Abstract
Nonribosomal peptide synthetases (NRPSs) catalyze the assembly line biosynthesis of peptide natural products that play important roles in microbial signaling and communication. These multidomain enzymes use an integrated carrier protein that delivers the growing peptide to the catalytic domains, requiring coordinated conformational changes that allow the proper sequence of synthetic steps. Recent structural studies of NRPSs have described important conformational states and illustrate the critical role of a small subdomain within the adenylation domains. This subdomain alternates between catalytic conformations and also serves as a linker domain, providing further conformational flexibility to enable the carrier to project from the core of NRPS. These studies are described along with remaining questions in the study of the structural dynamics of NRPSs.
The modular basis for nonribosomal peptide synthesis
The term natural products describes a wide variety of chemicals that are not involved in basal metabolism and are not necessary for microbial growth in rich media. Natural products are secreted into the environment where they play important roles in the communication within a species, between different species, and between microorganisms and the hosts in which they reside. Production of these compounds allows cells to adapt to new environments and to compete against other organisms. This latter property confers important biological functions that have been exploited in the development of natural products into important pharmaceutically active molecules [1,2].
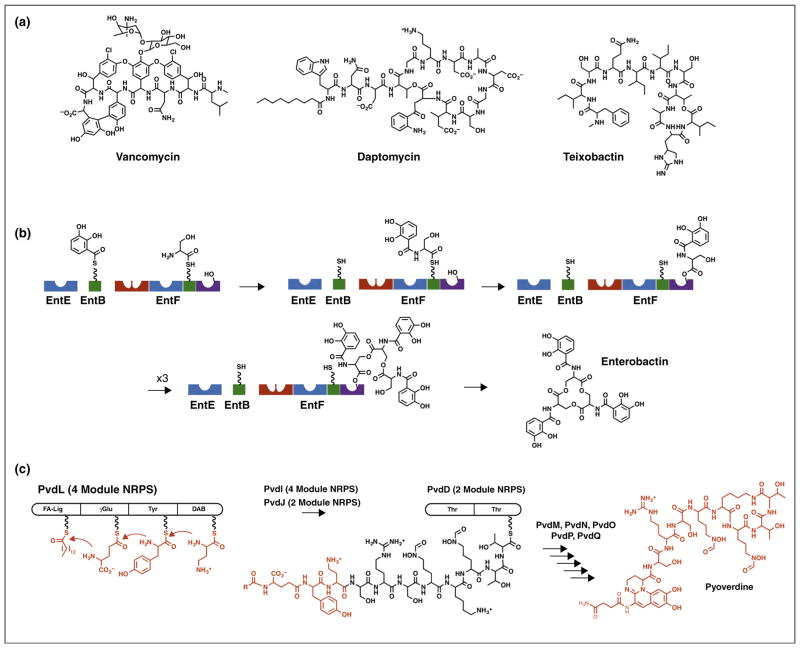
Nonribosomal peptide synthetases (NRPSs) produce a dazzling array of peptide natural products (Figure 1a). A distinctive modular architecture of the NRPS enzymes, elucidated nearly two decades ago, has fostered much interest in understanding these large, multidomain protein machines [3,4]. NRPSs load substrates onto an integrated peptidyl carrier protein (PCP) that migrates between different catalytic domains for individual steps in the synthesis (Figure 1b). This catalytic strategy has been likened to an ‘assembly line’ as the nascent peptide is extended and modified as it passes from module to module. Some very large fungal NRPS proteins contain the entire multimodular machinery in a single polypeptide chain. The fungus Trychoderma virens, for example, uses a single 20 925-residue NRPS to produce an 18-residue peptide [5]. More commonly, NRPS clusters span several proteins that range from single domains to multiple modules (Figure 1c).
Figure 1.
Biosynthetic assembly lines of NRPS proteins. (a) NRPS products vancomycin, daptomycin, and teixobactin demonstrate the diversity of NRPS products. (b) Protein domains catalyze steps in the biosynthesis of the siderophore enterobactin. Blue adenylation domains load 2,3-dihydroxybenzoic acid (EntE) or serine (EntF) onto partner PCP domains (green) in intermolecular or intramolecular reactions. The red condensation domain of EntF then catalyzes the transfer of DHB from the loaded EntB carrier to form the amide with serine on EntF. This amide is then transferred to a catalytic serine within the (purple) thioesterase domain. After three cycles of synthesis, the thioesterase domain catalyzes release of the mature enterobactin molecule. The carrier domain of EntF must therefore sequentially visit the active sites of the adenylation, condensation, and thioesterase domains. (c) The pyoverdine biosynthetic pathway involves four NRPS proteins. The four-module PvdL protein initiates synthesis with the modules that direct incorporation of myristic (or myristoleic) acid, glutamic acid, tyrosine, and 2,4-diaminobutyrate. The tetrapeptide is passed to and extended by neighboring NRPSs PvdI and PvdJ, not shown, and ultimately to the two-module NRPS PvdD. The terminal thioesterase domain of PvdD releases the pyoverdine molecule through a cyclization step, where additional enzymes complete the maturation to produce pyoverdine.
