Abstract
Glucan phosphatases are functionally conserved at the enzymatic level, dephosphorylating glycogen in animals and starch in plants. The human glucan phosphatase laforin is the founding member of the family and it is comprised of a carbohydrate binding module (CBM) domain followed by a dual specificity phosphatase (DSP) domain. Plants encode for two glucan phosphatases: Starch EXcess4 (SEX4) and Like Sex Four2 (LSF2). SEX4 contains a DSP domain followed by a CBM domain, while LSF2 contains a DSP domain and lacks a CBM. This review demonstrates how glucan phosphatase function is conserved and highlights how each family member employs a unique mechanism to bind and dephosphorylate glucan substrates.
Introduction
Animals and plants both utilize glucans comprised of α-1,4 linked glucose chains with α-1,6 branch points as energy storage macromolecules. Glycogen is water-soluble and is utilized by opisthokonts (e.g. animals and fungi) and most prokaryotes, while starch is water-insoluble and is utilized by Archaeplastida (e.g. algae and land plants) and eukaryotes with Archaeplastida-derived plastids. Glycogen and starch each can be enzymatically modified by the addition of phosphate and this phosphoryl group links research on a human epilepsy with plant starch metabolism.
Glycogen and Lafora disease
Glycogen, the major mammalian storage carbohydrate, has a critical function in energy metabolism [1]. Glycogen is typically modeled with an average of 13 glucose residues per chain and successive layers of α-1,6 branches up to 12 tiers total (Figure 1A). This unique organization allows cells to store up to ≈55,000 glucose units/molecule in a water-soluble form allowing rapid glucose mobilization. Recent discoveries from the study of Lafora disease have enabled identification of glycogen phosphorylation as an important regulatory mechanism with direct implications for human disease.
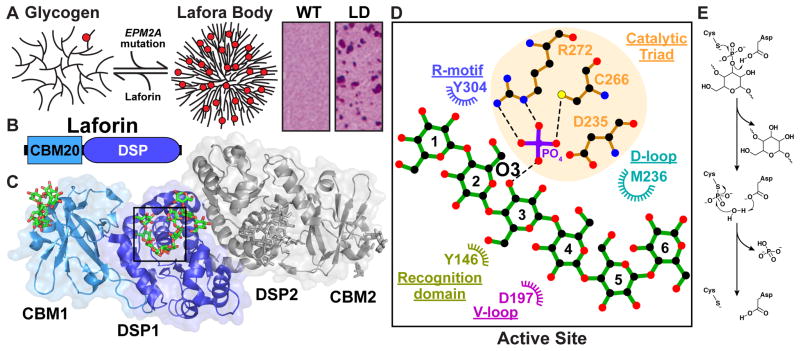
Figure 1.
(a) Glycogen model depicting 4 of the possible 12 tiers. Red spheres represent phosphate moieties. Laforin is a glycogen phosphatase encoded by the EPM2A gene. Mutations in EPM2A result in the hyperphosphorylation of glycogen that changes the normal branching pattern of glycogen and results in a Lafora body (LB). LBs are inclusions present in neurons of LD patients and stain Periodic acid-Schiff (PAS) positive. The micrographs are PAS stain slices of wildtype (WT) and Epm2a−/− (LD) mice hippocampus. Modified from [58] with permission. (b) Laforin is composed of an N-terminal carbohydrate binding module (CBM) and a C-terminal dual-specificity phosphatase (DSP) domain. (c) The crystal structure of the laforin dimer bound to maltohexaose and phosphate (PDB: 4RKK). The CBM and DSP of one subunit are shown in light blue and dark blue, respectively; the second subunit is shown in grey. The quaternary arrangement of the two subunits results in a CBM-DSP-DSP-CBM architecture. Structure figures were generated using Pymol [59]. (d) Ligplot+ generated representation of hydrophobic and hydrophilic protein-glucan interactions at the DSP domain active site [60]. Maltohexaose is shown in green and hydrogen bonds are shown in black. Glucose moieties are numbered from the non-reducing end to the reducing end. Shared glucan phosphatase-family DSP motifs contributing to glucan binding and their corresponding key residues are displayed. Multiple elements of the laforin DSP domain contribute to glucan binding at the active site. Both the recognition domain and the variable loop (V-loop) interact with the convex side of the glucan chain with residues of the R-motif, catalytic triad, and D-loop all interacting with the concave face. (e) The reaction mechanism for glucan dephosphorylation is proposed based on the well-characterized mechanism of PTPs [8,61]. The first step of catalysis involves nucleophilic attack by the sulfur atom of the active site Cys, which is believed to exist in thiolate form. The highly conserved Asp functions as a general acid, donating its proton to the substrate leaving group, and a cysteinyl-phosphate intermediate is formed. In the second step of catalysis, the same Asp functions as a a general base, accepting a proton from an attacking water molecule, which hydrolyzes the phosphoenzyme intermediate.
In 1911, Gonzalo Lafora, a student of Cajal and Alzheimer, reported cases of teen-age myoclonus epilepsy with dementia and elegantly described “amyloid bodies” in patient neurons (Figure 1A) [2,3]. While amyloid now is used to refer to proteinaceous deposits, it originally referred to any material that stained in a manner similar to starch. The disease identified by Lafora was later called Lafora disease (LD) and the starch-like inclusions Lafora bodies (LBs).
LD is a fatal, autosomal recessive, neurodegenerative epilepsy. The majority of cases are caused by mutations in the epilepsy, progressive myoclonus 2A (EPM2A) gene [4]. EPM2A encodes the bimodular protein laforin that contains a carbohydrate binding module-family 20 (CBM20) domain followed by a dual specific phosphatase (DSP) domain (Figure 1B) [4–6]. CBMs are a divergent family of domains that allow proteins to bind an array of different carbohydrates with the CBM20 family being specific for glycogen and starch [7]. DSPs belong to the protein tyrosine phosphatase (PTP) superfamily, a family of ~105 phosphatases that dephosphorylate proteinaceous or non-proteinaceous substrates regulating multiple signaling cascades [8]. Driven by the observation that many of the enzymes involved in glycogen metabolism are regulated by reversible phosphorylation, including the 1992 Nobel Prize for the discovery of reversible phosphorylation of glycogen phosphorylase, it was initially hypothesized that laforin’s function was to dephosphorylate an enzyme(s) involved in glycogen metabolism.
