Abstract
Simulations are a key tool in molecular ecology for inference and forecasting, as well as for evaluating new methods. Due to growing computational power and a diversity of software with different capabilities, simulations are becoming increasingly powerful and useful. However, the widespread use of simulations by geneticists and ecologists is hindered by difficulties in understanding these softwares’ complex capabilities, composing code and input files, a daunting bioinformatics barrier, and a steep conceptual learning curve. skeleSim (an R package) guides users in choosing appropriate simulations, setting parameters, calculating genetic summary statistics, and organizing data output, in a reproducible pipeline within the R environment. skeleSim is designed to be an extensible framework that can ‘wrap’ around any simulation software (inside or outside the R environment) and be extended to calculate and graph any genetic summary statistics. Currently, skeleSim implements coalescent and forward-time models available in the fastsimcoal2 and rmetasim simulation engines to produce null distributions for multiple population genetic statistics and marker types, under a variety of demographic conditions. skeleSim is intended to make simulations easier while still allowing full model complexity to ensure that simulations play a fundamental role in molecular ecology investigations. skeleSim can also serve as a teaching tool: demonstrating the outcomes of stochastic population genetic processes; teaching general concepts of simulations; and providing an introduction to the R environment with a user-friendly graphical user interface (using shiny).
Keywords: population genetics, simulations, the coalescent, forward-time, conservation genetics, open-source, null model, power-analysis
INTRODUCTION
Simulations of genetic and environmental processes have diverse uses in ecology and evolutionary biology research (Hoban 2014), as well as applications in agriculture and aquaculture, public health and conservation. In the past decade, simulations have been increasingly popular for: inferring the historical processes that resulted in current patterns in molecular data (Marino et al. 2013; Jombart et al. 2014); predicting the molecular genetic outcomes of complex future processes (Hedrick 1995; Bruford et al. 2010); testing the statistical performance of population genetic inference methods under different demographic scenarios (Girod et al. 2011; Hoban et al. 2013a); and evaluating spatial sampling strategies to infer the generating landscape process for spatial genetic patterns (Oyler-McCance et al. 2013; Lotterhos & Whitlock 2015). To generalize, simulations are used to create many in silico genetic datasets of individuals and populations which could have been produced under a given model of a real system. Someone using simulations will often wish to model a range of scenarios, such as the different degrees of hybridization, or different population- or species-divergence times. Summaries of datasets generated under these scenarios can then be compared to quantitatively establish which model is most consistent with real data, to generate hypotheses or predictions, to explore model sensitivity to particular parameters (e.g. population sizes), and to learn about the fundamentals of population genetics or decide on an appropriate sampling strategy, study method, or management approach.
Dozens of software packages that implement simulations of demography, ecology, genetics, spatial processes, behavior, adaptation, interspecies interactions, and more now exist (Hoban et al. 2011; Peng et al. 2013). These software vary in complexity (Carvajal-Rodriguez & Antonio 2008; Hoban et al. 2011), providing a wide range of options that make many simulators highly useful and flexible. Flexibility, however, often comes at a cost: many simulators require substantial investment in learning complex user interfaces and commands, the preparation of custom code and input files, and in-depth immersion and experimentation to explore suitable model space. Also required for the use of any simulator are relatively strong bioinformatics skills to prepare a series of simulation scenarios, produce many genetic datasets, analyze the data for various genetic summary statistics, and organize this output. Despite an increasing number of tutorials, books, workshops, and articles aimed at bioinformatic training for biologists (Haddock & Dunn 2011; Münkemüller et al. 2012), acquiring the knowledge and skills necessary to use simulation software continues to be a barrier for many potential users.
A user-friendly interface and analysis pipeline that guides a user through the steps of setting up a model, choosing analyses, running a simulation, and visualizing results would help circumvent obstacles that prevent population geneticists from using simulations in their research. Although several tools have made progress towards this goal, each has limitations. For instance: MODELER4SIMCOAL2 (Antao et al. 2007) provides a graphical interface to help write simulation input files for simcoal, including complex demographies; and PopPlanner (Ewing & Hermisson 2010; Ewing et al. 2015) is a graphical tool which can be used to construct ms (Hudson 2002) and msms (Ewing & Hermisson 2010) command lines that model various scenarios. The downside of these software, however, is that they are specific to only one simulator, and they only construct the simulations and do not organize or analyze the datafiles. An prime example of an end-to-end solution is LandGenReport (Gruber & Adamack 2015), a comprehensive R package that implements the multiple steps of landscape genetic analysis (see (Segelbacher et al. 2010) or (Manel et al. 2003) for an overview of landscape genetics) in one framework (Manel et al. 2003; Segelbacher et al. 2010; Gruber & Adamack 2015). Other examples of user-friendly genetic-simulation software include: SPOTG (Hoban et al. 2013b) and PowSim (Ryman et al. 2006), which are graphical and command-line interfaces that perform simulations and calculate statistical power of different sampling strategies; and onesamp (Tallmon et al. 2008) that uses coalescent simulations to infer effective population size (Ne). While these software help users analyze results, each is designed for a specific use of simulations, restricting users to certain scenarios and summary statistics. In contrast, coala (Staab & Metzler 2016) is a recent R package that can wrap several coalescent simulators, standardising the input and output files across programs and offering calculation of summary statistics. Such software that enables users to apply a single, convenient framework across different simulators would be very useful. In addition, such a software would ideally be built in an extensible, flexible way to enable the use of any simulator (both coalescent and forward-time), for many applications of simulations, for different genetic markers (eg. sequence, microsatellites, single-nucleotide polymorphisms), and for a variety of current and potentially future genetic analyses.