Crystallographic and NMR studies of the NRPS domains provide insight into the PCP domain and the core catalytic domains [6•,7]. The PCP is composed of four α-helices, a structural and functional homolog of the carrier proteins that are involved in fatty acid and polyketide synthesis [8]. The second helix contains a conserved serine residue that is post-translationally modified with a phosphopantetheine (PPant) derived from coenzyme A. The terminal thiol of this cofactor binds the amino acyl and peptidyl intermediates during peptide synthesis. Migration of the PCP delivers the substrates and intermediates to neighboring catalytic domains during the biosynthetic process.
In addition to a PCP domain, each NRPS module contains additional catalytic domains. N-terminal initiation modules contain an adenylation domain that catalyzes a two-step reaction to load the amino acid substrate onto the PPant [9]. Internal extension modules contain this adenylation domain but also include a condensation domain that transfers the upstream amino acid or peptide onto the amino acid encoded by that module. Finally, termination modules contain an additional thioesterase or reductase domain that catalyzes the final release of the product through hydrolysis, reduction, or cyclization. Other domains have been identified within NRPS modules to catalyze N-methylation, epimerization, cyclization, oxidation/reduction, or additional chemical modifications to the amino acid or peptide intermediates [10,11].
This assembly line architecture therefore requires that, within an internal module, a carrier protein sequentially visits three domains to first, be loaded with an amino acid specified by that module, second, receive the upstream peptide, and third, deliver the extended peptide to the downstream module. The inclusion of additional modifying domains requires further coordination to ensure the proper peptide product. Recent studies provide views of the features that govern the NRPS structural mechanism. While important issues remain — including the interactions of NRPS modules [12•], of substrates with PCP domains [13,14], and of substrates with the catalytic residues in domain active sites [15•] — this review will focus on the conformational dynamics of NRPSs. Specifically, we will explore the structural features that dictate transport of the substrates loaded onto the PCP between different catalytic domains.
Combining NRPS domains to build a module
Several reviews have cataloged the structures of individual NRPS domains [6•,7,16]. Briefly, the condensation domains are composed of two lobes that surround an active site. The symmetrical architecture allows the entry of the two PCPs from opposite sides to deliver the upstream and downstream substrates to the peptide bond forming site. Condensation domains have been observed in multiple conformations suggesting an opening and closing upon the bound substrates [17,18]. The adenylation domains, which contain a large N-terminal and a small C-terminal subdomain, undergo a domain rearrangement that is used to catalyze the two step reaction. First, the amino acid is activated with ATP to form an acyladenylate and pyrophosphate. Release of pyrophosphate from the active site enables a 140° rotation of the C-terminal subdomain to adopt a second orientation that is used to catalyze the second reaction [9]. This rotation results in a translation of ~15 Å of the C-terminal residue of the adenylation domain. Originally identified in the homologous acyl-CoA synthetases [19,20], this domain alternation conformational change has been confirmed through structural and functional studies of additional freestanding and multidomain NRPS proteins [21–25]. As the PCP domain is positioned immediately downstream of the adenylation domain, this change of orientation of the C-terminal subdomain influences the location of the PCP domain and, as described below, is a critical feature to shuffle the peptide intermediates between catalytic domains. Finally, structures of the terminal thioesterase [26,27] or reductase [28–30] domains demonstrate that these enzymes belong to larger families of enzymes that catalyze similar chemistry.
A complete understanding of the NRPS structural mechanism requires more than a knowledge of the individual catalytic domains. Toward this end, structures of multi-domain proteins have identified the features that govern the productive interactions between the PCP and their partner catalytic domains. In particular, structures of the PCP with thioesterase domains [31,32] or with adenylation domains [33–35] or with the auxiliary epimerization domain [36] demonstrate the importance of the residues that surround the pantetheinylation site of the carrier protein for recognition by the catalytic domains. Given the inherent flexibility of multidomain NRPSs, several strategies have been adopted to structurally characterize these proteins [6•].
