HER2−. For the mass spectrometric data, enrichment of signaling pathways was determined by submitting the average log2 fold-change in protein abundance between the HER2-high and HER2-low samples to the pre-ranked function of the Broad Institute’s GSEA software using gene sets in the Pathway Interaction Database (PID) and KEGG as curated in version 4 of the Broad Institute’s MSigDB (http://www.broadinstitute.org/gsea/msigdb/). Pathway enrichment for the RNA-Seq of the CTCs was done the same way with the exception that the full RPM matrix for the CTCs and the HER2+ versus HER2− distinction was input to the GSEA software instead of log2 fold-change.
Quantitative Proteomics
CTC Cell pellets were re-suspended in lysis buffer containing 75 mM NaCl, 50 mM HEPES (pH 8.5), 10 mM sodium pyrophosphate, 10 mM NaF, 10 mM β-glycerophosphate, 10 mM sodium orthovanadate, 10 mM phenylmethanesulfonylfluoride, Roche Complete Protease Inhibitor EDTA-free tablets, and 3 % sodium dodecyl sulfate. Cells were lysed by passing them 10 times through a 21-gauge needle, and the lyses were prepared for analysis on the mass spectrometer essentially as described previously33. Briefly, reduction and thiol alkylation were followed by purifying the proteins using MeOH/CHCl3 precipitation. Protein digest was performed with Lys-C and trypsin, and peptides were labeled with TMT-10plex reagents (Thermo Scientific)34 and fractionated by basic pH reversed phase chromatography. Multiplexed quantitative proteomics was performed on an Orbitrap Fusion mass spectrometer (Thermo Scientific) using a Simultaneous Precursor Selection (SPS) based MS3 method35. MS2 spectra were assigned using a SEQUEST-based proteomics analysis platform36. Based on the target-decoy database search strategy37 and employing linear discriminant analysis and posterior error histogram sorting, peptide and protein assignments were filtered to a false discovery rate (FDR) of < 1 %36. Peptides with sequences that were contained in more than one protein sequence from the UniProt database were assigned to the protein with most matching peptides36. TMT reporter ion intensities were extracted as that of the most intense ion within a 0.03 Th window around the predicted reporter ion intensities in the collected MS3 spectra. Only MS3 with an average signal-to-noise value larger than 40 per reporter ion as well as with an isolation specificity33 larger than 0.75 were considered for quantification. A two-step normalization of the protein TMT-intensities was performed by first normalizing the protein intensities over all acquired TMT channels for each protein based on the median average protein intensity calculated for all proteins. To correct for slight mixing errors of the peptide mixture from each sample, a median of the normalized intensities was calculated from all protein intensities in each TMT channel, and protein intensities were normalized to the median value of these median intensities.
Protein interactions were extracted from the String database (High confidence score > 0.7)38. Overlapping proteins were assigned to the pathway with the greatest number of proteins, and enriched PID pathways were ranked by log10(p-value) to nearest thousandth. Mass spectrometry raw data has been deposited in the MassIVE proteomics data repository under the accession number MSV000079419.
Drug Screens
Drugs were obtained from the MGH Center for Molecular Therapeutics and are listed in Supplemental Table 6 and were chosen due to common clinical use for treatment of breast cancer or unique targeting of epigenetic/stem cell pathways. One thousand cells were seeded in tumor sphere media in 384-well ultra-low attachment plates in triplicate wells on duplicate plates 24 hours prior to the addition of drugs. Three independent drug concentrations centered on the reported IC50 were used (Supplemental Table 6). Cell viability was assayed 6-days following drug exposure with CellTiter-Glo (Promega) and was normalized to corresponding untreated controls39.