Here we provide an overview of skeleSim, a new R package that will help molecular ecologists create and use simulations for a wide variety of purposes. We have aimed for maximum flexibility, power, guidance, and user-friendliness. We envision that this software will be used in teaching, research, and applied-science situations, such as biodiversity conservation and management. We implement our software in R because it is freely available, works on all operating systems, has powerful statistical and graphing functions, and is open-source, allowing users to access, modify, and extend code and package capability. R is commonly used in ecology, evolution, and epidemiology across all stages of career development because it offers a supportive learning environment (with provisioning of vignettes and tutorials) and an active and responsive community. These features of R, and a recently developed user-friendly graphical interface, called ‘shiny’ (Chang et al. 2015), will enable molecular ecologists to make use of, and learn with skeleSim regardless of their coding ability or knowledge base.
MATERIALS AND METHODS
Software features
skeleSim is a ‘control panel’ or ‘wrapper’ to enable the use of existing simulation software, without need for the user to create input files or write an informatics pipeline. The process is summarized as follows. A user enters parameters such as population sizes and migration rates, and makes choices regarding demographic events. skeleSim will take these choices and create input files or input code, and then simulate the various scenarios requested by the user. Next, skeleSim will calculate a suite of genetic summary statistics and graphically display these according to user-specified choices. The current version encompasses most widely used statistics in population genetics, with the capacity to add many more, including user-defined statistics. skeleSim will also organize the results into a convenient list object in R, so that statistics, replicates, and scenarios can be easily accessed, subsetted, analyzed, and summarized.
skeleSim has two desirable features that do not exist in current simulation software. First, skeleSim implements a series of automatic validation steps at multiple stages to ensure smooth operation of the simulation engine once the user is ready to run replicates (Figure 1). Classic problems that users of simulation software experience include: small formatting or text errors, incorrect file paths, missing files, and parameters that are incompatible or prevent convergence, all of which can cause the software to crash. These issues take time to identify and resolve. Few simulation software have such extensive internal controls to ensure the user has entered valid parameters to create feasible and realistic models with the possibility of coalescence within basic time and memory limits. Second, we have created an R S4 class for each simulation that will store data, results, and the parameters of all scenarios, facilitating complete documentation of all simulations run.
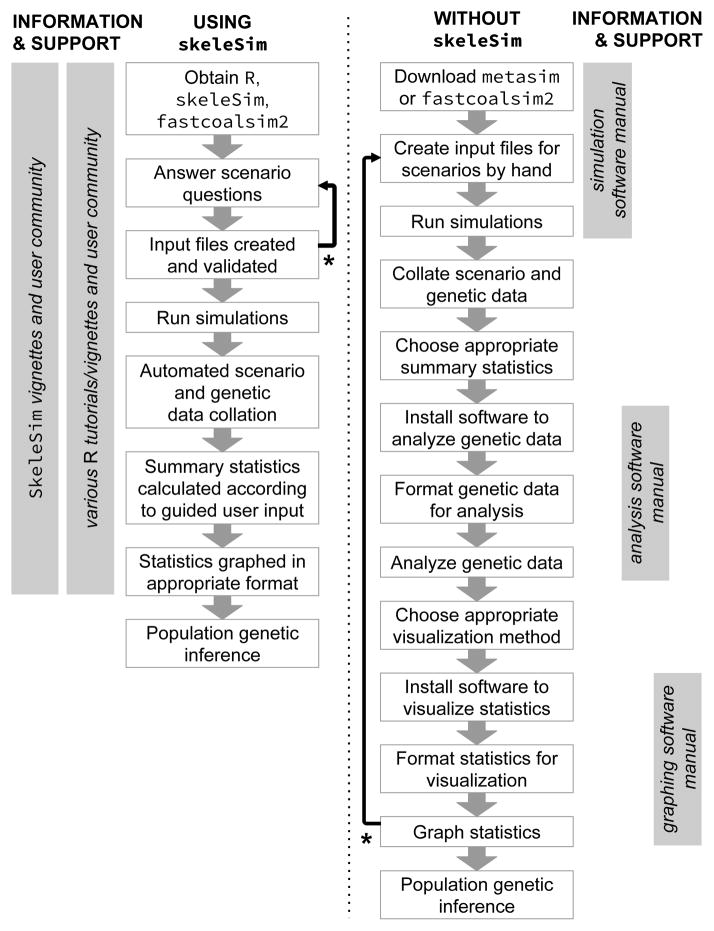
Figure 1. An outline of the steps required to undertake a population genetic simulation study, and the sources of information and support available to the user when using skeleSim versus the standalone metasim or fastsimcoal software.