Some NRPSs also contain additional domains that do not appear to catalyze reactions however they are responsible for activating other domains. The most common are MbtH-like proteins (MLPs) that are small ~8 kDa proteins that activate adenylation domains [37]. The structures of MLPs with truncated adenylation domains [38] or with complete NRPS modules [39••] identify a conserved binding motif but no obvious channel of communication between the interface and the catalytic residues. While most MLPs are independent proteins, some are covalently joined to adenylation domains in multidomain proteins. Additional adenylation domain activators have also been observed in other multidomain NRPSs [40,41]. Finally, other domains have been identified that serve as scaffolds for the interaction with additional catalytic partners. Certain glycopeptide antibiotic biosynthetic pathways contain a non-functional condensation domain homolog that recruits a P450 protein for amino acid cross-linking [42,43].
To generate a complete movie of the full catalytic cycle, it is necessary to obtain structural information for complete NRPS modules in multiple conformational states. The landmark structure of SrfA-C [17], a terminal module from the surfactin biosynthesis pathway, set the stage for the understanding of modular NRPSs. Several recent papers [39••,44••,45••] provided new views that lead to a new model for the modular architecture of NRPS enzymes.
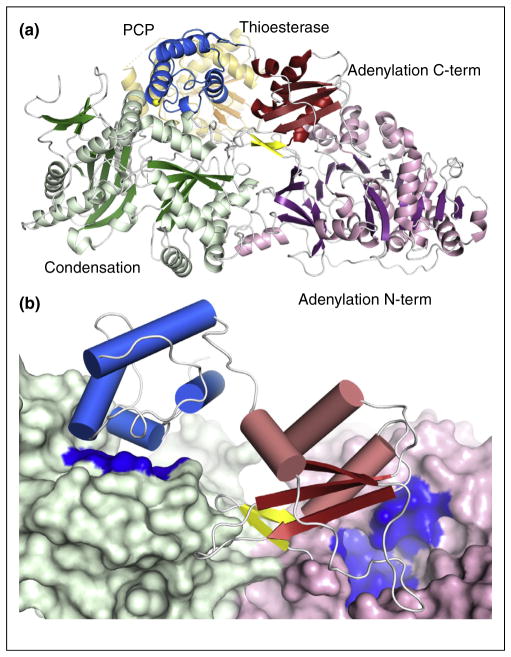
SrfA-C contains condensation, adenylation, PCP, and thioesterase domains, representing all of the core domains [17]. The protein showed several important features (Figure 2). The two largest domains, the 430-residue condensation and the 510-residue adenylation domain display a significant interface between them. The PCP was poised to interact with the condensation domain. The binding sites of the PCP with the condensation domain and the adenylation domain are both located on the same side of the protein, suggesting that a simple movement, perhaps triggered by the subdomain rotation of the adenylation domain [9], could transfer the PCP between the two sites.
Figure 2.
Structure of SrfA-C, the terminal module from the surfactin biosynthesis pathway. (a) The four domains of SrfA-C are highlighted including as well the two subdomains of the adenylation domain. The A8 loop, which initiates the C-terminal subdomain, is shown as yellow strands while Ser1003, the site of pantetheinylation is highlighted with a yellow sphere. The thioesterase domain is shown in orange and is positioned mostly behind the core of the protein in this orientation. In this conformation, the PCP is positioned near the condensation domain. (b) In this view of SrfA-C, the condensation domain and N-terminal subdomain of the adenylation domain are both shown in surface representation. Highlighted in blue on the surface are the entries to the active sites. This figure demonstrates how both entrances are present on the same face of the core of the protein and how rotation of the C-terminal subdomain and the PCP can allow the PCP to alternate between the two active sites.