Surprisingly, insights from starch metabolism led to the discovery of laforin’s function. Austin and coworkers demonstrated that LBs are glucose polymers more similar to starch than glycogen, and identified ~10-fold more phosphate in LBs than in normal glycogen [9,10]. Dixon and colleagues were struck by the realization of the aberrant glycogen metabolism in LD patients, the increased phosphate on LBs, and that the mutated EPM2A gene encoded a phosphatase with an unknown substrate. They demonstrated that laforin could remove phosphate from glucans and hypothesized that laforin is a glycogen phosphatase [11]. Subsequent work demonstrated that laforin does remove phosphate from glycogen, that glycogen becomes hyperphosphorylated in the absence of laforin activity, that this hyperphosphorylated glycogen becomes a LB, and that LBs are the pathogenic cause of LD (Figure 1A) [12–16]. Recent structural studies have elucidated the basis for the function of laforin as a glucan-specific phosphatase, and provided important insights into the molecular basis of LD.
Laforin possesses a cooperative tetramodular glucan-binding platform
We determined the structure of the human laforin dimer bound to maltohexaose and phosphate to 2.4 Å (Figure 1C) [17]. Maltohexaose is a linear, six-glucose polysaccharide with α-1,4 glycosidic linkages, representative of a linear glycogen chain. Laforin engages a maltohexaose chain within its CBM and DSP domain active site via aromatic and hydrophilic residues.
The laforin CBM and DSP domains form an integrated tertiary structure with >1200 Å2 of interfacial surface area. This interdomain interface is a hot spot for disease mutations, suggesting domain coupling is critical for laforin function. Further, laforin was analyzed by multiple biophysical techniques to confirm that it is a dimer in solution. Laforin forms an antiparallel dimer with a tetramodular CBM-DSP-DSP-CBM architecture (Figure 1C). The four glucan binding sites are oriented on one face of the dimer. Binding experiments demonstrated that shorter oligosaccharide chains, such as maltohexaose, bind without any observed cooperativity, consistent with the multiple distinct chains bound in the crystal structure. Strikingly, longer chains showed a single cooperative high-affinity binding. Importantly, the disease mutation (laforin-F321S) was shown to disrupt dimerization and abolish cooperative glucan binding. Thus, structural insights allowed definition of the molecular basis for laforin substrate binding and the important role of the unique laforin tertiary and quaternary structure, allowing it to function as a specific phosphoglucan phosphatase.
The glucan bound at the active site is enveloped by DSP domain motifs that coordinate binding to the phosphoglucan product (Figure 1D). DSP catalytic domains are ≈150 residues and are defined by several conserved motifs with the signature catalytic triad DX30CX5R forming the phosphate-binding site [18]. The catalytic Cys (laforin C266) sits at the base of the active site cleft and functions as a nucleophile during catalysis (Figure 1E). The phosphate is stabilized by hydrogen bonds to a conserved arginine (R272) with D235 serving as the general acid/base allowing formation and cleavage of enzyme-phosphoryl intermediate. The laforin DSP motifs possess aromatic and hydrophilic residues that create an active site channel that binds a glucan chain and positions it for dephosphorylation.
Reversible phosphorylation of transitory starch
In plants, starch phosphorylation was first suggested in 1890 and it was later shown that the concentration of starch phosphate depends on the plant origin, with tuber starch containing 30-fold more phosphate than cereal starch [19–21]. The biological function of this phosphate was realized with the discovery of a starch phosphorylating enzyme called Glucan, Water Dikinase (GWD/SEX1/R1) [22–26]. Arabidopsis plants mutated in the gene encoding GWD have a Starch EXcess phenotype since they synthesize starch, but the starch is not efficiently degraded [25,26].
While starch and glycogen are both comprised of similar glycosidic-linked glucose, the frequency of branching and chain length are quite different, with longer chains and fewer branches, yielding water-insoluble starch. Starch is composed of α-1,4 linked amylose that contains few branches and amylopectin that contains α-1,6 linked branch points, but the amylopectin chains are longer and contain fewer branches than in glycogen [27,28]. It is hypothesized that adjacent amylopectin glucose chains interact to form helices and organize into crystalline lamellae, resulting in the expulsion of water, and together with amylose make starch an energy-dense and stable glucose storage molecule [27,28]. Transitory starch is synthesized during the day and degraded during the night, however its crystalline nature hinders breakdown by glycolytic amylases. This crystallinity prevents starch catabolism and requires a molecular mechanism for amylases to release the stored energy cache.
Starch phosphate was initially considered a contaminant or metabolic mistake, but later found to be covalently attached to only the amylopectin fraction [29]. It is now understood that plants utilize reversible glucan phosphorylation as a key mechanism to modulate the helical nature of starch surface glucan chains [30,31]. Similar screens that identified GWD also identified the Starch EXcess4 (SEX4) gene that we subsequently demonstrated encodes a glucan phosphatase (Figure 2A) [26,32,33].
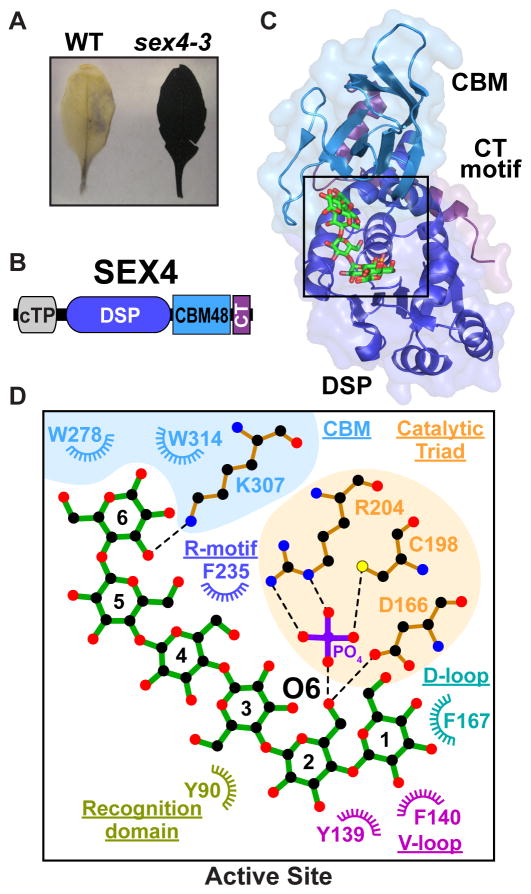
Figure 2.