Using the R platform, skeleSim provides centralized support for the user throughout the decision-making and technical steps involved in a population genetic simulation study. Furthermore, skeleSim enables the validity of a population genetic scenario to be checked early (indicated by * and retrograde arrows), avoiding considerable time investment in unnecessary data formatting, analysis and visualization.
Installation
skeleSim is organized as a standard R package such that normal installation of the package will also install most packages on which it depends, such as strataG, apex, adegenet, pegas, ggplot2 and rmetasim (Jombart et al.; Archer et al. Submitted to Molecular Ecology, 2016; Strand 2002; Jombart 2008; Wickham 2009; Paradis 2010). fastsimcoal2 is a separate executable which must be manually installed (Excoffier & Foll 2011; Excoffier et al. 2013). Links and instructions are provided in the ‘Installing’ vignette in skeleSim.
Currently implemented engines
skeleSim can interface with simulation engines written either as native R packages (e.g. rmetasim), or pre-compiled system executables (e.g. fastsimcoal2), making it flexible and extensible. Currently, we have built skeleSim to support a forward-time simulator (metasim, (Strand 2002)) and a coalescent simulator (fastsimcoal2, (Excoffier et al. 2013)). metasim and fastsimcoal2 are two of the most powerful and widely used simulation software available, and enable complex simulations (Table 1). Both allow simulation of a multiple-population system of arbitrary population sizes and migration rates, through potentially long periods of time (tens to thousands of generations). Also, both software allow multiple types of genetic markers for which the user specifies mutation rates (e.g. sequence data, microsatellite, and SNP data). metasim is especially advantageous because it allows for simulating complex life history and demography, such as multiple life history stages and stage / age specific mortality and reproduction rates. fastsimcoal2 is well suited for complex series of events in history such as bottlenecks, divergence events, and admixture among populations, and allows realistic genomic features such as linkage among markers, long sequences, and recombination rates. Though both software have detailed user guides to help the user navigate their high flexibility and detailed code, there is still a steep learning curve for these software, making them especially suitable for skeleSim. skeleSim helps users to access the functionality of metasim and fastsimcoal2 with little prior knowledge of the features and parameter options they provide. Furthermore, the skeleSim wrapper functions allow more advanced users to construct, run, and analyze complex scenarios with a reproducible pipeline. Thus, skeleSim caters to the wide range of skill levels found within the molecular ecology community, and may facilitate skill development at any stage of a molecular ecologist’s career.
Table 1.
A comparison of the functionality of the simulation software accessed by skeleSim, simcoal and metasim.
| Aspect | metasim | simcoal |
|---|---|---|
| Algorithm focused on: | Individuals | Lineages |
| Population | Population and individual based, migration controlled by migration matrix, migration can be different for each sex or stage | Population and sample based, migration controlled by migration matrix |
| Lifecycle / Stages | User defined number of stages, each with user defined survival rates | Single stage, Wright-Fisher (all individuals live for one time step) |
| Population growth rate | Arises through reproduction matrices, up to a hard carrying capacity at which point individuals are removed at random | User defines exponential growth rate (positive or negative) |
| Mating | Random mating within population; proportion selfing? | Random mating within population |
| Migration | Movement of either male gametes or offspring | Movement of proportion of the population adults |
| “Events” allowed | Change in migration rates, demographic matrices; harder to code, and not currently implemented in skeleSim | Population fission/fusion, change in population sizes & migration rates; very easy to change |
| Mutation rate | Sequence-wide mutation rate, stepwise mutation model for microsatellites | Substitution rate for sequences, stepwise mutation model for microsatellites |
| Recombination among loci | No | Yes |
| Natural selection | None | None |
| Other features of interest | Tracks previous generation pedigree (parent/offspring relations) and population of origin for individuals | Can define linkage between markers and thus construct chromosomal segments |
Choosing between coalescent and forward-time simulation
The user can either directly select a forward-time or coalescent simulation or allow the software to provide guidance on simulator selection. Generally, simulators are classified into one of two categories: coalescent (or backward) and forward-time (Hoban et al. 2011). Most forward-time simulations are individual-based, while coalescent simulators follow genetic lineages backwards in time. As a broad rule, coalescent simulators are suited for organisms with simple life histories, but complex demographic histories (e.g. multiple population divergences, complex changes in population size) and large populations (large meaning effective size of tens of thousands). Forward-time simulations are best suited for organisms in which life history is important (e.g. variance in reproduction among individuals, age structure, age-based migration) and large population sizes and long-time scales are not needed. An additional difference is computational speed: coalescent simulators can produce replicate simulations much faster than forward-time simulators and are typically preferred when complex demography is not required in a simulation. Note that there are also hybrid simulators such as the recent MetaPopGen (Andrello & Manel 2015), which are forward-time but follow genotype frequencies rather than individuals. The distinctions among simulation categories are discussed in detail elsewhere (Carvajal-Rodriguez & Antonio 2008; Liu et al. 2008; Hoban et al. 2011).
