Abstract
Duplications are the primary force by which new gene functions arise and provide a substrate for large-scale structural variation. Analysis of thousands of genomes shows that humans and great apes have more genetic differences in content and structure over recent segmental duplications than any other euchromatic region. Novel human-specific duplicated genes, ARHGAP11B and SRGAP2C, have recently been described with a potential role in neocortical expansion and increased neuronal spine density. Large segmental duplications and the structural variants they promote are also frequently stratified between human populations with a subset being subjected to positive selection. The impact of recent duplications on human evolution and adaptation is only beginning to be realized as new technologies enhance their discovery and accurate genotyping.
INTRODUCTION
In this review, we will summarize recent advances in our understanding of primate segmental duplications (SDs; defined as large tracts of sequence (>1 kbp) sharing >90% identity [1]) and their impact in contributing to new genes and functions within the human lineage. Mutation by duplication has two, very different, consequences on an evolving genome [2,3]. First, it creates genetic redundancy liberating functional DNA from ancestral selective constraint. This can lead to the birth of novel genes and regulatory elements either instantaneously through duplicative transpositions or by subsequent mutational tinkering of the paralogous copy [4]. Second, by dint of its high sequence identity, duplication provides the substrate for subsequent rearrangement through the process of non-allelic homologous recombination [5]. This mutational process is dynamic because the presence of SDs further increases the probability of subsequent rounds of duplications as a result of larger and more abundant tracts of identical sequences [6–8]. Duplications of genic material thus have the potential to radically change structure and content over extremely short periods of times. Here, we focus on gene innovation by duplication and emerging data regarding its importance to human adaptation.
Nonrandom evolution of great ape segmental duplication
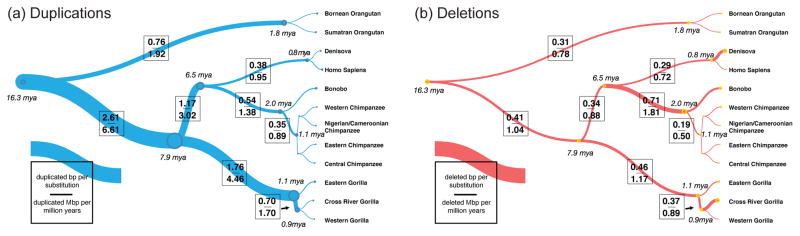
The accumulation of SDs in the human–great ape lineage has been nonrandom in both time and space [9]. Comparative and sequence-identity analyses support a threefold excess of duplications in the common ancestor of human and great apes lineage (7–16 mya) in contrast to deletions, which have occurred in a more clocklike manner during evolution (Figure 1) [10]. The intrachromosomal burst and to a lesser extent interchromosomal duplications have been associated with particular segments identified as core duplicons [11,12] (Table 1). Using a repeat-graph approach, 24 core duplicons (~15 kbp) were originally identified as segments overrepresented in 437 duplication blocks within the human genome. Subsequent phylogenetic analyses revealed that the cores represent the focal point for the serial accumulation of SDs, resulting in increasingly larger duplication blocks (>250 kbp in size) composed of mosaic duplicated segments where evolutionarily younger duplicated segments are located at increasing distances from the core.
Figure 1. Primate rates of duplication and deletion.
Rates of fixed (a) duplication and (b) deletion are shown as a function of the number of substitutions along each branch of the great ape phylogeny. Branch widths are scaled proportionally to the number of duplicated base pairs per substituted base pair based on analysis of 97 human/ape genomes. A burst of duplicated base pairs appears to have occurred in the common ancestral branch leading to humans and African great apes, where duplicated base pairs were added at 2.6-fold the rate of substitution. In contrast, the rate of deletion in the great ape lineage is more clocklike along all branches (mean of 0.32 deleted base pairs per substitution) with the exception of the chimpanzee–bonobo ancestral lineage, where an approximate twofold increase in the rate of deletion is observed (0.71 deleted base pairs per substitution). Adapted from [10].
Table 1.