Mouse Xenograft Assays and Drug Treatment
In compliance with ethical regulations and approved by the animal protocol (IACUC 2010N000006), six-week old female NSG (NOD. Cg-Prkscsdid Il2rgtm1Wjl/SzJ) mice from Jackson Laboratories were anesthetized with isofluorane, and GFP-LUC labeled CTCs (200,000, 20,000 and/or limiting dilutions as low as 200 cells) or 50:50 mixed CTCs (GFP-LUC+/HER2+: Untagged/HER2−, and the converse) were injected into the 4th right mammary fat pad. A 90-day release 0.72mg estrogen pellet (Innovative Research of America) was implanted subcutaneously behind the neck of each mouse. Tumor growth was monitored weekly by in vivo imaging using IVIS Lumina II (PerkinElmer) following intraperitoneal injection (150 μl/animal) of D-Luciferin substrate (Sigma). For in vivo drug sensitivity testing, Paclitaxel (10mg/kg) was administered weekly via IV injection for four consecutive weeks. Notch inhibitors (Notch2) LY-411575 (10mg/kg) or (Notch2) RO429097 (10mg/kg) were administered daily (5 days on/2 days off) via oral gavage in 2% solvent (2% sodium caroboxymethyl cellulose) for four consecutive weeks. No animal randomization or blinding was used for these mouse studies. All animal studies used 8 mice per condition to ensure sufficient statistical power.
Extended Data
Extended Data Fig. 1. Advanced ER+/HER2− breast cancer patients harbor discrete HER2+ and HER2− subpopulations.
(a) CTCs freshly isolated from 19 ER+/HER2− breast cancer patients were stained with HER2 (yellow) and EpCAM (green) and imaged using imaging flow cytometry. Bar graph shows the number of HER2+ (black) and HER2− (white) CTCs (median 22% HER2+ CTCs, range 4–58%). Supplemental Table 1 provides HER2+/HER2− ratios and each patient’s clinical history. (b) scRNA-seq for ERBB2 expression at multiple time-points showing acquisition of HER2+ CTCs (Brx-82, Brx-42) over the course of progressive disease. Single asterisk (*) denotes patient expiration. Rx = Sacituzumab (IMMU-132); Rx1 = Vinorelbine + Trastuzumab; Rx2 = Eribulin. (c) Distinct HER2+ and HER2− CTCs from 13 TNBC patients determined by scRNA-seq (HER2− ≤ 0 read per million (RPM); HER2+ > 153, range 33–463). (d) HER2 FISH analysis of metastatic tumors from patients, Brx-42, Brx-82 and Brx-142, shows no amplification of ERBB2 compared to HER2-amplified control (Supplemental Table 1 for tumor source data). HER2 (red); chromosome enumeration probe 17 (CEP17) (cyan); Scale bar: 10 μm. Representative image from 5 independent fields are shown. (e) Bright field and IF (DAPI, blue; HER2, green) images of CTC lines, Brx-42, Brx-82 and Brx-142, demonstrate heterogeneity in HER2 expression. Scale bar: 100 μm (bright field); 20 μm (IF). Representative image from 3 independent fields are shown. (f) FACS analysis shows two distinct HER2+ and HER2− subpopulations in the CTC line Brx-42 (at initiation) compared to HER2− control. Representative data of two independent experiments are shown. (g) HER2 FISH analysis of the HER2+ and HER2− subpopulations from CTC lines Brx-42, Brx-82 and Brx-142 shows that the ERBB2 is not amplified. HER2-amplified SKBR3 cells shown as control. HER2 (red); CEP17 (green); Scale bar: 10 μm. Representative images from 5 independent fields are shown.
Extended Data Fig. 2. HER2+ and HER2− subpopulations exhibit distinct functional properties.
(a) Increased expression of the proliferation marker Ki67 (red) in the HER2+ subpopulation of CTC line Brx-142 (T-test p < 0.0001), compared with HER2− subpopulation, with no change in cleaved-caspase 3 (red). HER2+ cells (green); scale bar: 20 μm. Representative image from five independent fields are shown. (b) FACS analysis for the apoptotic marker Annexin V-FITC shows no difference in apoptosis between the HER2+ and HER2− subpopulations of FACS-purified CTC line Brx-142. Representative data from two independent experiments are shown. (c) Tumors initiated by HER2+ or HER2− CTCs (Brx-82: 200,000 cells) orthotopically injected into the mammary fat pad show differential growth rates; n=8. (d) Metastatic frequency of HER2+ and HER2− cultured CTCs (Brx-82: p=0.05 and Brx-142: p=0.009) following orthotopic injection; n=8. (e) Limiting dilution experiments demonstrate comparable tumor-initiating ability from 200 HER2+ and HER2− cultured CTCs (Brx-82, Brx-142); n=8.