(a) Arabidopsis leaves stained with iodine to reveal starch excess phenotypes. While wild-type plants contain little transitory starch at the end of night and do not stain, leaves of sex4-3 plants stain darkly, indicating high starch content. (b) SEX4 contains a chloroplast-targeting peptide (cTP) at its N-terminus, followed by a DSP domain, CBM, and a C-terminal (CT) motif. (c) The crystal structure of SEX4 lacking the cTP (residues 1–89) bound to maltoheptaose and phosphate (PDB: 4PYH). SBS, surface binding site. (d) Ligplot+ generated representation of protein-glucan interactions at the SEX4 active site. Glucose moieties are numbered from the non-reducing end to the reducing end. The four glucose moieties at the non-reducing end of the glucan chain interact with key residues of the DSP domain with the recognition domain, variable loop (V-loop), D-loop, catalytic triad and R-motif all providing important contacts. SEX4 has an integrated active site bridging DSP and CBM domains and the two glucose moieties at the reducing end interact with key residues of the CBM domain, which is highlighted with a blue background.
SEX4 – integrated DSP-CBM glucan binding platform
SEX4 is comprised of a chloroplast targeting peptide (cTP), a DSP domain, a CBM48, and a carboxy-terminal (CT) motif (Figure 2B). We determined the structure of Arabidopsis SEX4 bound to phosphate (2.4 Å) and maltoheptaose and phosphate (1.65 Å) (Figure 2C) [34,35]. The bound maltoheptaose and phosphate at the SEX4 active site represent the SEX4 glucan product. The SEX4 DSP and CBM domains interact with 457 Å2 of interfacial surface area via multiple highly conserved motifs. Intriguingly, the SEX4 tertiary structure closely juxtaposes the CBM and DSP glucan binding sites, forming a continuous substrate binding surface involving both domains. This binding surface encompasses aromatic and hydrophilic residues from the CBM and a number of aromatic residues within the DSP domain that form a glucan-binding platform unique for SEX4 (Figure 2D). The SEX4 DSP domain utilizes a number of DSP motif residues to coordinate glucan binding at the active site, but this coordination is dependent on CBM residues. Mutation of CBM residues W278 or W314 to Ala dramatically decreased SEX4 binding to starch and abolished SEX4 starch dephosphorylation [34]. Despite these differences, there are commonalities within the DSP domains of laforin and SEX4 that interact with the glucan, with similar residues from multiple phosphatase signature motifs directly contributing to specific glucan engagement in both proteins (Figure 2D).
Multiple glucan dikinases and phosphatases
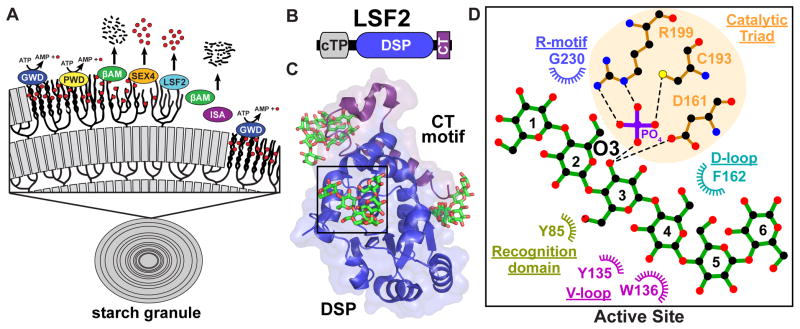
After the discovery of GWD, a second starch dikinase was discovered that only acted on phosphorylated starch and was called phosphoglucan, water dikinase (PWD/GWD3) [36,37]. Elegant biochemical experiments demonstrated that GWD phosphorylates the hydroxyl group at the C6 position and this phosphorylation triggers PWD to phosphorylate the C3 position of starch glucose units [23,36,37]. This phosphorylation introduces steric hindrance that disrupts the amylopectin helices and results in localized solubilization of the starch surface glucans (Figure 3A) [38–40]. Once solubilized, classes of amylases efficiently release maltose and glucose for transport into the cytoplasm [41,42]. While glucan phosphorylation is required for efficient starch degradation, phosphate moieties present an additional problem to overcome. β-amylases (BAMs), the primary starch glycolytic enzymes, degrade glucose chains up to a phosphate group, but BAMs are unable to continue past a phosphate [43]. Thus, glucan phosphatase activity is needed to remove the phosphate and reset this cycle (Figure 3A).
Figure 3.
(a) Transitory starch breakdown requires the action of dikinases, phosphatases, and amylases. Glucan, water dikinase (GWD) transfers the β-phosphate of ATP to the C6 position of glucose moieties within the starch lamellae. Phosphoglucan, water dikinase (PWD) then phosphorylates the C3 position of glucose residues. Phosphorylation leads to the disruption of the helices and solubilization of the starch surface, giving β-amylases (βAM) access to degrade the linear chains. SEX4 and LSF2 then remove the C6- and C3-bound phosphate. Dephosphorylation permits β-amylases to continue to degrade the linear chains. Isoamylase (ISA) is required for debranching within the amorphous layer. The cycle resets at the next layer of the starch granule. Red circles represent phosphates. Black dashes and lines represent maltose and glucose. (b) LSF2 is comprised of a chloroplast-targeting peptide (cTP), DSP domain and carboxy-terminal motif (CT), but lacks a CBM. (c) The crystal structure of LSF2 bound to maltohexaose and phosphate (PDB: 4KYR). In addition to phosphate, a single maltohexaose molecule was found at the active site (shown in green). Maltohexaose molecules were also found at two surface binding sites (SBSs) distant from the LSF2 active site. (d) Ligplot+ generated representations of protein-glucan interactions at the LSF2 active site. Glucose moieties are numbered from the non-reducing end to the reducing end. The glucan chain interacts with key residues of the DSP domain with the recognition domain, variable loop (V-loop), D-loop, catalytic triad and R-motif all providing important contacts.