Examples of genes/gene families mapped to core duplicons.
| clade | gene | significant expression | subcellular localization | Description | possible function | example disease-associated genomic hotspot |
|---|---|---|---|---|---|---|
| chr1 | NBPF | soft tissue | cytoplasm | neuroblastoma breakpoint gene family, DUF1220 | transcription factor regulated by NF-κB | 1q21.1: Neuroblastoma, ASD, ID, Schizophrenia |
| chr2 | RANBP2 | testis | nuclear pore | RANBP2-like and GRIP domain- containing 5 isoform | Ran GTPase binding | 2q13: ID |
| chr7_2 | PMS2L5 | ubiquitous | nuclear | postmeiotic segregation increased 2-like 5 | DNA mismatch repair | 7q11.23: Williams-Beuren syndrome, ASD, ID |
| chr7_2 | SPDYE1 | testis | unknown | speedy/RINGO cell cycle regulator family member E1 | cell cycle regulator | 7q11.23: Williams-Beuren syndrome, ASD, ID |
| chr7_3 | DPY19L2 | testis | unknown | dpy-19 like 2 | spermatogenesis | none |
| chr9_1 | SPATA31A1 | exclusively in testis | unknown | SPATA31 subfamily A, member 1 | unknown | none |
| chr9_2 | ZNF790 | ubiquitous | nuclear | zinc-finger protein 790 | DNA binding | none |
| chr11/chr2 | TRIM51 | mammary gland | unknown | tripartite motif-containing 51 | unknown | 2q11.2: ID, ADHD |
| chr15 | GOLGA | exclusively in testis | unknown | golgin-like protein, golgi autoantigen, golgin subfamily a | DNA binding | 15q13.3: ASD, ID, Schizophrenia, Epilepsy |
| chr16 | NPIP | ubiquitous | nuclear membrane | nuclear pore complex interacting protein, morpheus gene family | unknown | 16p11.2: ASD, ID, Schizophrenia, Epilepsy |
| chr17_1 | LRRC37A | ubiquitous | unknown | leucine-rich repeat, c114 SLIT-like testicular protein | ATP-dependent peptidase activity | 17q21.31: ID |
| chr17_2 | TBC1D3 | testis | cytoplasm | TBC1 domain family member | cell growth and proliferation | 17q12: ASD, ID, Schizophrenia |
| M1 | OR7E | unknown | unknown | olfactory receptor 7E pseudogenes | receptors mediating sense of smell | 8p23.1: ID |
| M5 | SMA | spinal cord | lysosome | spinal muscular atrophy associated gene | hydrolase activity | 5q13.2: Spinal Muscular Atrophy |
| M6 | CCDC127 | ubiquitous | unknown | coiled-coiled domain containing 27 | unknown | none |
ASD: autism spectrum disorder; ID: intellectual disability and associated developmental delay, ADHD: attention-deficit and hyperactivity disorder. Adapted from [11].
There is evidence that the cores have been reused in different great ape lineages [13] and are preferential sites of rearrangements leading to large-scale inversion polymorphisms [14,15]. Interestingly, many of these cores are enriched in rapidly evolving human–great ape gene families [16–23]. Though the function of most of these gene families has not yet been determined, biochemical investigations into a few (e.g., TBC1D3) have suggested an association with cell proliferation [24]; others (e.g., DUF1220 or NBPF) have been implicated in changes of brain size [21,25,26] while some (e.g., NPIP) show remarkable signatures of positive selection [19]. At the periphery of these core-mediated duplication blocks lie most of the human-specific segmental duplications (HSDs) [27,28]. HSDs are restricted to specific regions of the genome and appear to be enriched in genes associated with brain development and neuronal apoptosis. Approximately 25 of these regions are now associated with recurrent chromosomal rearrangements and neurodevelopmental disease [29–31] suggesting that there has been a negative fitness effect with the propagation of these elements during evolution.
Segmental duplication and the evolution of a larger human brain
Over the last four years, a few interesting examples have highlighted the potential functional impact of SDs with respect to the evolution of the human brain. Of the three recently described examples, two—SRGAP2C and ARHGAP11B—are the direct consequence of HSDs [32–35]. It is noteworthy that the third, a recently described human-accelerated regulatory region (HARE5) of Wnt receptor FZD8 (frizzled-8) [36], is part of an 18 kbp primate-specific SD on chromosome 10 that likely arose after divergence of Old and New World primates, although the duplication of the locus occurred long before the burst of functional single-base-pair substitutions on the human lineage.