Interface
skeleSim is designed primarily to be used in the shiny graphical user interface. Guidance on installing skeleSim and calling skeleSimGUI() is provided in the skeleSim vignette ‘Installing’. The vignette ‘Simulations’ provides an overview of the steps and describes the processes, labelling, and construction of files that occur behind the graphical interface. The interface itself is organized in sequential tabs, each with their own description that guides a user through necessary steps. First, the user may choose whether they wish to run fastsimcoal2 or rmetasim, or the user may receive guidance on this choice through a series of questions (‘Help Choosing Simulator’ tab, e.g. does the simulation require an organism with complex life history, what are the computational and time limits of the user’s system). Next, the user defines: general parameters in the ‘General Conf’ tab, including a title and selecting types of genetic summary statistics; scenarios in the ‘Scenario Conf’ tab, including the number of study populations and the type of locus; and simulator-specific parameters (e.g. either ‘Rmetasim Params’ tab or ‘FastSimCoal Params’ tab currently). In each case, the skeleSim interface presents labelled text boxes and drop-down menus of the required and optional parameters (with some further explanation). As it is common practice to change one parameter per scenario for comparisons, a user can define additional scenarios by simply modifying the first scenario. A given study may examine two or up to dozens of scenarios, depending on its complexity and purposes (see (Hoban et al. 2011) for more guidance on designing scenarios). Once the parameters of each scenario are saved, the user is able to run the simulation. The second-to-last tab of the skeleSim interface is ‘Results’. In this tab, users can upload their simulation output results to quickly visualize the genetic summary statistics for each simulated scenario and compare results among scenarios. The last tab of the interface is ‘Current ssClass’. This tab allows the user to visualise how the skeleSim S4 class object in R is altered by options and operations executed within the interface, helping to familiarize the naive R user with object-oriented coding conventions.
Architecture
All parameters of each scenario and results (including full output of all replicates, and all genetic summary statistics) are contained in a single S4 class object. Users parameterize the object using the shiny web browser interface, or for more experienced users, directly via R code. This object can be saved at any time and re-loaded in the shiny interface, or in any R environment (e.g. to run later on a different personal computer or a server).
All primary functions in skeleSim receive this S4 class object as their single argument, thus providing the function with all information about the simulation. Functions can also add information to this object in predefined slots and return modified versions of the object to the workspace. In this manner, the course of the simulations, from parameter specification to simulation output and analysis, is fully captured. This ensures that the results of analyses will be permanently linked to the parameters and models used to produce the data, a relatively novel feature in simulation software.
Summary statistics
We have included numerous genetic summary statistics within skeleSim to describe simulation outputs. To help guide the selection of appropriate summary statistics, users are presented with suites of summary statistics nested under categories that align with hypotheses relating to: alpha diversity or population-specific measures (‘Locus Statistics’, e.g. number of alleles, m-ratio), beta diversity or population-pairwise measures (‘Pairwise Statistics’, e.g. FST, nucleotide divergence), and global measures (‘Global Statistics’, e.g. global FST). Analysis options can be customised by advanced users, by nesting further summary statistic options under the existing categories, or creating a new category. Routines for calculating population genetic statistics are sourced from existing R packages including strataG (Archer et al. Submitted to Molecular Ecology, 2016) and adegenet (Jombart 2008) that offer interoperability and complementary analysis options for population geneticists. A full list of genetic summary statistics available in the current version of skeleSim are described in Appendix 1.