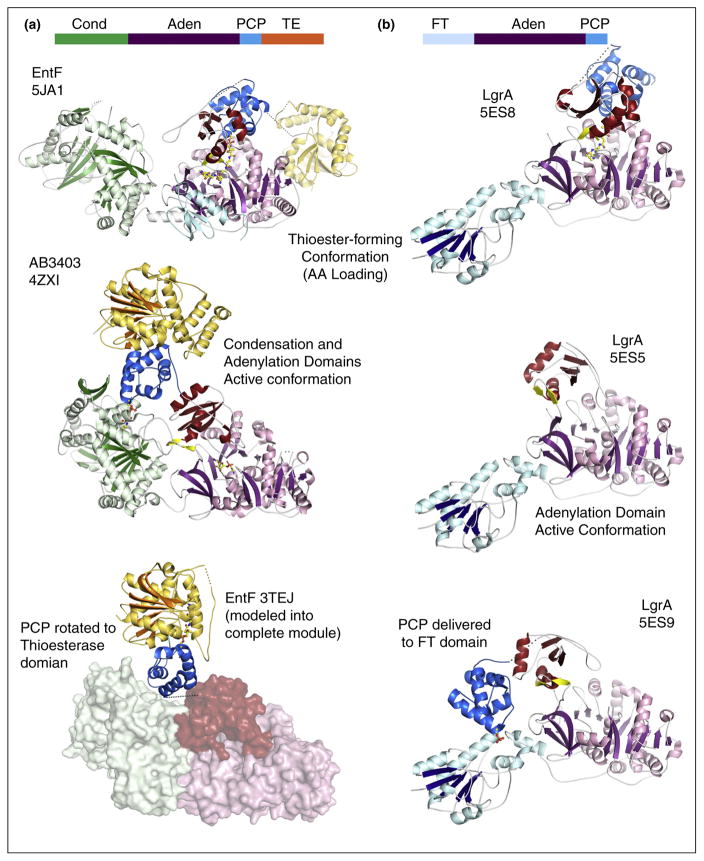
This has now been demonstrated with two structures of additional termination modules containing the same domain architecture as SrfA-C [44••]. The structure of holo-AB3403 (Figure 3), an NRPS from an uncharacterized biosynthetic system in Acinetobacter baumannii, was similar to the SrfA-C structure as the PCP was interacting with the condensation domain. Importantly, the pantetheine cofactor entered the active site of the condensation domain, illustrating subtle differences in the orientation of the PCP from the position adopted in SrfA-C. Given the flexibility of the pantetheine cofactor, this could reflect true differences in the functional interaction between the two proteins or may represent the need for a slight reorientation of the PCP to achieve the catalytically competent interaction.
Figure 3.
Recent structures of multidomain NRPS proteins. Structures of (a) termination modules AB3403 and EntF and (b) LgrA in multiple conformations illustrate the dynamics used to deliver the PCP to multiple catalytic active sites. The top of each panel shows the domain architecture with condensation (Cond), adenylation (Aden), PCP, thioesterase (TE), and formyltransferase (FT) indicating the position of the domains within the protein. The terminal modules adopt the adenylation conformation (Panel a, middle) to carry out the initial adenylation of the amino acid substrate. Rotation of the C-terminal subdomain delivers the PCP to the adenylation domain for covalent loading (thiolation) step (Panel a, top). Return of the protein to the adenylate-forming conformation simultaneously delivers the PCP to the condensation domain (Panel a, middle) where the loaded PCP is positioned to receive the upstream substrate for peptide bond formation. Rotation of the PCP into the thioesterase domain (Panel a, bottom) would deliver the PCP here for removal of the loaded peptide. The bottom panel illustrates the functional complex of the PCP and thioesterase domain from EntF (PDB 3TEJ) superimposed onto the position of the thioesterase domain of AB3403. In the structural cycle of the LgrA protein, the same two initial steps, adenylation (Panel b, middle) and thioester forming (Panel b, top) are illustrated in crystal structures. The third state illustrates the delivery of the loaded PCP to the formyltransferase domain for modification of the loaded substrate (Panel b, bottom). The extension of the C-terminal subdomain (red) of the adenylation domain enables this functional interaction.
In contrast to SrfA-C and AB3403, structures of the E. coli holo-EntF termination module [39••,44••] offered a new view of an NRPS module that identifies the structural changes that guide the NRPS domain dynamics. The EntF structure used a mechanism-based inhibitor that bound in the active site of the adenylation domain, mimicking the seryladenylate intermediate [46]. Upon approach of the PCP and the pantetheine thiol, a covalent bond traps the thioester-forming step. The two views offered by AB3403 and EntF identified the conformations adopted by a holo-NRPS module during two critical steps — the loading of an amino acid onto the PCP in EntF and the peptide bond forming step in AB3403. These structures confirm that the transfer of the PCP between these two domains accompanies the rotation of the small C-terminal subdomain as suggested by the structural features in the homologous acyl-CoA and aryl-CoA synthetases [9]. The domain alternation movement of the C-terminal subdomain and the accompanying movement of the loaded PCP has a natural effect of delivering the substrate directly to the upstream condensation domain to await the approach of the upstream PCP that harbors the preceding amino acid or peptide. Interestingly, as the pantetheine is directed into the condensation domain in the AB3403 structure, the adenylation domain adopts the adenylate-forming conformation, illustrating that two domains can simultaneously adopt their active conformation, improving the efficiency of the catalytic cycle.