SRGAP2C
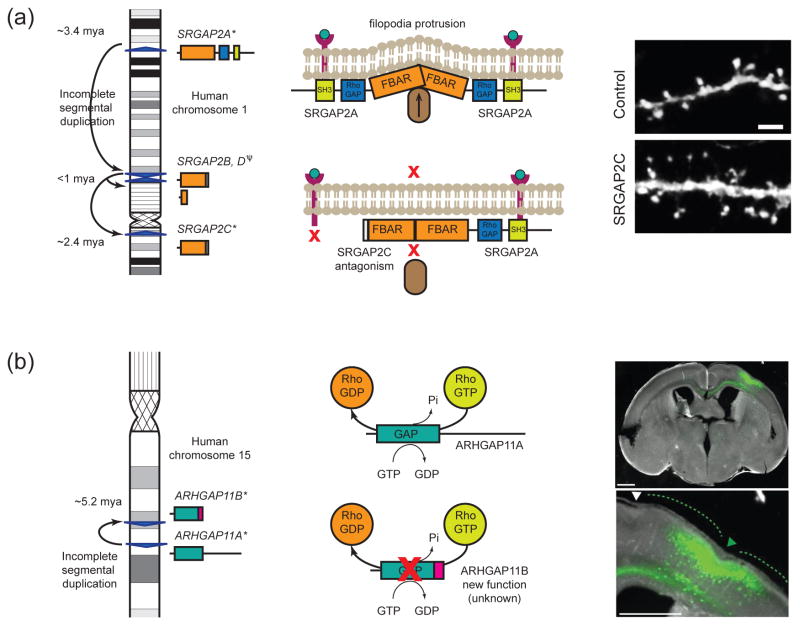
The cortical developmental gene SRGAP2 (Slit-Robo Rho GTPase-activating protein 2) has duplicated three separate times uniquely in the human lineage with its paralogs dispersing across 85 Mbp on chromosome 1 (Figure 2a) [33]. Ancestral SRGAP2A is capable of homodimerizing via an FBAR domain at the cell surface where it induces membrane protrusions required for neuronal migration and morphogenesis in mammals [37]. Evolutionary reconstructions showed that a 258 kbp SD transposed from chromosome 1q32.1 to 1q21.1 ~3.4 million years ago (mya) where it spawned a truncated SRGAP2B, including the first 9 of 22 exons and putative regulatory sequences of the ancestral copy. Larger secondary duplications of SRGAP2B created an additional truncated “granddaughter” paralog SRGAP2C (at chromosome 1p12 ~2.4 mya). Despite it being one of the most recent paralogs, SRGAP2C is fixed for copy number in all modern humans—unlike its predecessor SRGAP2B. This truncated paralog is co-expressed with the full-length ancestral SRGAP2A in the germinal layers of human embryonic cortex, where neural progenitors divide to produce postmitotic neurons, and the cortical plate, where neurons undergo terminal differentiation and synaptogenesis [32]. Transient overexpression of SRGAP2C in culture and in vivo leads to human-specific features, including neoteny of dendritic spine maturation, promotion of longer spines at a greater density, and sustained radial migration in the developing mouse neocortex. Human-specific SRGAP2C, which lacks RhoGAP and SH3 functional domains, dimerizes with the ancestral SRGAP2A via its partial FBAR domain, in effect usurping functional full-length proteins from homodimerizing and assembling at the cell surface. This antagonistic function may have been present at the birth of SRGAP2C as a result of the incomplete SD that excluded the RhoGAP and SH3 functional domains. It is intriguing that these SRGAP2 duplicates arose 2–3 mya, the estimated divergence time of Australopithecus to Homo and immediately prior to the paleontological estimate of human neocortex expansion [38]. This raises the exciting possibility that the SRGAP2C duplication played an important role in neural adaptive changes specific to the hominin lineage.
Figure 2. Gene duplication and neuroanatomical adaptations.
(a) SRGAP2A encodes a 1,071 aa protein with three protein domains shown as boxes: FBAR (orange), RhoGAP (blue), and SH3 (green). An incomplete SD from chromosome 1q32.1 created SRGAP2B at 1q21.1 encoding a partial FBAR domain (458 aa) and seven unique residues [33]. A subsequent duplication from SRGAP2B created SRGAP2C at 1p12.1 and SRGAP2D at 1q21.1 (which later partially deleted and represents a pseudogene). SRGAP2A homodimerizes and assembles at the cell membrane surface via its SH3 domain and induces filopodia protrusion by interacting with a protein complex including F-actin (brown oval) [37]. Truncated SRGAP2C is capable of heterodimerizing with SRGAP2A via its FBAR domain but lacks the RhoGAP and SH3 domains in turn antagonizing the function of the ancestral gene by not allowing it to assemble at the cell membrane surface. SRGAP2C expression induces long thin spines in mouse-cultured cortical neurons that phenocopies SRGAP2A deficiency in mice. Pictured are segments of dendrites from cortical neurons (20DIV) expressing EGFP alone (control) or EGFP and SRGAP2C (SRGAP2C) imaged two days after transfection. Reprinted with permission from [32]. (b) ARGHAP11A encodes a 1023 aa protein with a RhoGAP domain (blue). It was partially duplicated at chromosome 15q13.3 resulting in a paralog ARHGAP11B encoding a truncated RhoGAP domain (220 aa) and 47 unique residues at the C terminus (pink box) [35]. Via its RhoGAP domain, ARHGAP11A and truncated alternative isoforms encoding 220 aa (not pictured) dephosphorylate Rho-GTP unlike ARHGAP11B, which does not exhibit RhoGAP activity. ARHGAP11B overexpression leads to an increase in basal progenitors in the mouse neocortex possibly inducing cortical folding. Pictured are coronal sections of an E18.5 mouse telencephali in utero electroporated at E13.5 with ARHGAP11B and GFP expression plasmids. Phase contrast and GFP fluorescence of one section along the rostro-caudal axis. Scale bars, 500 μm. Green and white dashed lines and triangles indicate gyrus- and sulcus-like structures in and adjacent to the electroporated area, respectively. Reprinted with permission from [34].