Extended Data Fig. 3. Dynamics of HER2+ and HER2− interconversion.
(a) FACS purified HER2+ and HER2− subpopulations from CTC line Brx-82 were monitored over 28 days to determine shifts in the composition of sorted populations. Representative data of two independent experiments are shown. (b) Growth curves for HER2+ (red) and HER2− (blue) FACS-purified single cell clones from CTC line Brx-142; Two-way ANOVA p-value < 0.0001; n=20. (c) IHC HER2 staining of tumor xenografts derived from unlabeled HER2− and HER2+ CTCs showing acquisition/loss of HER2 (brown), respectively. Arrows indicate regions of HER2 acquisition/loss. Representative image from at least 5 independent fields; n=8. ER+/HER2− and HER2-amplified breast cancers are shown below as controls. (d) Low magnification (landscape) view of HER2 IHC staining of tumor xenografts derived from mixed HER2+ and HER2− CTC cultures containing either GFP-tagged HER2+/HER2− cells (high magnification images are shown in Figure 2f). Upper panel: representative GFP-tagged HER2− cells give rise to GFP+/HER2+ cells (GFP: cytoplasmic red stain, HER2: cell surface brown stain). Lower panel: GFP-tagged HER2+ cells produce GFP+/HER2− cells. Scale bar: 100 μm.
Extended Data Fig. 4. Proteomic and scRNA-Seq analysis of HER2+ versus HER2− cells.
(a–b) MS-based whole cell proteome profiles (6349 proteins) comparing HER2+ and HER2− populations from CTC lines (Brx-42, Brx-82, Brx-142). Matched HER2+ versus HER2− proteomic differences show significant linear correlation (Pearson correlation coefficient = 0.71 between Brx-82 and Brx-42; Pearson correlation coefficient = 0.64 between Brx-142 and Brx-42); NI = Normalized Intensity; n=2 per cell line are shown. (c) Phospho-RTK array of HER2+ and HER2− populations of CTC cell lines Brx-142 and Brx-82 show increased phosphorylation of RTKs in the HER2+ population. Numbers denote: 1. HER2; 2. HER3; 3. HER4; 4. INSR; 5. EPHA1; 6. EPHA2; 7. AXL. Representative data from two independent experiments are shown. (d) Volcano plot depicts genes enriched in HER2+ (red) and HER2− (blue) individual CTCs isolated from patients Brx-42 and Brx-82 and analyzed by scRNA-seq; n=22. (e) Venn diagram showing overlap of genes and proteins derived from scRNA-seq (Brx-42, Brx-82) and quantitative proteomics of HER2+ CTCs, respectively.
Extended Data Fig. 5. 55-panel drug screen shows differential drug sensitivities exhibited by HER2+ versus HER2− subpopulations.
(a) Heat map showing percent cell viability (represented as decimal) after 6 days drug treatment of the HER2+ and HER2− subpopulations derived from CTC lines Brx-142 and Brx-82. Red and blue represent high and low drug sensitivities, respectively; n=6. (b) Lapatinib sensitivity of HER2+ (red) and HER2− (blue) subpopulations of CTC line Brx-82. MDA-231 (TNBC) and SKBR3 (HER2-amplified) are shown as controls. (c) Chemosensitivity of HER2+ (red) and HER2− (blue) subpopulations of CTC line Brx-142. MDA-231 and SKBR3 are shown as controls. (d) Sensitivity of HER2+ (red) and HER2− (blue) subpopulations of CTC line Brx-142 to Notch inhibition with Notchi1 (BMS-708163) and Notchi2 (RO4929097). MDA-231 and SKBR3 cells are shown as controls. (a–d) Represent at least two independent experiments for each condition; n=6.