An elegant recent study describes several states of LgrA, a multidomain NRPS from the linear gramicidin synthetase NRPS system [45••]. The protein contains a formyltransferase domain upstream of the adenylation-PCP architecture that modifies the valine substrate. LgrA was captured in multiple open and catalytic states in which the PCP was interacting with both the formylation and adenylation domains. Unlike the prior examples of large modules that require comparisons between different proteins, the LgrA structures present the relevant catalytic states within a single protein.
Dynamics within NRPS modules
These structures of multidomain NRPSs can now be pieced together to understand the changes that occur within and between domains that allow transport of the PCP (Figure 3). The domain rotation of the C-terminal subdomain of the adenylation domain is exploited in the migration of the PCP between the condensation domain, where it was observed in AB3403 and SrfA-C, and the adenylation domain, as in EntF (Figure 3a).
These two states are also observed in the holo-LgrA structures, which demonstrate the adenylate-forming and thioester-forming conformation for the adenylation domain (Figure 3b). However, the LgrA structures also demonstrate an unexpected role for this C-terminal subdomain [45••]. As the PCP is delivered to the upstream formylation domain, the C-terminal subdomain pulls away from the adenylation domain and into an extended open conformation [45••]. The 180° rotation allows the PCP to translate 50–60 Å to reach the formylation domain. Here, the adenylation C-terminal subdomain is acting in a new capacity as a linker domain. The necessity of different surfaces of this domain to be exposed to solvent in the two primary catalytic states of the adenylation reaction apparently also allows the entire domain to extend away from the core of the protein. This feature could be exploited to deliver the PCP to other internal or external domains.
To date, the relative positions of the two subdomains of the adenylation domain remains the most dramatic movement within an NRPS domain. The two lobes of the condensation domain also exhibit an opening and closing that likely is used to bind and release the pantetheine bound substrates and may position catalytic residues in the active site [18]. As the condensation domain is generally the first domain of an internal NRPS module, this conformational change at the boundary between modules may facilitate delivery of the upstream PCP.
Comparison of the interfaces between the condensation and adenylation N-terminal subdomain in SrfA-C, AB3403, and EntF demonstrate that flexibility also exists between these domains [44••]. This may offer additional dynamic features that enable the delivery of the PCP. Additionally, the dramatically different positions of the thioesterase domains of SrfA-C, AB3403, and EntF, as well as the limited interaction between this domain and the core of the module, suggests that there may be significant flexibility in the linker that joins the PCP and thioesterase domains. This linker could introduce another layer of dynamic flexibility into the NRPS modular architecture that may be exploited in the delivery of the PCP to neighboring domains. From the current structures, the downstream thioesterase domain interacts only weakly with the core of the module and the positions adopted in the structures may be imposed by the constraints of the crystal lattice. In fact, the changes in the relative orientations of the three major domains of EntF were supported by the use of single particle electron microscopy studies of holo-EntF [44••], which adopted multiple states, ranging from linear to compact domain arrangements. Additional structural studies with more NRPS architectures will determine if the limited interactions between a module core and other downstream domains or modules is a common structural feature off all NRPSs.
Seeking a complete understanding of the structural dynamics of NRPSs
While these recent studies have shown great promise toward understanding the structural features that govern the interactions of multidomain NRPSs, important questions remain. Primary among them is the characterization of the remaining states in the catalytic cycle. Superposition of the PCP-thioesterase crystal structure [32] onto the position of the thioesterase domain in AB3403 (Figure 3a) shows that a simple rotation may allow delivery of the peptide to the final thioesterase domain.
An obvious next structural target is of a multimodule protein. The model of the PCP-condensation didomain from TycC [47] in which the PCP pantetheinylation site directed away from the condensation domain served as the foundation for a proposal that the multimodular NRPSs may exist in a helical arrangement in which the condensation and adenylation domains form a central axis around which the remaining domains reside [12•]. Structures of multidomain NRPS proteins that demonstrate the relationship between a module and the downstream domains or modules are needed to support this model and will provide insight into the nature of the additional features that guide the PCP to the downstream module.
This is an exciting time in the understanding of NRPS structural biology, with important new structures appearing regularly that provide clues to the necessary movements that dictate these fascinating assembly line proteins. Understanding the biosynthesis of natural products raises the potential to identify or engineer new biosynthetic pathways for new drugs [48].
Acknowledgments
Work in my lab is supported by grants from the National Institutes of Health (GM116957 and AI116998) and the National Science Foundation (1158169).