ARHGAP11B
Another potentially functionally relevant HSD gene is ARHGAP11B, the product of an incomplete duplication of ARHGAP11A (Rho-type GTPase-activating protein 11A) shortly after the chimpanzee and human lineages diverged ~5.2 mya (Figure 2b) [35]. It was one of the initiating events that led to the formation of large complex HSDs responsible for mediating recurrent rearrangements contributing to 15q13.3 microdeletion syndrome associated with intellectual disability, epilepsy and schizophrenia [39]. Notably, all events were associated with the chromosome 15 core duplicon containing GOLGA [35]. The truncated ARHGAP11B lacks the terminal 756 aa residues of ARHGAP11A and instead encodes a modified carboxyl terminus consisting of 47 functionally distinct residues. Unlike the full-length ARHAP11A or a shorter 250 aa isoform, the resulting 267 aa protein does not exhibit RhoGAP activity based on a RhoA/Rho-kinase-based cell transfection assay. ARHGAP11B may have evolved a completely novel function [34].
Performing an RNA-seq assay of cortical progenitor subpopulations in human and mouse, Florio and colleagues identified only ARHGAP11A when searching for transcripts unique to humans that show >10-fold expression difference between basal radial glial cell populations—isolated from the subventricular zone—compared with neuronal cells [34]. In utero electroporation and expression of the gene in E13.5 mouse embryos resulted in a ~30% increase in the thickness of the subventricular zone due to greater basal radial glial mitotic divisions. Further, microinjection of ARHGAP11B into E13.5 mouse apical radial glial cells showed a subsequent increase in basal radial glial cells where the progeny lost apical contact (delamination). Remarkably, examination of the mouse brain slices at E18.5 demonstrated in half of the cases an increase in the cortical plate area and neocortical folding reminiscent of the gyrification common among primate species. These findings are consistent with the hypothesis that an increased number of radial glial neuron progenitor cells, possibly through increased symmetric cell divisions, would lead to a neuronal expansion in the cortical layers of the developing brain. While potentially exciting, caution should be exercised until transgenic mice are constructed that replicate these findings. There is concern that both microinjection as well as electroporation may have induced artefacts associated with cell proliferation.