Like Sex Four2 – surface binding sites
The characterization of SEX4 led to the discovery of an additional plant glucan phosphatase called Like Sex Four2 (LSF2) [44]. LSF2 is unique in that it is comprised of only a cTP, a DSP domain, and CT motif (Figure 3B). This CT motif is a conserved feature unique to plant glucan phosphatases. It forms a conserved α-helical sub-domain that integrates into the DSP domain and is essential for protein folding, stability, and function [34,35]. Although LSF2 lacks a CBM domain, it still efficiently binds and dephosphorylates starch [44]. The structures of Arabidopsis LSF2 revealed that LSF2 engages a glucan chain at the DSP active site and additional chains via two surface binding sites (SBSs) nearly 20 Å away from the active site (Figure 3C) [45]. SBSs are an emerging theme in carbohydrate binding proteins and have, for example, been shown to have diverse critical roles in amylase function [46,47]. SBSs are difficult to identify via bioinformatics due to their heterogeneity and are typically identified via structural studies. The LSF2 active site and two SBSs each utilize aromatic and hydrophilic residues to coordinate glucan binding. Combinatorial mutations of the active site and SBSs showed additive decreases in LSF2 starch binding and dephosphorylation, indicating that the sites function synergistically [45,48]. The specific role(s) of the multiple SBSs of LSF2 is an active area of research, the observed synergistic function could be due to altered dynamics from proximity effects, distinct binding preferences, or global effects on glucan stability. The LSF2 active site is similar to laforin in that the DSP domain alone coordinates glucan binding using many of the same elements found in both laforin and SEX4 (Figure 3D). These elements combine to form an aromatic channel within an extended phosphatase active site to position the glucan for dephosphorylation.
Glucan phosphatase active site topology and substrate specificity
The DSP clade, containing the glucan phosphatases, is very sequence divergent at the amino acid level yet DSPs maintain a prototypical αβα PTP fold [18]. Tyrosine-specific PTPs utilize a narrow and deep catalytic cleft to accommodate pTyr and to exclude shorter side chains (Figure 4A). The catalytic cleft of DSPs that dephosphorylate pTyr, pSer, and pThr is shallow to accommodate both short and long side chains. Alternatively, the lipid phosphatases and glucan phosphatases possess a hybrid active site to accommodate the unique phospholipid or phosphoglucan (Figure 4A). Each glucan phosphatase examined to date possesses conserved structural elements with an active site rich in aromatic and hydrophilic residues uniquely structured to accommodate glucan substrates [17,34,35,45,49–51]. Further, the glucan phosphatase CX5R active site loop possesses a signature motif of Cys, hydrophilic, Ala, Gly, long chain aliphatic, Gly, Arg (CζAGψGR), specific to glucan phosphatases (Figure 4A) [49].
Figure 4.
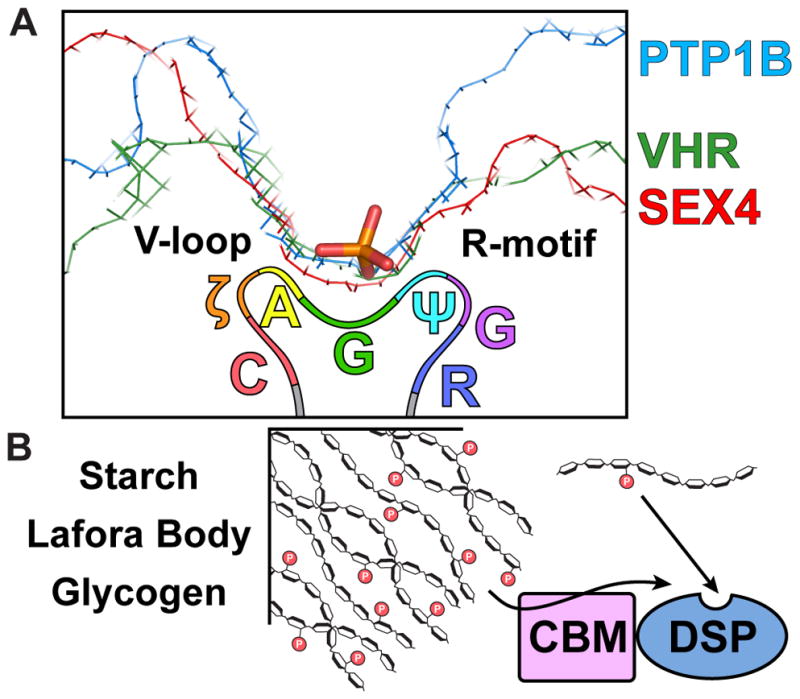
(a) The tyrosine phosphatase PTP1B (PDB: 2HNP) displays a narrow and deep catalytic cleft for dephosphorylating pTyr and excluding pSer/pThr [62]. The human DSP VHR (PDB: 1VHR) possesses a narrow and shallow catalytic cleft for dephosphorylating pTyr and pSer/pThr [63]. The glucan phosphatase SEX4 (PDB: 4PHY) has a hybrid catalytic cleft suited for phosphoglucan substrates [34]. Phosphate is displayed in orange. The variable loop (V-loop) and R-motif are DSP domain elements that flank the active site cleft. The active site consensus motif for a glucan phosphatase is depicted below the active site. This consensus motif is: Cys, hydrophilic, Ala, Gly, long chain aliphatic, Gly, Arg (CζAGψGR). (b) Soluble phospho-oligosaccharides are dephosphorylated directly by the DSP domain. Complex polyglucan substrates (starch and glycogen) must be engaged by an ancillary binding platform (i.e. CBM or SBS) so that dephosphorylation can occur by the DSP domain.
While the active site topology of the glucan phosphatases has common elements, they have key differences in the orientation of the glucan in their active site that determines their position specific activity. Structural data have defined a physical basis for the observed position specificity. Examination of the glucan phosphate in the laforin active site demonstrated that the O2 and O3 groups of Glc3 are 3.8 Å and 2.7 Å from the phosphate in the laforin active site with the O6 position 7.7 Å away, consistent with the observed O3-specific activity of laforin (Figure 1D) [17,49]. Conversely, in the SEX4 DSP active site, the O6 hydroxyl of Glc2 interacts with phosphate at a distance of 2.6 Å with the O3 hydroxyl 7.1 Å away and points into the solvent, consistent with the observed O6-specific activity of SEX4 (Figure 2D) [34]. At the LSF2 active site, the O3 hydroxyl of Glc3 directly interacts with the phosphate at a distance of 2.4 Å compared to 7.0 Å for the O6 position, consistent with the observed O3-specific activity of LSF2 (Figure 3D) [45]. Further, the conformation and orientation of the glucan in the active site is significantly different, with laforin displaying a highly curved glucan conformation while SEX4 and LSF2 display a more helical glucan conformation but with opposite orientations [50,51]. Thus, each glucan phosphatase engages phospho-glucans at their active site in an orientation-specific manner, providing a physical mechanism for their position-specific activity.