Example use: Forecasting case study
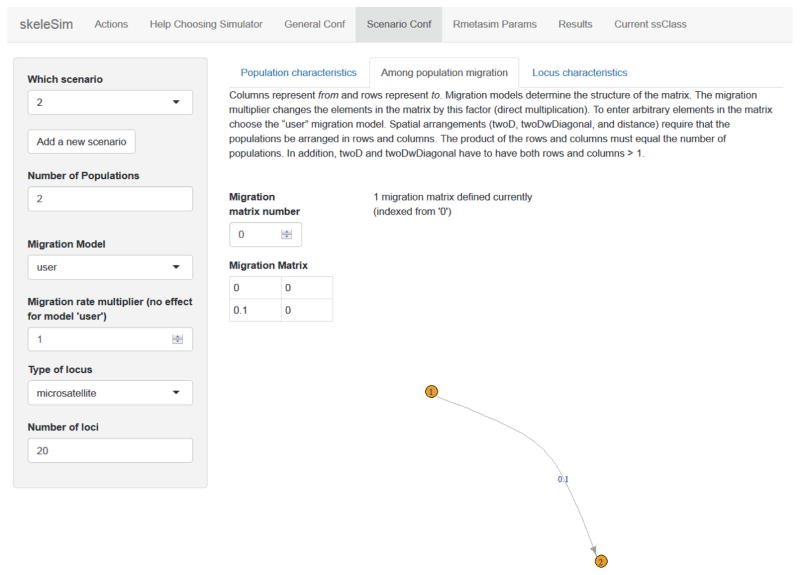
Example case studies for the use of skeleSim are provided as vignettes that are downloaded with the package. This ‘Forecasting’ vignette demonstrates how simulations may be used to forecast possible outcomes of rare-species management. Conservation managers are often faced with decisions about corridors or translocations to link two populations. A user may want to implement a simulation in which two populations of different sizes are either disconnected (Scenario 1) or connected via gene flow (Scenario 2). Using the skeleSim interface, the two scenarios are constructed by the user to differ only by an asymmetric migration rate (0.10) from the larger population to the smaller population in Scenario 2, to mimic translocation by conservation managers (Figure 2). These scenario parameters can be saved and the simulations for both can then be executed simultaneously.
Figure 2. The ‘Scenario Conf’ tab of the skeleSim user interface.
In this tab the simulation scenarios are defined by the user. Migration rate and directionality are defined by the user in the matrix, and a matching population graph is automatically populated in the interface. This population graph corresponds to Scenario 2 of the ‘Forecasting’ example (see main text and skeleSim vignettes).
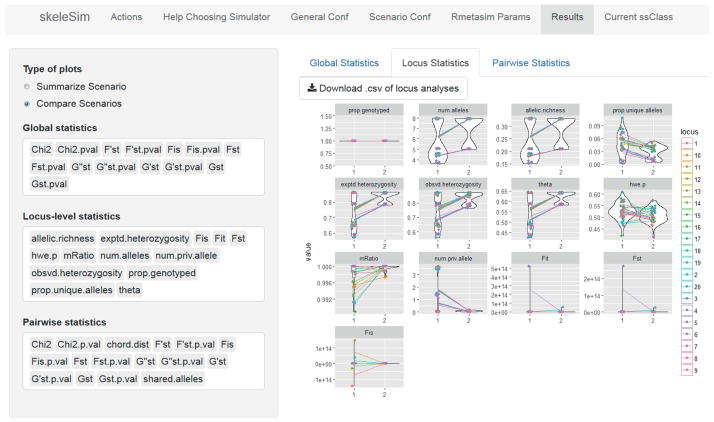
In this example, where one population is quite small and the other large, the user may be interested in whether this level of gene flow (Scenario 2) sufficiently maintains genetic diversity and counteracts drift in the small population, and whether the small population’s unique diversity is swamped by gene flow from the larger population. The results tab of the skeleSim interface allows the user to immediately compare results among scenarios. In this example, the user will quickly see that by comparing the ‘Locus statistics’ for Scenario 1 and 2, that the smaller population has a greater number of alleles (num.alleles), higher allelic richness (allelic.richness), and observed heterozygosity (obsvd.heterozygosity) in Scenario 2, where there is migration from the larger population (Figure 3; see Appendix 1 for further explanation of analyses). However, by observing the ‘Global statistics’ the users will also see that, intuitively, the global population structure (e.g. FST) is reduced in Scenario 2, as a consequence of both the proportion of unique alleles (prop.unique.alleles in ‘Locus Statistics’) in the smaller population being reduced by gene flow, and the number of private alleles (num.priv.alleles in ‘Locus Statistics’) found in Population 1 also being decreased in Scenario 2. Further examples are provided as additional vignettes.
Figure 3. The ‘Results’ tab of the skeleSim user interface.
Following the completion of simulations, users can upload and visualize the results for the genetic summary statistics they elected. In this ‘forecasting’ example (see main text and skeleSim vignettes), the results for the ‘Locus-level statistics’ from having no migration among populations (Scenario 1) and migration from the larger population to the smaller population (Scenario 2) can be observed. The rapid visualization of simulation results enabled by skeleSim facilitates prompt decision-making for conservation, and any subsequent scenario modifications if necessary.
DISCUSSION
skeleSim occupies a unique niche that has long been neglected in simulation software, which is a user-friendly, streamlined interface for the entire simulation process, from setting up the models, to documenting pipelines, to obtaining organized results. It is often difficult for a user to know which parameters are required, whether their code or input files will run successfully, and how to interpret error messages. While some simulation software incorporate built-in analyses (e.g. metasim; cdpop (Landguth & Cushman 2010)), and some have visually accessible interfaces (e.g. Pedagog, (Coombs et al. 2010)), few software have both, and none have been designed and structured in order to facilitate the entire process of using simulations (perhaps with the exception of some Approximate Bayesian Computation packages). Many simulation software are stand-alone programs, which adds an additional step of organizing and importing data into other software, such as R, where one can use statistical and graphical approaches to draw inference from the simulations.