Variation and selection within the human populations
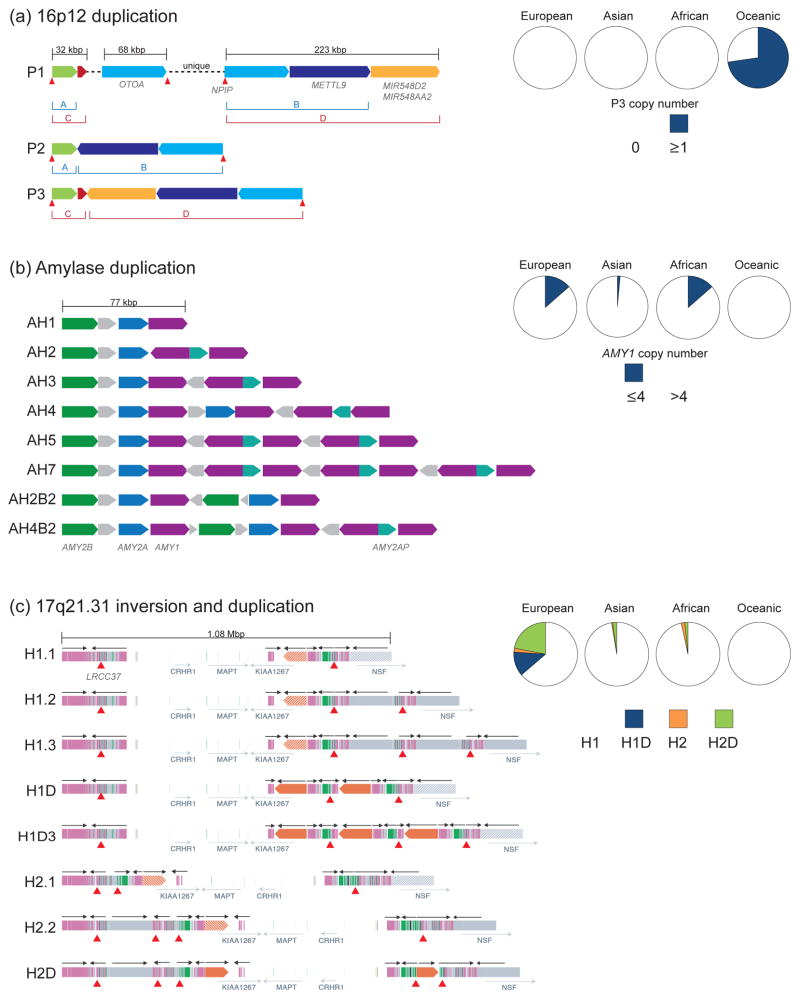
SDs show extensive copy number variation in the human species [27,28,40–44] and contribute more variant base pairs between any two individuals than any other single source, including single-nucleotide polymorphisms (SNPs), indels and structural variants [45]. Genomic analysis of a diversity panel of 236 sequenced human genomes identified 1,036 copy number variants (CNVs) stratified between human populations [45]. Using the Vst statistic, analogous to Fst for multiallelic or multicopy states, found that duplications were 1.8 times more likely than deletions to exhibit population stratification but less likely to be tagged by adjacent SNPs (R2 = 0.03 Vst of duplications vs. Fst of flanking SNPs) making them more difficult to detect. This is in agreement with other studies showing the majority of multicopy duplications correlating less well with flanking SNP genotypes [43,46]. Among this set were five duplications identified in Oceanic populations that were shared with the archaic Denisovans. A subset of these constituted a larger 255 kbp SD mapping to chromosome 16p12.1 (Figure 3a). Using shared SNPs, this SD was determined to have arisen in Denisovans ~400 thousand years ago (kya) followed by introgression into the Oceanic ancestral population ~40 kya, where it has since nearly fixed either through positive selection or genetic drift. The duplication is associated with the chromosome 16 core duplicon containing NPIP [19], appears to be exclusive to living Oceanic human populations and represents the first example of an SD arising in a sibling species of humans that was later introduced to our lineage likely by introgression. While interesting, is there any evidence that SDs and their associated structural variants may be substrates for more recent selection and adaptation?
Figure 3. Population diversity of duplications in modern human populations.
SDs are indicated as colored blocks with directionality shown. Pie charts show frequencies of paralogs/alleles across European (E), Asian (As), African (Af), and Oceanic (O) populations. Core duplicons (red triangles) are depicted below structural paralogs or haplotypes. (a) A ~255 kbp Oceanic-specific SD (P3) was identified and shown to be made up of disperse segments from chromosome 16p12.1. The duplication is present in the genome of the ancient hominin, Denisova, but not observed in any other modern human populations possibly as a result of introgression back into the human lineage. The Oceanic-specific P3 formed as a result of interspersed duplications of P1 (represented in the human reference genome GRCh38) and P2 and included the NPIP core duplicon. P3 frequency was estimated based on genome sequence read-depth from the Human Genome Diversity Project (HGDP) cohort (E = 59, As = 45, Af = 36, O = 21) [41]. The P3 duplication has been identified in all Papuan individuals. (b) Diverse duplication structures exist at the amylase locus, with at least eight predicted haplotypes representing varying copies of AMY2B, AMY2A, AMY1, and AMY2AP (figure adapted from [50]). Overall copy number estimates of AMY1 in 1000 Genomes Project [41] and HGDP cohorts [44] were calculated and low-copy (CN ≤ 4) versus high-copy (CN > 4) frequencies were determined (E = 145, As = 204, Af = 133, O = 21). European and African populations show overall lower copy numbers of AMY1 compared with Asian and Oceanic. (c) Structural diversity of the chromosome 17q21.31 haplotype (figure adapted from [54]). Various forms of the directly orientated haplotype (H1) have been identified, including European-enriched haplotypes that show duplications of the promoter and first exon of KANSL1 (H1D). The inverted haplotype (H2) exists in a simpler form (H2.1) found among the San Khoisan and in more complex duplicated forms in European/Mediterranean haplotypes, including a smaller duplication of the promoter and first exon of KANSL1 (H2D). Allele frequencies of H1, H1D, H2, and H2D are shown based on sequence data from 1000 Genomes Project and HGDP cohorts (E = 628, As = 733, Af = 820, O = 27) [54]. H2.1 is predicted to represent the ancestral haplotype (2.3 mya), H1 is now the dominant haplotype worldwide, and the increased frequencies of H1D, H2, and H2D in European populations are the result of positive selection or extraordinary genetic drift. This is remarkable in light of the fact that the European H2 haplotype is predisposed to microdeletion due to the accumulation of directly oriented SDs. The less complex pattern of SDs observed among African H2.1 allele carriers suggest that the ancestral H2 haplotype is not predisposed to disease [54].