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Walsh CT. A chemocentric view of the natural product inventory. Nat Chem Biol. 2015;11:620–624. doi: 10.1038/nchembio.1894. [DOI] [PubMed] [Google Scholar]
- 2.Walsh CT, Fischbach MA. Natural products version 2. 0: connecting genes to molecules. J Am Chem Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crosa JH, Walsh CT. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marahiel MA, Stachelhaus T, Mootz HD. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 5.Wiest A, Grzegorski D, Xu BW, Goulard C, Rebuffat S, Ebbole DJ, Bodo B, Kenerley C. Identification of peptaibols from trichoderma virens and cloning of a peptaibol synthetase. J Biol Chem. 2002;277:20862–20868. doi: 10.1074/jbc.M201654200. [DOI] [PubMed] [Google Scholar]
- 6•.Miller BR, Gulick AM. Structural biology of nonribosomal peptide synthetases. Methods Mol Biol. 2016;1401:3–29. doi: 10.1007/978-1-4939-3375-4_1. This review describes structures of NRPS domains and contains a table that lists all reported crystal structures as of early 2014. The review also highlights strategies that have been used to limit NRPS dynamics to enable structural studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strieker M, Tanovic A, Marahiel MA. Nonribosomal peptide synthetases: structures and dynamics. Curr Opin Struct Biol. 2010;20:234–240. doi: 10.1016/j.sbi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 8.Crosby J, Crump MP. The structural role of the carrier protein — active controller or passive carrier. Nat Prod Rep. 2012;29:1111–1137. doi: 10.1039/c2np20062g. [DOI] [PubMed] [Google Scholar]
- 9.Gulick AM. Conformational dynamics in the acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walsh CT, Chen H, Keating TA, Hubbard BK, Losey HC, Luo L, Marshall CG, Miller DA, Patel HM. Tailoring enzymes that modify nonribosomal peptides during and after chain elongation on NRPS assembly lines. Curr Opin Chem Biol. 2001;5:525–534. doi: 10.1016/s1367-5931(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 11.Labby KJ, Watsula SG, Garneau-Tsodikova S. Interrupted adenylation domains: unique bifunctional enzymes involved in nonribosomal peptide biosynthesis. Nat Prod Rep. 2015;32:641–653. doi: 10.1039/c4np00120f. [DOI] [PubMed] [Google Scholar]
- 12•.Marahiel MA. A structural model for multimodular NRPS assembly lines. Nat Prod Rep. 2016;33:136–140. doi: 10.1039/c5np00082c. This review presents a potential architecture for multimodular NRPSs that are proposed to adopt a helical conformation. [DOI] [PubMed] [Google Scholar]
- 13.Goodrich AC, Harden BJ, Frueh DP. Solution structure of a nonribosomal peptide synthetase carrier protein loaded with its substrate reveals transient, well-defined contacts. J Am Chem Soc. 2015;137:12100–12109. doi: 10.1021/jacs.5b07772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaremko MJ, Lee DJ, Opella SJ, Burkart MD. Structure and substrate sequestration in the pyoluteorin type II peptidyl carrier protein PltL. J Am Chem Soc. 2015;137:11546–11549. doi: 10.1021/jacs.5b04525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Bloudoff K, Alonzo DA, Schmeing TM. Chemical probes allow structural insight into the condensation reaction of nonribosomal peptide synthetases. Cell Chem Biol. 2016;23:331–339. doi: 10.1016/j.chembiol.2016.02.012. The structure of a condensation domain is shown bound to active site probes that were covalently attached to an engineered residue within the pantetheine binding tunnel. [DOI] [PubMed] [Google Scholar]
- 16.Koglin A, Walsh CT. Structural insights into nonribosomal peptide enzymatic assembly lines. Nat Prod Rep. 2009;26:987–1000. doi: 10.1039/b904543k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanovic A, Samel SA, Essen LO, Marahiel MA. Crystal structure of the termination module of a nonribosomal peptide synthetase. Science. 2008;321:659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 18.Bloudoff K, Rodionov D, Schmeing TM. Crystal structures of the first condensation domain of CDA synthetase suggest conformational changes during the synthetic cycle of nonribosomal peptide synthetases. J Mol Biol. 2013;425:3137–3150. doi: 10.1016/j.jmb.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulick AM, Lu X, Dunaway-Mariano D. Crystal structure of 4-chlorobenzoate:Coa ligase/synthetase in the unliganded and aryl substrate-bound states. Biochemistry. 2004;43:8670–8679. doi: 10.1021/bi049384m. [DOI] [PubMed] [Google Scholar]
