Figure 4. pH sensing in BinAB crystals.
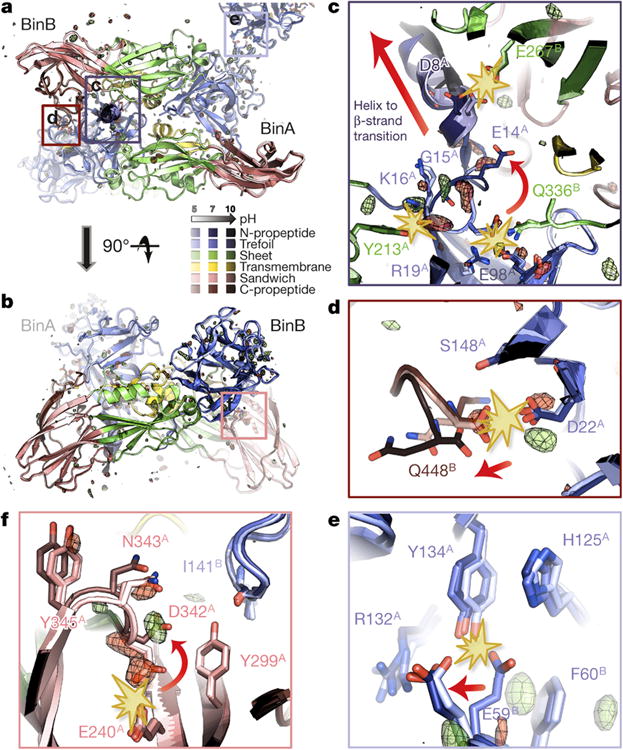
a-f, BinA and BinB cartoons are coloured by subdomain. The pH 5, pH 7 and pH 10 structures are increasingly shaded. Symmetry-related molecules are coloured similarly. The Fo[pH10] - Fo[pH7] map is superimposed at ± 3.5 σ, with positive and negative peaks shown in green and red, respectively. Panels (a) and (b) show orthogonal views of the BinAB dimer. The Fo-Fo map reveals four regions (c, d, e, f) of the BinAB crystal that are perturbed by elevated pH, probably reflecting early events in crystal dissolution. Rupture of H-bonds and conformational changes are highlighted by starbursts and arrows, respectively. (c); Deprotonation of Asp8, Glu14 and Tyr213 triggers a helix to extended β-strand conformational change in BinA N-term propeptide, increasing its accessibility to proteolytic cleavage. (d); Deprotonation of BinB Gln448 terminal carboxylate breaks its H-bond with BinA Asp22, weakening the BinA-BinB dimer interface and making the C-terminal propeptide of BinB available for proteolysis. (e); Deprotonation of BinA Tyr134 and His125 results in the rupture of their H-bonds with BinB Glu59. Loss of these contacts directly weakens the lattice. (f); Negative electrostatic repulsion pushes Asp342 away from Glu240 in BinA. This rearrangement could be an early step in the transformation into a pore owing to Asp342's location at the junction between the three PFD subdomains.