Amylase and adaptation to a starch-rich diet
Recent duplications of the salivary amylase gene (AMY1) are frequently cited as an example of adaptive evolution in the human lineage [47,48]. Expansion of AMY1 and the concomitant increase in salivary enzyme production may improve our ability to digest starch-rich foods, a potentially beneficial trait related to the diet of early modern humans as they switched from a hunter-gatherer to a primarily agricultural society ~10 kya. Genotyping modern humans using qPCR and microarray, Perry and colleagues determined that copy number ranged from 2–15, noting stark differences between populations with low and high starch diets [47]. Further comparisons in closely related nonhuman primates and archaic hominins show these extreme expansions of gene copy are unique to modern humans and not seen in Neanderthal and Denisovan genomes [45]. More refined copy number estimates and insights into the structural variation of this locus are beginning to emerge [45,49,50], with diverse haplotypes identified ranging in size from 77 kbp to ~200 kbp including variability (2–6 copies) of nearby pancreatic amylase genes (AMY2B and AMY2A) (Figure 3b) [50].
17q21.31 duplication and inversion
The expansion of chromosome 17q21.31 duplications and the associated inversion polymorphism represent another potential example of recent human adaptation. Stefansson and colleagues originally reported a 900 kbp inversion polymorphism flanked by SDs enriched in European and Mediterranean populations [51]. Within the Icelandic population [51] and in European-Americans [52], the inverted haplotype (H2) has been associated with increased fecundity of H2 carrier females and an overall increase in global recombination, potentially explaining the increase in frequency. Subsequent sequencing and characterization of the predominant H2 duplicated (H2D) haplotype in individuals of European descent showed that the H2 haplotypes were virtually identical having expanded ~17–48 kya [53,54]. The high European allele frequency (~36%) of such a young haplotype was consistent with the original suggestion of positive selection. This finding was even more remarkable in light of the fact that the H1 and H2 haplotypes diverged ~2–3 mya and the H2 haplotype predisposes to recurrent microdeletions associated with Koolen-de Vries syndrome [55,56] and, thus, subject to some modest negative selection. A total of eight complex human structural haplotypes have been characterized to date ranging in size from 1–1.5 Mbp with considerable expansions of SDs occurring in the last million years of human evolution [53,54] (Figure 3c). The LRRC37A core duplicon resides near many of the breakpoints, including the inversion, flanking SDs, and disease-associated microdeletion [57]. Although the molecular basis for the selection of the European H2 locus is not known, it is interesting that the region contains several genes important in neural function, including MAPT—associated with several neurodegenerative disorders including Alzheimer’s disease, CRHR1—a cortical-releasing hormone receptor, and KANSL1—the gene responsible for the Koolen-de Vries syndrome [58]. It is interesting to note that the H2 haplotype was recently associated with increased intracranial volume indicating larger brain size [59].