Microenvironments of soluble and insoluble glucans
Structure guided mutagenesis revealed that glucan-binding residues within the CBM domain of laforin or SEX4 are largely responsible for glucan engagement and residues within the DSP domain are essential in defining specificity independent of the CBM [17,34]. LSF2 lacks a CBM domain, instead employing SBSs for starch binding, and its DSP domain also determines substrate specificity [45].
These mutagenesis studies coupled with biochemical assays also uncovered that glucan phosphatase activity is different depending on the glucan in question. As highlighted above, there is a gradation of water solubility among glucans with oligosaccharides being most water-soluble, then glycogen, amylopectin and starch being water-insoluble. In laforin, SEX4, and LSF2 the DSP domain is necessary and sufficient to dephosphorylate water-soluble phosphopolyglucans, e.g. oligosaccharides and amylopectin (Figure 4B) [49,52]. These glucans lack the higher-ordered crystalline structure of starch and instead the phosphoglucose within them is freely accessible in solution to the glucan phosphatase active site. Alternatively, ancillary binding domains, either a CBM domain or SBSs, are necessary for the activity against more insoluble glucans and these domains are necessary for dephosphorylation [49].
Conclusions
Reversible glucan phosphorylation has broad biological importance, from a role in a fatal, neurodegenerative epilepsy to starch metabolism. Misregulation of glucan (de)phosphorylation in humans and plants both results in the formation of water insoluble non-degradable glucans. There are striking similarities and differences between the animal and plant pathways as well as among the glucan phosphatases. Intriguingly, the starch excess phenotype used to identify plant dikinases and glucan phosphatases is analogous to the cellular phenotype of LBs in LD patients. Indeed, human laforin could partially compensate for the starch excess phenotype in sex4-mutant plants [32]. The glucan phosphatases are all capable of dephosphorylating phosphoglucans utilizing similar active site topology, but they utilize different binding platforms to engage the glucan chains to obtain different substrate specificities.
Our understanding of the molecular basis for reversible starch phosphorylation in plants has made significant progress with the structural studies of SEX4 and LSF2 bound to simple model substrates. Future studies are key to defining the function of the glucan phosphatases in the heterogeneous microenvironments of their natural substrates, including long and short oligosaccharide chains, phosphorylated and non-phosphorylated chains, branch points, and helical structure. While glucan phosphatases have been extensively studied, little is known regarding the function of glucan dikinases on a molecular level. They possess multiple CBM domains and a unique dikinase domain. Further, while GWD clearly functions during starch degradation, recent studies have highlighted a role for GWD in starch synthesis, though the exact function is unclear. Additionally, it is not known when during the diurnal cycle starch dikinases and phosphatases are active or inactive, nor is it known how these activities are regulated. Further studies on the molecular basis for enzymes that phosphorylate and dephosphorylate glucan substrates are important for defining their function and specificity. Active research is focusing on identifying additional glucan kinase and phosphatase enzymes in all kingdoms of life. Additionally, molecular studies not only inform on the underlying biology, but also open new avenues to utilize this important chemical modification of glucans for industrial application.
Less is known regarding the regulation and function of glycogen phosphorylation. This fact is surprising given that glycogen metabolism is a mainstay in biochemistry textbooks and four Nobel Prizes have been awarded in the glycogen field [1]. Glycogen clearly contains monoester phosphate on the C2, C3, and C6 positions and laforin is definitively a glycogen phosphatase, but the origin of that phosphate is unknown [53–55]. Some suggest that C2 and perhaps C3 phosphate is due to a metabolic error by glycogen synthase [54,56,57]. Alternatively, others have proposed that a kinase similar to GWD phosphorylates glycogen, but no candidate has been reported [53]. Whatever the source of the phosphate, it is clear that glycogen hyperphosphorylation is pathological and results in LD. Intriguingly, increasing evidence indicates that glycogen phosphorylation may also have an important physiological role [62]. It is possible that glycogen phosphorylation on different hydroxyls may have distinct physical and functional roles, similar to phosphoinositols where different phosphorylation positions signal different fates or downstream signaling events.
Highlights.
Glucan phosphatase activity is conserved in plants and animals.
Mutations in the human glucan phosphatase laforin result in a fatal disease.
Laforin utilizes glucan-binding platforms in the CBM and DSP domains.
SEX4 employs an integrated CBM-DSP glucan binding channel to dephosphorylate starch.
LSF2 uses surface binding sites and the DSP active site to dephosphorylate starch.