We have designed skeleSim to fill this gap and provide a resource for both teaching and research. As a teaching tool, the skeleSim GUI lends a gentle introduction to the R environment. It also provides a framework for learners to understand and compare the most common genetic markers (currently microsatellite, sequence, and polymorphism data). As a resource for researchers, skeleSim caters to the growing number of molecular ecologists who use R. Executing simulations within the familiar R environment will enable easy cross-application of coding skills and use of learning resources and for a already familiar to this community (Figure 1). Furthermore, by ‘wrapping’ existing software in a common interface with streamlined terminology and inference output, population geneticists and molecular ecologists will be able to confidently switch between simulators. As the field of molecular ecology expands, skeleSim fulfills a need of making software tools and analyses accessible to a wide audience.
skeleSim will lessen the initial time and knowledge needed to start doing simulations. By helping to bring genetic simulators to a wider audience, we ultimately hope simulation tools will be better understood and more widely used in ecology and evolutionary biology. Simulations complement and strengthen empirical investigations at multiple stages, from planning a study to interpreting results to applying models and data in a predictive fashion for forecasting (Hoban 2014). Their use will enable greater power and rigor of studies in molecular ecology (Epperson et al. 2010; Balkenhol & Landguth 2011; Andrew et al. 2013). skeleSim aims to remove the ‘black box’ perception associated with many population genetic and simulation software. skeleSim parameters are permanently attached to the data itself without the need for outside documentation, which is more easily lost or corrupted. The parameters, the data, and the software for running new simulations are thus easily shareable and completely transparent. We see skeleSim as a platform that will make simulations more usable to groups such as conservation practitioners, scientists in public health and agriculture, and educators.
skeleSim is designed to be extended to new situations and simulators. Outside developers can add wrappers for new simulators following the examples set by the wrappers for fastsimcoal and rmetasim. That being said, skeleSim will be most easily extensible to other simulation software that have similar models and parameters to those that we have implemented, such as kernelpop (Strand & Niehaus 2007), cdpop, and nemo (Guillaume & Rougemont 2006), and coalescent software like ms. External developers can also add additional analyses to skeleSim. For example, currently, linkage disequilibrium and estimates of effective population size are not implemented as these analyses are computationally intensive and not as suitable for large-scale simulations. Nonetheless, these may be important outcomes in some studies. The analytical result slot of the skeleSim S4 class object is an R list that is currently structured to hold summary statistics for each population, pairs of populations, and globally (i.e. all populations). New statistics that fit one of those categories can be easily added to the current analytical functions or a new category can be created to store summaries that do not fit these categories, such as linkage disequilibrium.
One aspect not implemented in the current version of skeleSim is natural selection. While future versions of skeleSim may wrap simulators that include natural selection, this release serves as a proof-of-concept package that can wrap different simulators while facilitating the entire simulation process from parameter selection to result visualization. Thus, the current version of skeleSim caters to the most common and tangible uses of population genetic simulations - generating ‘null’ distributions of statistics - the statistics that we expect to occur as a product of the neutral processes of mutation, drift and migration. These null distributions have immense utility for inference of demographics and for identifying ‘outlier’ loci that do not fall within such distributions.
skeleSim is a dynamic package that we hope will grow in capability based on feedback from users and additions from other developers. We have concentrated initial development of the package on helping users to produce null distributions of statistics under scenarios based on varying the most common demographic parameters. In order to rapidly develop this proof-of-concept package, we have not incorporated some useful features, such as introducing genotyping or sequencing error into the simulated data, analyzing empirical user-contributed data alongside simulated data, or allowing a user to specify starting conditions for the simulations, such as the distribution of genotype frequencies. In addition to generating null distributions, we envision expanding skeleSim to include modules to examine the statistical power of various tests as well as to conduct performance testing of analytical methods. Users are encouraged to fork the skeleSim code from GitHub and suggest or contribute to updates, new analyses, and new simulators.
Supplementary Material
Acknowledgments
This project originated at the Population Genetics in R Hackathon, which was held in March 2015 at the National Evolutionary Synthesis Center (NESCent) in Durham, NC, with the goal of addressing interoperability, scalability, and workflow challenges for the population genetics package ecosystem in R. The authors were participants in the hackathon, and are indebted to the event organizers (T. Jombart, S. Manel, E. Paradis, and H. Lapp), other participants, and NESCent (NSF #EF-0905606) for hosting and supporting the event. Ongoing development of this resource was supported by the National Institute of Mathematical and Biological Synthesis (NIMBioS) through a funded short-term visit. CMP was supported by funding from the NIH: T32GM007092 and F30AI109979. LL was supported by an Allan Wilson Centre for Molecular Ecology and Evolution Postdoctoral Fellowship and a Rutherford Foundation New Zealand Postdoctoral Fellowship.