Summary & Future Directions
Although the selective disadvantage of interspersed SDs is well established, examples of their selective benefit have been slower to emerge. Examples such as SRGAP2C and ARHGAP11B are interesting because, in both cases, the SDs were incomplete and potentially functional at birth due to truncations facilitating antagonistic interactions with the parental gene (SRGAP2A) or leading to rapid neofunctionalization (ARHGAP11B). Examples such as the 17q21.31 inversion and the amylase gene family highlight the continued importance of SDs to more recent adaptations. Despite these intriguing examples, several challenges remain. First, many of the regions are still not properly assembled in human and other great ape genomes due to their high sequence identity and association with other larger more complex duplications. Although the catalog of human SDs is nearly complete [41], recent estimates suggest that as much as 40 Mbp of euchromatic sequence may be missing from the current human reference due to structural variation [60] of which at least 4.2 Mbp are duplicated (>3 copies) and are copy number polymorphic [45]. This has meant that underlying genes have only been discovered through targeted efforts via sequencing of clones, full-length cDNA, and de novo genome assembly methods using long-read methods to correct the reference genome and distinguish paralogs [61–63]. Second, our understanding of the patterns of human variation in these regions is currently in its infancy. An important step forward will be the development of robust genotyping assays capable of inferring both the sequence content and structure of these regions [28,42,50]. Finally, functional studies assaying duplications are not well established. While CRISPR/Cas9 technology has facilitated knockouts within human induced pluripotent stem cells (iPSCs), the high degree of sequence identity between paralogs makes such an undertaking nontrivial often promoting the formation of large-scale rearrangements [64]. Though less straightforward due to the megabase pairs of highly rearranged sequence frequently associated with HSDs, genome editing can also be used to “knock in” HSDs in species where these genes do not exist. Generating the genomic context to establish equivalence and proof of functional effect in vivo has not yet been achieved for any HSD. Nevertheless, efforts can be made to “humanize” chimpanzee iPSCs as well as mice. The ultimate test of function lies in identifying mutations of HSD genes associated with human disease (e.g., natural gene knockouts in humans). Current whole-genome sequencing efforts will make it possible to assay variation in a number of disease cohorts. Though limitations exist when using short reads to assay variation between highly similar paralogs, recent advances in synthetic long read methods via barcoding (e.g., 10X Genomics) may prove effective in improving variant calling and characterizing the complexity of structural change. Notwithstanding these challenges, the emerging data suggest a disproportionate role for SDs not only in human disease but also human evolution.
Acknowledgments
We thank Brad Nelson, Karyn Meltz Steinberg, Francesca Antonacci, Peter Sudmant, Stuart Cantsilieris, and Tonia Brown for helpful comments on the manuscript. We apologize to authors whose work we could not cite given space considerations in this mini-review. This work was supported, in part, by U.S. National Institutes of Health (NIH) grants R00 NS083627 from NINDS (M.Y.D) and R01 HG002385 from NHGRI (E.E.E). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haldane JBS. The causes of evolution. London, New York etc: Longmans, Green and co; 1932. [Google Scholar]
- 3.Ohno S. Evolution by gene duplication. London, New York: Allen & Unwin; Springer-Verlag; 1970. [Google Scholar]
- 4.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 5••.Carvalho CM, Lupski JR. Mechanisms underlying structural variant formation in genomic disorders. Nat Rev Genet. 2016;17:224–238. doi: 10.1038/nrg.2015.25. This review provides an excellent resource of currently known mechanisms of structural variant formation leading to SDs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis EB. Pseudoallelism and gene evolution. Cold Spring Harb Symp Quant Biol. 1951;16:159–174. doi: 10.1101/sqb.1951.016.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Sturtevant AH. The Effects of Unequal Crossing over at the Bar Locus in Drosophila. Genetics. 1925;10:117–147. doi: 10.1093/genetics/10.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller HJ. Bar Duplication. Science. 1936;83:528–530. doi: 10.1126/science.83.2161.528-a. [DOI] [PubMed] [Google Scholar]