Acknowledgments
Research reported in this publication was supported by the National Institute of Neurological Disorders And Stroke of the National Institutes of Health under Award Numbers R01NS070899 and P01NS097197, Kentucky Science and Engineering Foundation grants KSEF-2268RDE-014 and KSEF-2971-RDE-017, a Mitzutani Foundation for Glycoscience award, and National Science Foundation grants IIA-1355438 and MCB-1252345. The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies. M.S.G. and C.W.V.K. are founders of OptiMol Enzymes LLC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS. Glycogen and its metabolism: some new developments and old themes. Biochem J. 2012;441:763–787. doi: 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lafora GR. Uber des Vorkommen amyloider KJrperchen im innern der Ganglienzellen. Virchows Arch f Path Anat. 1911;205:295. [Google Scholar]
- 3.Lafora GR, Gluck B. Beitrag zur histopathologie der myoklonischen epilepsie. Z Ges Neurol Psychiatr. 1911;6:1–14. [Google Scholar]
- 4.Gentry MS, Dixon JE, Worby CA. Lafora disease: insights into neurodegeneration from plant metabolism. Trends Biochem Sci. 2009;34:628–639. doi: 10.1016/j.tibs.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Minassian BA, Lee JR, Herbrick JA, Huizenga J, Soder S, Mungall AJ, Dunham I, Gardner R, Fong CY, Carpenter S, et al. Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat Genet. 1998;20:171–174. doi: 10.1038/2470. This manuscript identifies EPM2A as the Lafora disease gene locus. [DOI] [PubMed] [Google Scholar]
- 6•.Serratosa JM, Gomez-Garre P, Gallardo ME, Anta B, de Bernabe DB, Lindhout D, Augustijn PB, Tassinari CA, Malafosse RM, Topcu M, et al. A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2) Hum Mol Genet. 1999;8:345–352. doi: 10.1093/hmg/8.2.345. This manuscript identifies EPM2A as the Lafora disease gene locus. [DOI] [PubMed] [Google Scholar]
- 7.Boraston AB, Bolam DN, Gilbert HJ, Daview GJ. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. The Biochemical Journal. 2004;382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 9.Sakai M, Austin J, Witmer F, Trueb L. Studies in myoclonus epilepsy (Lafora body form). II. Polyglucosans in the systemic deposits of myoclonus epilepsy and in corpora amylacea. Neurology. 1970;20:160–176. doi: 10.1212/wnl.20.2.160. [DOI] [PubMed] [Google Scholar]
- 10.Yokoi S, Austin J, Witmer F. Isolation and characterization of Lafora bodies in two cases of myoclonus epilepsy. Journal of Neuropathology and Experimental Neurology. 1967;26:125–127. [PubMed] [Google Scholar]
- 11••.Worby CA, Gentry MS, Dixon JE. Laforin: A dual specificity phosphatase that dephosphorylates complex carbohydrates. J Biol Chem. 2006;281:30412–30418. doi: 10.1074/jbc.M606117200. These authors describe laforin as the first glucan phosphatase by showing that laforin dephosphorylates phospho-glucans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duran J, Gruart A, Garcia-Rocha M, Delgado-Garcia JM, Guinovart JJ. Glycogen accumulation underlies neurodegeneration and autophagy impairment in Lafora disease. Hum Mol Genet. 2014;23:3147–3156. doi: 10.1093/hmg/ddu024. [DOI] [PubMed] [Google Scholar]
- 13.Tagliabracci VS, Girard JM, Segvich D, Meyer C, Turnbull J, Zhao X, Minassian BA, Depaoli-Roach AA, Roach PJ. Abnormal metabolism of glycogen phosphate as a cause for Lafora disease. J Biol Chem. 2008;283:33816–33825. doi: 10.1074/jbc.M807428200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Tagliabracci VS, Turnbull J, Wang W, Girard JM, Zhao X, Skurat AV, Delgado-Escueta AV, Minassian BA, Depaoli-Roach AA, Roach PJ. Laforin is a glycogen phosphatase, deficiency of which leads to elevated phosphorylation of glycogen in vivo. Proc Natl Acad Sci USA. 2007;104:19262–19266. doi: 10.1073/pnas.0707952104. This manuscript demonstrates that Lafora bodies from laforin-deficient mice have increased covalently linked phosphate compared to glycogen and showed that recombinant laforin dephosphorylates glycogen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valles-Ortega J, Duran J, Garcia-Rocha M, Bosch C, Saez I, Pujadas L, Serafin A, Canas X, Soriano E, Delgado-Garcia JM, et al. Neurodegeneration and functional impairments associated with glycogen synthase accumulation in a mouse model of Lafora disease. EMBO Mol Med. 2011;3:667–681. doi: 10.1002/emmm.201100174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vilchez D, Ros S, Cifuentes D, Pujadas L, Valles J, Garcia-Fojeda B, Criado-Garcia O, Fernandez-Sanchez E, Medrano-Fernandez I, Dominguez J, et al. Mechanism suppressing glycogen synthesis in neurons and its demise in progressive myoclonus epilepsy. Nat Neurosci. 2007;10:1407–1413. doi: 10.1038/nn1998. [DOI] [PubMed] [Google Scholar]
- 17••.Raththagala M, Brewer MK, Parker MW, Sherwood AR, Wong BK, Hsu S, Bridges TM, Paasch BC, Hellman LM, Husodo S, et al. Structural Mechanism of Laforin Function in Glycogen Dephosphorylation and Lafora Disease. Molecular Cell. 2015;57:261–272. doi: 10.1016/j.molcel.2014.11.020. This manuscript reports the structure of laforin bound to glucan phosphate product, revealing its tertiary and quaternary structure and demonstrating mechanistically how laforin dephosphorylates glycogen as well as defining the mechanism by which subsets of LD mutations disrupt laforin function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alonso A, Rojas A, Godzik A, Mustelin T. In: The dual-specific protein tyrosine phosphatase family. Hohmann S, editor. Vol. 5. Berlin: Springer; 2003. [Google Scholar]
- 19.Coehn A. Über elektrische Wanderung von Kolloiden. Z Electrochem. 1897;4:63–67. [Google Scholar]
- 20.Blennow A, Bay-Smidt AM, Olsen CE, Moller BL. The distribution of covalently bound phosphate in the starch granule in relation to starch crystallinity. Int J Biol Macromol. 2000;27:211–218. doi: 10.1016/s0141-8130(00)00121-5. [DOI] [PubMed] [Google Scholar]
- 21.Carciofi M, Shaik SS, Jensen SL, Blennow A, Svensson JT, Vincze É, Hebelstrup KH. Hyperphosphorylation of cereal starch. Journal of Cereal Science. 2011;54:339–346. [Google Scholar]