Footnotes
- Source code and current development version available from GitHub (github.com/christianparobek/skeleSim)
- Vignettes ship with the package, and additionally can be accessed in markdown form at (https://github.com/christianparobek/skeleSim/blob/master/vignettes)
- Stable version of skeleSim and vignettes will be available from CRAN
- All authors contributed to the idea conception, coding, testing, and manuscript writing.
References
- Andrello M, Manel S. MetaPopGen: an r package to simulate population genetics in large size metapopulations. Molecular ecology resources. 2015;15:1153–1162. doi: 10.1111/1755-0998.12371. [DOI] [PubMed] [Google Scholar]
- Andrew RL, Bernatchez L, Bonin A, et al. A road map for molecular ecology. Molecular ecology. 2013;22:2605–2626. doi: 10.1111/mec.12319. [DOI] [PubMed] [Google Scholar]
- Antao T, Beja-Pereira A, Luikart G. MODELER4SIMCOAL2: a user-friendly, extensible modeler of demography and linked loci for coalescent simulations. Bioinformatics. 2007;23:1848–1850. doi: 10.1093/bioinformatics/btm243. [DOI] [PubMed] [Google Scholar]
- Archer FI, Adams PE, Schneiders B. strataG: An R package for manipulating, summarizing, and analyzing population genetic data. doi: 10.1111/1755-0998.12559. Submitted to Molecular Ecology, 2016. [DOI] [PubMed]
- Balkenhol N, Landguth EL. Simulation modelling in landscape genetics: on the need to go further. Molecular ecology. 2011;20:667–670. doi: 10.1111/j.1365-294x.2010.04967.x. [DOI] [PubMed] [Google Scholar]
- Bruford MW, Ancrenaz M, Chikhi L, et al. Projecting genetic diversity and population viability for the fragmented orang-utan population in the Kinabatangan floodplain, Sabah, Malaysia. Endangered species research. 2010;12:249–261. [Google Scholar]
- Carvajal-Rodriguez A, Antonio C-R. Simulation of Genomes: A Review. Current genomics. 2008;9:155–159. doi: 10.2174/138920208784340759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Cheng J, Allaire J, Xie Y, McPherson J. Shiny: web application framework for R. 2015. [Google Scholar]
- Coombs JA, Letcher BH, Nislow KH. pedagog: software for simulating eco-evolutionary population dynamics. Molecular ecology resources. 2010;10:558–563. doi: 10.1111/j.1755-0998.2009.02803.x. [DOI] [PubMed] [Google Scholar]
- Epperson BK, McRae BH, Scribner K, et al. Utility of computer simulations in landscape genetics. Molecular ecology. 2010;19:3549–3564. doi: 10.1111/j.1365-294X.2010.04678.x. [DOI] [PubMed] [Google Scholar]
- Ewing G, Hermisson J. MSMS: a coalescent simulation program including recombination, demographic structure and selection at a single locus. Bioinformatics. 2010;26:2064–2065. doi: 10.1093/bioinformatics/btq322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing GB, Reiff PA, Jensen JD. PopPlanner: visually constructing demographic models for simulation. Frontiers in genetics. 2015;6:150. doi: 10.3389/fgene.2015.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Foll M. fastsimcoal: a continuous-time coalescent simulator of genomic diversity under arbitrarily complex evolutionary scenarios. Bioinformatics. 2011;27:1332–1334. doi: 10.1093/bioinformatics/btr124. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laurent E, Isabelle D, et al. Robust Demographic Inference from Genomic and SNP Data. PLoS genetics. 2013;9:e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girod C, Vitalis R, Leblois R, Freville H. Inferring Population Decline and Expansion From Microsatellite Data: A Simulation-Based Evaluation of the Msvar Method. Genetics. 2011;188:165–179. doi: 10.1534/genetics.110.121764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber B, Adamack AT. landgenreport: a new r function to simplify landscape genetic analysis using resistance surface layers. Molecular ecology resources. 2015;15:1172–1178. doi: 10.1111/1755-0998.12381. [DOI] [PubMed] [Google Scholar]
- Guillaume F, Rougemont J. Nemo: an evolutionary and population genetics programming framework. Bioinformatics. 2006;22:2556–2557. doi: 10.1093/bioinformatics/btl415. [DOI] [PubMed] [Google Scholar]
- Haddock SHD, Dunn CW. Practical Computing for Biologists. Sinauer Associates Incorporated; 2011. [Google Scholar]
- Hedrick PW. Gene Flow and Genetic Restoration: The Florida Panther as a Case Study. Conservation biology: the journal of the Society for Conservation Biology. 1995;9:996–1007. doi: 10.1046/j.1523-1739.1995.9050988.x-i1. [DOI] [PubMed] [Google Scholar]
- Hoban S. An overview of the utility of population simulation software in molecular ecology. Molecular ecology. 2014;23:2383–2401. doi: 10.1111/mec.12741. [DOI] [PubMed] [Google Scholar]