- 20.Gulick AM, Starai VJ, Horswill AR, Homick KM, Escalante-Semerena JC. The 1. 75 Å crystal structure of acetyl-coa synthetase bound to adenosine-5′-propylphosphate and coenzyme A. Biochemistry. 2003;42:2866–2873. doi: 10.1021/bi0271603. [DOI] [PubMed] [Google Scholar]
- 21.Osman KT, Du L, He Y, Luo Y. Crystal structure of bacillus cereus d-alanyl carrier protein ligase (DltA) in complex with ATP. J Mol Biol. 2009;388:345–355. doi: 10.1016/j.jmb.2009.03.040. [DOI] [PubMed] [Google Scholar]
- 22.Yonus H, Neumann P, Zimmermann S, May JJ, Marahiel MA, Stubbs MT. Crystal structure of DltA. Implications for the reaction mechanism of non-ribosomal peptide synthetase adenylation domains. J Biol Chem. 2008;283:32484–32491. doi: 10.1074/jbc.M800557200. [DOI] [PubMed] [Google Scholar]
- 23.Zettler J, Mootz HD. Biochemical evidence for conformational changes in the cross-talk between adenylation and peptidyl-carrier protein domains of nonribosomal peptide synthetases. FEBS J. 2010;277:1159–1171. doi: 10.1111/j.1742-4658.2009.07551.x. [DOI] [PubMed] [Google Scholar]
- 24.Drake EJ, Nicolai DA, Gulick AM. Structure of the EntB multidomain nonribosomal peptide synthetase and functional analysis of its interaction with the ente adenylation domain. Chem Biol. 2006;13:409–419. doi: 10.1016/j.chembiol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 25.Miller BR, Sundlov JA, Drake EJ, Makin TA, Gulick AM. Analysis of the linker region joining the adenylation and carrier protein domains of the modular nonribosomal peptide synthetases. Proteins. 2014;82:2691–2702. doi: 10.1002/prot.24635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruner SD, Weber T, Kohli RM, Schwarzer D, Marahiel MA, Walsh CT, Stubbs MT. Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE. Structure (Camb) 2002;10:301–310. doi: 10.1016/s0969-2126(02)00716-5. [DOI] [PubMed] [Google Scholar]
- 27.Samel SA, Wagner B, Marahiel MA, Essen LO. The thioesterase domain of the fengycin biosynthesis cluster: a structural base for the macrocyclization of a non-ribosomal lipopeptide. J Mol Biol. 2006;359:876–889. doi: 10.1016/j.jmb.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 28.Kinatukara P, Patel KD, Haque AS, Singh R, Gokhale RS, Sankaranarayananan R. Structural insights into the regulation of NADPH binding to reductase domains of nonribosomal peptide synthetases: a concerted loop movement model. J Struct Biol. 2016;194:368–374. doi: 10.1016/j.jsb.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Wyatt MA, Mok MC, Junop M, Magarvey NA. Heterologous expression and structural characterisation of a pyrazinone natural product assembly line. Chembiochem. 2012;13:2408–2415. doi: 10.1002/cbic.201200340. [DOI] [PubMed] [Google Scholar]
- 30.Barajas JF, Phelan RM, Schaub AJ, Kliewer JT, Kelly PJ, Jackson DR, Luo R, Keasling JD, Tsai SC. Comprehensive structural and biochemical analysis of the terminal myxalamid reductase domain for the engineered production of primary alcohols. Chem Biol. 2015;22:1018–1029. doi: 10.1016/j.chembiol.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 31.Frueh DP, Arthanari H, Koglin A, Vosburg DA, Bennett AE, Walsh CT, Wagner G. Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase. Nature. 2008;454:903–906. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Zheng T, Bruner SD. Structural basis for phosphopantetheinyl carrier domain interactions in the terminal module of nonribosomal peptide synthetases. Chem Biol. 2011;18:1482–1488. doi: 10.1016/j.chembiol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell CA, Shi C, Aldrich CC, Gulick AM. Structure of PA1221; a nonribosomal peptide synthetase containing adenylation and peptidyl carrier protein domains. Biochemistry. 2012;51:3252–3263. doi: 10.1021/bi300112e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sundlov JA, Gulick AM. Structure determination of the functional domain interaction of a chimeric nonribosomal peptide synthetase from a challenging crystal with noncrystallographic translational symmetry. Acta Crystallogr D Biol Crystallogr. 2013;69:1482–1492. doi: 10.1107/S0907444913009372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sundlov JA, Shi C, Wilson DJ, Aldrich CC, Gulick AM. Structural and functional investigation of the intermolecular interaction between NRPS adenylation and carrier protein domains. Chem Biol. 2012;19:188–198. doi: 10.1016/j.chembiol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen WH, Li K, Guntaka NS, Bruner SD. Interdomain and intermodule organization in epimerization domain containing nonribosomal peptide synthetases. ACS Chem Biol. 2016;11:2293–2303. doi: 10.1021/acschembio.6b00332. [DOI] [PubMed] [Google Scholar]