Extended Data Fig. 6. NOTCH1 expression and activity in HER2− CTCs.
(a) Western blot analysis of HER2+ and HER2− subpopulations from CTC lines BRx-142 and Brx-82 show increased NOTCH1 in HER2− cells. γ-tubulin is shown as control. IF analysis and scRNA-seq of NOTCH1 (red) and HER2 (green) shows inversely correlated expression in CTC lines (Brx-142, Brx-82). (b) Ectopic expression of constitutively-active Notch intercellular domain (ICD) or NRF2 results in increased expression of the Notch1 ligand JAG1 but does not alter HER2 expression. Representative data of two independent experiments are shown; S.E.M (error bars). (c) siRNA-mediated inhibition of HER2 in Brx-42 HER2+ CTCs, and lapatinib-mediated inhibition of HER2 in SKBR3 cells results in dose dependent increases in the expression of genes involved in Notch signaling (NOTCH1, JAG1, DLL1, HES1, HEY1, HEY2). Representative data of two independent experiments are shown; S.E.M (error bars). (d) Inhibition of HER2 using lapatinib or siRNA knockdown in Brx-82 HER2+ CTCs increases the expression of NRF2-driven cytoprotective genes downstream of Notch. Representative data of two independent experiments are shown; S.E.M. (error bars). (e) Quantitation of the interconversion of HER2+ cells from single cell clones into 5–9-cell and >10-cell clusters following treatment with 10mM H2O2; T-test p-value < 0.05; n=10. (f) Paclitaxel treatment of mice with tumors derived from Brx-142 FACS purified HER2+ CTCs, demonstrating a reduction in CTCs and no change in HER2− CTC counts; T-test p <0.05; NS=Not Significant. (g) Paclitaxel treatment of mice with mammary xenografts derived from parental CTC line Brx-142 showing initial tumor response, followed by recurrent tumor growth. IHC analysis and quantitation of the recurrent tumor shows greatly reduced HER2+ (brown stain) cell composition in the Paclitaxel drug treated (T, 3-weeks post-treatment) tumor compared with the untreated tumor (U) and the recovered tumor (R, 5-weeks post-treatment). Bar indicates duration of drug treatment (Rx). Scale bar = 100 μm; Two-way ANOVA p-value < 0.0001; n=8. Representative images from 5 independent fields per tumor are shown and quantified; T-test p <0.001. (h) Dual GFP (red, cytoplasmic stain) and HER2 (brown, cell surface stain) IHC of tumor xenografts derived from mixed GFP-tagged HER2+ and untagged HER2− CTC cultures demonstrating enhanced conversion from GFP+/HER2+ to GFP+/HER2− following 4-weeks paclitaxel treatment; T-test p-value < 0.0001; n=8. Scale bar: 100 μm. Arrows indicate interconverting cells. Representative images from 5 independent fields per tumor are shown. (i) Mouse tumor xenografts derived from the CTC cell line Brx-142 treated with a combination of the Notchi3 (LY-414575) and paclitaxel shows diminished tumor relapse; n=8. Bar indicates treatment duration.
Acknowledgments
We thank the patients who participated in this study. This work was supported by NIH CA009361, the Howard Hughes Medical Institute, the Breast Cancer Research Foundation, the National Foundation for Cancer Research (DAH) and Wellcome Trust 102696 (CB), NIH Quantum 2U01EB012493 (MT, DAH), T32 CA009361, Susan G. Komen Foundation PDF16376429 (NVJ), K12 5K12CA087723 (AB) and T32GM007753 (RYE). We thank D. Dombrowski (NIH 1S100D1016372-01) for expert flow cytometry.