- Hoban S, Bertorelle G, Gaggiotti OE. Computer simulations: tools for population and evolutionary genetics. Nature reviews Genetics. 2011;13:110–122. doi: 10.1038/nrg3130. [DOI] [PubMed] [Google Scholar]
- Hoban SM, Gaggiotti OE, Bertorelle G. The number of markers and samples needed for detecting bottlenecks under realistic scenarios, with and without recovery: a simulation-based study. Molecular ecology. 2013a;22:3444–3450. doi: 10.1111/mec.12258. [DOI] [PubMed] [Google Scholar]
- Hoban S, Sean H, Oscar G, Giorgio B ConGRESS Consortium. Sample Planning Optimization Tool for conservation and population Genetics (SPOTG): a software for choosing the appropriate number of markers and samples. Methods in ecology and evolution / British Ecological Society. 2013b;4:299–303. [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Jombart T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- Jombart T, Cori A, Didelot X, et al. Bayesian reconstruction of disease outbreaks by combining epidemiologic and genomic data. PLoS computational biology. 2014;10:e1003457. doi: 10.1371/journal.pcbi.1003457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T, Kamvar ZN, Schliep K, Archer FI, Harris R. apex: Phylogenetic Methods for Multiple Gene Data. [Google Scholar]
- Landguth EL, Cushman SA. cdpop: A spatially explicit cost distance population genetics program. Molecular ecology resources. 2010;10:156–161. doi: 10.1111/j.1755-0998.2009.02719.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Athanasiadis G, Weale ME. A survey of genetic simulation software for population and epidemiological studies. Human genomics. 2008;3:79–86. doi: 10.1186/1479-7364-3-1-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotterhos KE, Whitlock MC. The relative power of genome scans to detect local adaptation depends on sampling design and statistical method. Molecular ecology. 2015;24:1031–1046. doi: 10.1111/mec.13100. [DOI] [PubMed] [Google Scholar]
- Manel S, Stéphanie M, Schwartz MK, Gordon L, Pierre T. Landscape genetics: combining landscape ecology and population genetics. Trends in ecology & evolution. 2003;18:189–197. [Google Scholar]
- Marino IAM, Benazzo A, Agostini C, et al. Evidence for past and present hybridization in three Antarctic icefish species provides new perspectives on an evolutionary radiation. Molecular ecology. 2013;22:5148–5161. doi: 10.1111/mec.12458. [DOI] [PubMed] [Google Scholar]
- Münkemüller T, Tamara M, Sébastien L, et al. How to measure and test phylogenetic signal. Methods in ecology and evolution / British Ecological Society. 2012;3:743–756. [Google Scholar]
- Oyler-McCance SJ, Valdez EW, O’Shea TJ, Fike JA. Genetic characterization of the Pacific sheath-tailed bat (Emballonura semicaudata rotensis) using mitochondrial DNA sequence data. Journal of mammalogy. 2013;94:1030–1036. [Google Scholar]
- Paradis E. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics. 2010;26:419–420. doi: 10.1093/bioinformatics/btp696. [DOI] [PubMed] [Google Scholar]
- Peng B, Chen H-S, Mechanic LE, et al. Genetic Simulation Resources: a website for the registration and discovery of genetic data simulators. Bioinformatics. 2013;29:1101–1102. doi: 10.1093/bioinformatics/btt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryman N, Nils R, Stefan P. POWSIM: a computer program for assessing statistical power when testing for genetic differentiation. Molecular ecology notes. 2006;6:600–602. doi: 10.1046/j.0962-1083.2001.01345.x. [DOI] [PubMed] [Google Scholar]
- Segelbacher G, Gernot S, Cushman SA, et al. Applications of landscape genetics in conservation biology: concepts and challenges. Conservation genetics. 2010;11:375–385. [Google Scholar]
- Staab PR, Metzler D. Coala: an R framework for coalescent simulation. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw098. [DOI] [PubMed] [Google Scholar]
- Strand AE. metasim 1.0: an individual-based environment for simulating population genetics of complex population dynamics. Molecular ecology notes. 2002;2:373–376. [Google Scholar]
- Strand AE, Niehaus JM. kernelpop, a spatially explicit population genetic simulation engine. Molecular ecology notes. 2007;7:969–973. [Google Scholar]
- Tallmon DA, Koyuk A, Luikart G, Beaumont MA. COMPUTER PROGRAMS: onesamp: a program to estimate effective population size using approximate Bayesian computation. Molecular ecology resources. 2008;8:299–301. doi: 10.1111/j.1471-8286.2007.01997.x. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.