- 37.Baltz RH. MbtH homology codes to identify gifted microbes for genome mining. J Ind Microbiol Biotechnol. 2014;41:357–369. doi: 10.1007/s10295-013-1360-9. [DOI] [PubMed] [Google Scholar]
- 38.Herbst DA, Boll B, Zocher G, Stehle T, Heide L. Structural basis of the interaction of MbtH-like proteins, putative regulators of nonribosomal peptide biosynthesis, with adenylating enzymes. J Biol Chem. 2013;288:1991–2003. doi: 10.1074/jbc.M112.420182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39••.Miller BR, Drake EJ, Shi C, Aldrich CC, Gulick AM. Structures of a nonribosomal peptide synthetase module bound to MbtH-like proteins support a highly dynamic domain architecture. J Biol Chem. 2016 doi: 10.1074/jbc.M116.746297. (in press). The crystal structure of the EntF NRPS module is shown in a complex with two auxilliary MbtH-like proteins. The EntF thioesterase domain adopts a new position compared to previous structures, highlighting the dynamic nature of the NRPS module. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kalb D, Lackner G, Rappe M, Hoffmeister D. Activity of alpha-aminoadipate reductase depends on the N-terminally extending domain. Chembiochem. 2015;16:1426–1430. doi: 10.1002/cbic.201500190. [DOI] [PubMed] [Google Scholar]
- 41.Boll B, Heide L. A domain of RubC1 of rubradirin biosynthesis can functionally replace MbtH-like proteins in tyrosine adenylation. Chembiochem. 2013;14:43–44. doi: 10.1002/cbic.201200633. [DOI] [PubMed] [Google Scholar]
- 42.Haslinger K, Peschke M, Brieke C, Maximowitsch E, Cryle MJ. X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis. Nature. 2015;521:105–109. doi: 10.1038/nature14141. [DOI] [PubMed] [Google Scholar]
- 43.Haslinger K, Brieke C, Uhlmann S, Sieverling L, Sussmuth RD, Cryle MJ. The structure of a transient complex of a nonribosomal peptide synthetase and a cytochrome p450 monooxygenase. Angew Chem Int Ed Engl. 2014;53:8518–8522. doi: 10.1002/anie.201404977. [DOI] [PubMed] [Google Scholar]
- 44••.Drake EJ, Miller BR, Shi C, Tarrasch JT, Sundlov JA, Allen CL, Skiniotis G, Aldrich CC, Gulick AM. Structures of two distinct conformations of holo-non-ribosomal peptide synthetases. Nature. 2016;529:235–238. doi: 10.1038/nature16163. Recent crystal structures of two holo-NRPS modules illustrate how conformational changes of the adenylation domain drive the delivery of the carrier protein between the condensation and adenylation domain active sites. Simultaneous catalytic conformations within these two domains are illustrated in the structure of AB3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Reimer JM, Aloise MN, Harrison PM, Schmeing TM. Synthetic cycle of the initiation module of a formylating nonribosomal peptide synthetase. Nature. 2016;529:239–242. doi: 10.1038/nature16503. Recent crystal structures of the LgrA initiation module illustrate multiple catalytic conformations and the ability of the adenylation C-terminal subdomain to function as a linker domain that extends from the core of the protein to deliver the PCP to the upstream formyltransferase domain. [DOI] [PubMed] [Google Scholar]
- 46.Qiao C, Wilson DJ, Bennett EM, Aldrich CC. A mechanism-based aryl carrier protein/thiolation domain affinity probe. J Am Chem Soc. 2007;129:6350–6351. doi: 10.1021/ja069201e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Samel SA, Schoenafinger G, Knappe TA, Marahiel MA, Essen LO. Structural and functional insights into a peptide bond-forming bidomain from a nonribosomal peptide synthetase. Structure. 2007;15:781–792. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 48.Winn M, Fyans JK, Zhuo Y, Micklefield J. Recent advances in engineering nonribosomal peptide assembly lines. Nat Prod Rep. 2016;33:317–347. doi: 10.1039/c5np00099h. [DOI] [PubMed] [Google Scholar]