- 22.Lorberth R, Ritte G, Willmitzer L, Kossmann J. Inhibition of a starch-granule-bound protein leads to modified starch and repression of cold sweetening. Nat Biotechnol. 1998;16:473–477. doi: 10.1038/nbt0598-473. [DOI] [PubMed] [Google Scholar]
- 23•.Ritte G, Heydenreich M, Mahlow S, Haebel S, Kotting O, Steup M. Phosphorylation of C6-and C3-positions of glucosyl residues in starch is catalysed by distinct dikinases. FEBS Lett. 2006;580:4872–4876. doi: 10.1016/j.febslet.2006.07.085. These authors demonstrated that GWD selectively catalyzes phosphorylation of C6 hydroxlys and PWD phosphorylates the C3 position. [DOI] [PubMed] [Google Scholar]
- 24••.Ritte G, Lloyd JR, Eckermann N, Rottmann A, Kossmann J, Steup M. The starch-related R1 protein is an alpha -glucan, water dikinase. Proc Natl Acad Sci USA. 2002;99:7166–7171. doi: 10.1073/pnas.062053099. This manuscript demonstrates that the protein known as GWD/R1/SEX1 is a glucan water dikinase that transfer the β-phosphate of ATP onto starch. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu TS, Kofler H, Hausler RE, Hille D, Flugge UI, Zeeman SC, Smith AM, Kossmann J, Lloyd J, Ritte G, et al. The Arabidopsis sex1 mutant is defective in the R1 protein, a general regulator of starch degradation in plants, and not in the chloroplast hexose transporter. Plant Cell. 2001;13:1907–1918. doi: 10.1105/TPC.010091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caspar T, Lin T-P, Kakefuda G, Benbow L, Preiss J, Somerville C. Mutants of Arabidopsis with Altered Regulation of Starch Degradation. Plant Physiol. 1991;95:1181–1188. doi: 10.1104/pp.95.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertoft E. Starch. Springer; 2015. Fine Structure of Amylopectin; pp. 3–40. [Google Scholar]
- 28.Streb S, Zeeman SC. Arabidopsis Book. 2012. Starch metabolism in Arabidopsis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blennow A. Starch. Springer; 2015. Phosphorylation of the starch granule; pp. 399–424. [Google Scholar]
- 30.Silver DM, Kotting O, Moorhead GB. Phosphoglucan phosphatase function sheds light on starch degradation. Trends Plant Sci. 2014;19:471–478. doi: 10.1016/j.tplants.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 31.Smirnova J, Fernie AR, Steup M. Starch. Springer; 2015. Starch degradation; pp. 239–290. [Google Scholar]
- 32••.Gentry MS, Dowen RH, 3rd, Worby CA, Mattoo S, Ecker JR, Dixon JE. The phosphatase laforin crosses evolutionary boundaries and links carbohydrate metabolism to neuronal disease. J Cell Biol. 2007;178:477–488. doi: 10.1083/jcb.200704094. This manuscript defines SEX4 as a glucan phosphatase, shows that laforin is conserved in an ancient class of protozoans, and demonstrates a functional conservation between laforin and SEX4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33•.Niittyla T, Comparot-Moss S, Lue W-L, Messerli G, Trevisan M, Seymour MDJ, Gatehouse JA, Villadsen D, Smith SM, Chen J, et al. Similar protein phosphatases control starch metabolism in plants and glycogen metabolism in mammals. J Biol Chem. 2006;281:11815–11818. doi: 10.1074/jbc.M600519200. These authors define the SEX4 locus, demonstrate that SEX4 is localized in chloroplasts, and show that SEX4 binds starch. [DOI] [PubMed] [Google Scholar]
- 34••.Meekins DA, Raththagala M, Husodo S, White CJ, Guo HF, Kotting O, Vander Kooi CW, Gentry MS. Phosphoglucan-bound structure of starch phosphatase Starch Excess4 reveals the mechanism for C6 specificity. Proc Natl Acad Sci USA. 2014;111:7272–7277. doi: 10.1073/pnas.1400757111. This paper reports the structure of SEX4 bound to glucan phosphate product to define how SEX4 binds and preferentially dephosphorylates starch at the C6-position, and engineers a SEX4 double mutant that reverses substrate specificity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35•.Vander Kooi CW, Taylor AO, Pace RM, Meekins DA, Guo HF, Kim Y, Gentry MS. From the Cover: Structural basis for the glucan phosphatase activity of Starch Excess4. Proc Natl Acad Sci USA. 2010;107:15379–15384. doi: 10.1073/pnas.1009386107. This manuscript reports the first structure of a glucan phosphatase, demonstrating the integrated CBM-DSP domain architecture employed by SEX4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baunsgaard L, Lutken H, Mikkelsen R, Glaring MA, Pham TT, Blennow A. A novel isoform of glucan, water dikinase phosphorylates pre-phosphorylated alpha-glucans and is involved in starch degradation in Arabidopsis. Plant J. 2005;41:595–605. doi: 10.1111/j.1365-313X.2004.02322.x. [DOI] [PubMed] [Google Scholar]
- 37.Kotting O, Pusch K, Tiessen A, Geigenberger P, Steup M, Ritte G. Identification of a novel enzyme required for starch metabolism in Arabidopsis leaves. The phosphoglucan, water dikinase. Plant Physiol. 2005;137:242–252. doi: 10.1104/pp.104.055954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blennow A, Engelsen SB. Helix-breaking news: fighting crystalline starch energy deposits in the cell. Trends Plant Sci. 2010;15:236–240. doi: 10.1016/j.tplants.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 39.Hansen PI, Larsen FH, Motawia SM, Blennow A, Spraul M, Dvortsak P, Engelsen SB. Structure and hydration of the amylopectin trisaccharide building blocks--Synthesis, NMR, and molecular dynamics. Biopolymers. 2008;89:1179–1193. doi: 10.1002/bip.21075. [DOI] [PubMed] [Google Scholar]
- 40.Hansen PI, Spraul M, Dvortsak P, Larsen FH, Blennow A, Motawia MS, Engelsen SB. Starch phosphorylation--maltosidic restrains upon 3′- and 6′-phosphorylation investigated by chemical synthesis, molecular dynamics and NMR spectroscopy. Biopolymers. 2009;91:179–193. doi: 10.1002/bip.21111. [DOI] [PubMed] [Google Scholar]