Footnotes
Author Contributions N.V.J., D.A.H. and S.M. conceived the project and provided project leadership. A.B. enrolled patients and provided clinical guidance. B.S.W. and S. R. performed the bioinformatics analyses. M.L. and Y.Z. assisted with animal experiments. T.K.S., M.L.O. and A.J.I. performed the mutational analysis and FISH. J.A.L, R.D., R.O. and R.Y.E. picked micromanipulated CTCs for scRNA-seq and assisted with molecular biology experiments. D.S. analyzed pathology specimens. C.B. and M.Y. helped with drug screens. S.R. and D.T.T. provided scRNA-seq support. W.H. and MB performed MS experiments and analysis. R. V. and M.T. collaboratively developed the CTC-iChip isolation of viable CTCs.
Supplementary Information is available in the online version of this paper.
Single-cell RNA-seq data have been deposited in the Gene Expression Omnibus under accession number GSE75367. Mass spectrometry raw data have been deposited in the MassIVE proteomics data repository under the accession number MSV000079419.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Arteaga CL, Engelman JA. ERBB Receptors: From Oncogene Discovery to Basic Science to Mechanism-Based Cancer Therapeutics. Cancer Cell. 2014;25:282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houssami N, Macaskill P, Balleine RL, Bilous M, Pegram MD. HER2 discordance between primary breast cancer and its paired metastasis: tumor biology or test artefact? Insights through meta-analysis. Breast Cancer Res Treat. 2011;129:659–674. doi: 10.1007/s10549-011-1632-x. [DOI] [PubMed] [Google Scholar]
- 3.Ozkumur E, et al. Inertial Focusing for Tumor Antigen-Dependent and -Independent Sorting of Rare Circulating Tumor Cells. Science Translational Medicine. 2013;5:179ra47–179ra47. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu M, et al. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345:216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Meth. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osipo C, et al. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a γ-secretase inhibitor. Oncogene. 2008;27:5019–5032. doi: 10.1038/onc.2008.149. [DOI] [PubMed] [Google Scholar]
- 7.Abravanel DL, et al. Notch promotes recurrence of dormant tumor cells following HER2/neu-targeted therapy. J Clin Invest. 2015;125:2484–2496. doi: 10.1172/JCI74883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2015;475:106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakabayashi N, et al. Notch-Nrf2 Axis: Regulation of Nrf2 Gene Expression and Cytoprotection by Notch Signaling. Molecular and Cellular Biology. 2014;34:653–663. doi: 10.1128/MCB.01408-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korkaya H, Wicha MS. HER-2, Notch, and Breast Cancer Stem Cells: Targeting an Axis of Evil. Clinical Cancer Research. 2009;15:1845–1847. doi: 10.1158/1078-0432.CCR-08-3087. [DOI] [PubMed] [Google Scholar]
- 11.Ithimakin S, et al. HER2 Drives Luminal Breast Cancer Stem Cells in the Absence of HER2 Amplification: Implications for Efficacy of Adjuvant Trastuzumab. Cancer Research. 2013;73:1635–1646. doi: 10.1158/0008-5472.CAN-12-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korkaya H, Paulson A, Iovino F, Wicha MS. HER2 regulates the mammary stem/progenitor cell population driving tumorigenesis and invasion. Oncogene. 2008;27:6120–6130. doi: 10.1038/onc.2008.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginestier C, et al. ALDH1 Is a Marker of Normal and Malignant Human Mammary Stem Cells and a Predictor of Poor Clinical Outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martz CA, et al. Systematic identification of signaling pathways with potential to confer anticancer drug resistance. Science Signaling. 2014;7:ra121. doi: 10.1126/scisignal.aaa1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pandya K, et al. Targeting both Notch and ErbB-2 signalling pathways is required for prevention of ErbB-2-positive breast tumour recurrence. British Journal of Cancer. 2011;105:796–806. doi: 10.1038/bjc.2011.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting Wnt, notch, and Hedgehog pathways. Nature Reviews Clinical Oncology. 2010;8:97–106. doi: 10.1038/nrclinonc.2010.196. [DOI] [PubMed] [Google Scholar]
- 17.Vanharanta S, Massagué J. Origins of Metastatic Traits. Cancer Cell. 2013;24:410–421. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawson DA, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526:131–135. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bidard FC, Pierga JY. Clinical Utility of Circulating Tumor Cells in Metastatic Breast Cancer. Journal of Clinical Oncology. 2015;33:1622–1622. doi: 10.1200/JCO.2014.57.9714. [DOI] [PubMed] [Google Scholar]
- 20.Schramm A, et al. The DETECT Study Program: Personalized treatment in advanced breast cancer based on circulating tumor cells (CTCs) ASCO Meeting Abstracts. 2015;33:TPS11109. [Google Scholar]
- 21.Ignatiadis M, et al. Abstract OT1-2-02: Trastuzumab in HER2-negative early breast cancer as adjuvant treatment for circulating tumor cells (CTCs) (Treat CTC) Cancer Research. 2015;75:OT1–2–02–OT1–2–02. [Google Scholar]
- 22.Amakye D, Jagani Z, Dorsch M. Unraveling the therapeutic potential of the Hedgehog pathway in cancer. Nature Medicine. 2013;19:1410–1422. doi: 10.1038/nm.3389. [DOI] [PubMed] [Google Scholar]
- 23.Kim EJ, et al. Pilot Clinical Trial of Hedgehog Pathway Inhibitor GDC-0449 (Vismodegib) in Combination with Gemcitabine in Patients with Metastatic Pancreatic Adenocarcinoma. Clinical Cancer Research. 2014;20:5937–5945. doi: 10.1158/1078-0432.CCR-14-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LoRusso PM, et al. Phase I Trial of Hedgehog Pathway Inhibitor Vismodegib (GDC-0449) in Patients with Refractory, Locally Advanced or Metastatic Solid Tumors. Clinical Cancer Research. 2011;17:2502–2511. doi: 10.1158/1078-0432.CCR-10-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krop I, et al. Phase I Pharmacologic and Pharmacodynamic Study of the Gamma Secretase (Notch) Inhibitor MK-0752 in Adult Patients With Advanced Solid Tumors. Journal of Clinical Oncology. 2012;30:2307–2313. doi: 10.1200/JCO.2011.39.1540. [DOI] [PubMed] [Google Scholar]
- 26.Miyamoto DT, et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 2015;349:1351–1356. doi: 10.1126/science.aab0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mohapatra G, et al. Glioma Test Array for Use with Formalin-Fixed, Paraffin-Embedded Tissue. The Journal of Molecular Diagnostics. 2010;8:268–276. doi: 10.2353/jmoldx.2006.050109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snuderl M, et al. Polysomy for Chromosomes 1 and 19 Predicts Earlier Recurrence in Anaplastic Oligodendrogliomas with Concurrent 1p/19q Loss. Clinical Cancer Research. 2009;15:6430–6437. doi: 10.1158/1078-0432.CCR-09-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolff AC, et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer. Journal of Clinical Oncology. 2006;25:118–145. doi: 10.1200/JCO.2006.09.2775. [DOI] [PubMed] [Google Scholar]
- 30.Zheng Z, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nature Medicine. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 31.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cibulskis K, et al. sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ting L, Rad R, Gygi SP, Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat Meth. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McAlister GC, et al. Increasing the Multiplexing Capacity of TMTs Using Reporter Ion Isotopologues with Isobaric Masses. Anal Chem. 2012;84:7469–7478. doi: 10.1021/ac301572t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAlister GC, et al. MultiNotch MS3 Enables Accurate, Sensitive, and Multiplexed Detection of Differential Expression across Cancer Cell Line Proteomes. Anal Chem. 2014;86:7150–7158. doi: 10.1021/ac502040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huttlin EL, et al. A Tissue-Specific Atlas of Mouse Protein Phosphorylation and Expression. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Meth. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 38.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Research. 2010;39:D561–D568. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]