- 41.Edner C, Li J, Albrecht T, Mahlow S, Hejazi M, Hussain H, Kaplan F, Guy C, Smith SM, Steup M, et al. Glucan, water dikinase activity stimulates breakdown of starch granules by plastidial beta-amylases. Plant Physiol. 2007;145:17–28. doi: 10.1104/pp.107.104224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hejazi M, Fettke J, Haebel S, Edner C, Paris O, Frohberg C, Steup M, Ritte G. Glucan, water dikinase phosphorylates crystalline maltodextrins and thereby initiates solubilization. Plant J. 2008;55:323–334. doi: 10.1111/j.1365-313x.2008.03513.x. [DOI] [PubMed] [Google Scholar]
- 43.Takeda Y, Hizukuri S. Studies on starch phosphate. 1981. Part. [Google Scholar]
- 44•.Santelia D, Kotting O, Seung D, Schubert M, Thalmann M, Bischof S, Meekins DA, Lutz A, Patron N, Gentry MS, et al. The phosphoglucan phosphatase like sex Four2 dephosphorylates starch at the C3-position in Arabidopsis. Plant Cell. 2011;23:4096–4111. doi: 10.1105/tpc.111.092155. These authors define the LSF2 protein as a glucan phosphatase that hydrolyzes phosphate from the C3-position and demonstrate that lsf2 mutant starch has elevated leves of C3-bound phosphate. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Meekins DA, Guo HF, Husodo S, Paasch BC, Bridges TM, Santelia D, Kotting O, Vander Kooi CW, Gentry MS. Structure of the Arabidopsis glucan phosphatase like sex four2 reveals a unique mechanism for starch dephosphorylation. Plant Cell. 2013;25:2302–2314. doi: 10.1105/tpc.113.112706. This paper describes the structure of Arabidopsis thaliana LSF2 both with and without phopho-glucan product bound, identifies two surface binding sites that coordinate starch binding, and defines the mechanism of LSF2 C3-specificity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cuyvers S, Dornez E, Delcour JA, Courtin CM. Occurrence and functional significance of secondary carbohydrate binding sites in glycoside hydrolases. Crit Rev Biotechnol. 2012;32:93–107. doi: 10.3109/07388551.2011.561537. [DOI] [PubMed] [Google Scholar]
- 47.Cockburn D, Nielsen MM, Christiansen C, Andersen JM, Rannes JB, Blennow A, Svensson B. Surface binding sites in amylase have distinct roles in recognition of starch structure motifs and degradation. Int J Biol Macromol. 2015;75:338–345. doi: 10.1016/j.ijbiomac.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 48.Wilkens C, Auger KD, Anderson NT, Meekins DA, Raththagala M, Abou Hachem M, Payne CM, Gentry MS, Svensson B. Plant alpha-glucan phosphatases SEX4 and LSF2 display different affinity for amylopectin and amylose. FEBS Lett. 2016;590:118–128. doi: 10.1002/1873-3468.12027. [DOI] [PubMed] [Google Scholar]
- 49.Meekins DA, Raththagala M, Auger KD, Turner BD, Santelia D, Kotting O, Gentry MS, Kooi CWV. Mechanistic Insights into Glucan Phosphatase Activity against Polyglucan Substrates. Journal of Biological Chemistry. 2015;290:23361–23370. doi: 10.1074/jbc.M115.658203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emanuelle S, Brewer MK, Meekins DA, Gentry MS. Unique carbohydrate binding platforms employed by the glucan phosphatases. Cell Mol Life Sci. 2016 doi: 10.1007/s00018-016-2287-x. [DOI] [PMC free article] [PubMed]
- 51.Meekins DA, Vander Kooi CW, Gentry MS. Structural Mechanisms of Plant Glucan Phosphatases in Starch Metabolism. FEBS J. 2016 doi: 10.1111/febs.13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dias DM, Furtado J, Wasielewski E, Cruz R, Costello B, Cole L, Faria TQ, Baaske P, Brito RM, Ciulli A, et al. Biophysical characterization of laforin-carbohydrate interaction. Biochem J. 2016;473:335–345. doi: 10.1042/BJ20141555. [DOI] [PubMed] [Google Scholar]
- 53.Nitschke F, Wang P, Schmieder P, Girard JM, Awrey DE, Wang T, Israelian J, Zhao X, Turnbull J, Heydenreich M, et al. Hyperphosphorylation of glucosyl c6 carbons and altered structure of glycogen in the neurodegenerative epilepsy lafora disease. Cell Metab. 2013;17:756–767. doi: 10.1016/j.cmet.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 54.Tagliabracci VS, Heiss C, Karthik C, Contreras CJ, Glushka J, Ishihara M, Azadi P, Hurley TD, DePaoli-Roach AA, Roach PJ. Phosphate incorporation during glycogen synthesis and Lafora disease. Cell Metab. 2011;13:274–282. doi: 10.1016/j.cmet.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DePaoli-Roach AA, Contreras CJ, Segvich DM, Heiss C, Ishihara M, Azadi P, Roach PJ. Glycogen phosphomonoester distribution in mouse models of the progressive myoclonic epilepsy, Lafora disease. J Biol Chem. 2015;290:841–850. doi: 10.1074/jbc.M114.607796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chikwana VM, Khanna M, Baskaran S, Tagliabracci VS, Contreras CJ, DePaoli-Roach A, Roach PJ, Hurley TD. Structural basis for 2′-phosphate incorporation into glycogen by glycogen synthase. Proc Natl Acad Sci USA. 2013;110:20976–20981. doi: 10.1073/pnas.1310106111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Contreras CJ, Segvich DM, Mahalingan K, Chikwana VM, Kirley TL, Hurley TD, DePaoli-Roach AA, Roach PJ. Incorporation of phosphate into glycogen by glycogen synthase. Arch Biochem Biophys. 2016;597:21–29. doi: 10.1016/j.abb.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ganesh S, Delgado-Escueta AV, Sakamoto T, Avila MR, Machado-Salas J, Hoshii Y, Akagi T, Gomi H, Suzuki T, Amano K, et al. Targeted disruption of the Epm2a gene causes formation of Lafora inclusion bodies, neurodegeneration, ataxia, myoclonus epilepsy and impaired behavioral response in mice. Hum Mol Genet. 2002;11:1251–1262. doi: 10.1093/hmg/11.11.1251. [DOI] [PubMed] [Google Scholar]
- 59.Schrodinger L. The PyMOL MOlecular Graphics System, Version 1.3. 2010. [Google Scholar]
- 60.Laskowski RA, Swindells MB. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 61.Kolmodin K, Aqvist J. The catalytic mechanism of protein tyrosine phosphatases revisited. FEBS Lett. 2001;498:208–213. doi: 10.1016/s0014-5793(01)02479-6. [DOI] [PubMed] [Google Scholar]
- 62.Barford D, Flint AJ, Tonks NK. Crystal structure of human protein tyrosine phosphatase 1B. Science. 1994;263:1397–1404. [PubMed] [Google Scholar]
- 63.Yuvaniyama J, Denu JM, Dixon JE, Saper MA. Crystal structure of the dual specificity protein phosphatase VHR. Science. 1996;272:1328–1331. doi: 10.1126/science.272.5266.1328. [DOI] [PubMed] [Google Scholar]