- 9.Marques-Bonet T, Kidd JM, Ventura M, Graves TA, Cheng Z, Hillier LW, Jiang Z, Baker C, Malfavon-Borja R, Fulton LA, et al. A burst of segmental duplications in the genome of the African great ape ancestor. Nature. 2009;457:877–881. doi: 10.1038/nature07744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sudmant PH, Huddleston J, Catacchio CR, Malig M, Hillier LW, Baker C, Mohajeri K, Kondova I, Bontrop RE, Persengiev S, et al. Evolution and diversity of copy number variation in the great ape lineage. Genome Res. 2013;23:1373–1382. doi: 10.1101/gr.158543.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Z, Tang H, Ventura M, Cardone MF, Marques-Bonet T, She X, Pevzner PA, Eichler EE. Ancestral reconstruction of segmental duplications reveals punctuated cores of human genome evolution. Nat Genet. 2007;39:1361–1368. doi: 10.1038/ng.2007.9. [DOI] [PubMed] [Google Scholar]
- 12.Ji X, Zhao S. DA and Xiao-two giant and composite LTR-retrotransposon-like elements identified in the human genome. Genomics. 2008;91:249–258. doi: 10.1016/j.ygeno.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson ME, Cheng Z, Morrison VA, Scherer S, Ventura M, Gibbs RA, Green ED, Eichler EE National Institute of Health Intramural Sequencing Center Comparative Sequencing P. Recurrent duplication-driven transposition of DNA during hominoid evolution. Proc Natl Acad Sci U S A. 2006;103:17626–17631. doi: 10.1073/pnas.0605426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonacci F, Kidd JM, Marques-Bonet T, Teague B, Ventura M, Girirajan S, Alkan C, Campbell CD, Vives L, Malig M, et al. A large and complex structural polymorphism at 16p12. 1 underlies microdeletion disease risk. Nat Genet. 2010;42:745–750. doi: 10.1038/ng.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zody MC, Jiang Z, Fung HC, Antonacci F, Hillier LW, Cardone MF, Graves TA, Kidd JM, Cheng Z, Abouelleil A, et al. Evolutionary toggling of the MAPT 17q21. 31 inversion region. Nat Genet. 2008;40:1076–1083. doi: 10.1038/ng.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giannuzzi G, Siswara P, Malig M, Marques-Bonet T, Program NCS, Mullikin JC, Ventura M, Eichler EE. Evolutionary dynamism of the primate LRRC37 gene family. Genome Res. 2013;23:46–59. doi: 10.1101/gr.138842.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bekpen C, Tastekin I, Siswara P, Akdis CA, Eichler EE. Primate segmental duplication creates novel promoters for the LRRC37 gene family within the 17q21. 31 inversion polymorphism region. Genome Res. 2012;22:1050–1058. doi: 10.1101/gr.134098.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popesco MC, Maclaren EJ, Hopkins J, Dumas L, Cox M, Meltesen L, McGavran L, Wyckoff GJ, Sikela JM. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- 19.Johnson ME, Viggiano L, Bailey JA, Abdul-Rauf M, Goodwin G, Rocchi M, Eichler EE. Positive selection of a gene family during the emergence of humans and African apes. Nature. 2001;413:514–519. doi: 10.1038/35097067. [DOI] [PubMed] [Google Scholar]
- 20.Ciccarelli FD, von Mering C, Suyama M, Harrington ED, Izaurralde E, Bork P. Complex genomic rearrangements lead to novel primate gene function. Genome Res. 2005;15:343–351. doi: 10.1101/gr.3266405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dumas LJ, O’Bleness MS, Davis JM, Dickens CM, Anderson N, Keeney JG, Jackson J, Sikela M, Raznahan A, Giedd J, et al. DUF1220-domain copy number implicated in human brain-size pathology and evolution. Am J Hum Genet. 2012;91:444–454. doi: 10.1016/j.ajhg.2012.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandepoele K, Van Roy N, Staes K, Speleman F, van Roy F. A novel gene family NBPF intricate structure generated by gene duplications during primate evolution. Mol Biol Evol. 2005;22:2265–2274. doi: 10.1093/molbev/msi222. [DOI] [PubMed] [Google Scholar]
- 23.Perry GH, Yang F, Marques-Bonet T, Murphy C, Fitzgerald T, Lee AS, Hyland C, Stone AC, Hurles ME, Tyler-Smith C, et al. Copy number variation and evolution in humans and chimpanzees. Genome Res. 2008;18:1698–1710. doi: 10.1101/gr.082016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stahl PD, Wainszelbaum MJ. Human-specific genes may offer a unique window into human cell signaling. Sci Signal. 2009;2:pe59. doi: 10.1126/scisignal.289pe59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25•.Zimmer F, Montgomery SH. Phylogenetic Analysis Supports a Link between DUF1220 Domain Number and Primate Brain Expansion. Genome Biol Evol. 2015;7:2083–2088. doi: 10.1093/gbe/evv122. This paper provides additional support that HSD expansions of DUF1220 could contribute to increased brain size in humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26•.Davis JM, Searles VB, Anderson N, Keeney J, Dumas L, Sikela JM. DUF1220 dosage is linearly associated with increasing severity of the three primary symptoms of autism. PLoS Genet. 2014;10:e1004241. doi: 10.1371/journal.pgen.1004241. The authors present a linear association with DUF1220 copy number and increasing severity of symptoms in 170 individuals with autism spectrum disorder. This represents the first study linking dosage of a core duplicon gene with a neurological disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fortna A, Kim Y, MacLaren E, Marshall K, Hahn G, Meltesen L, Brenton M, Hink R, Burgers S, Hernandez-Boussard T, et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:E207. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sudmant PH, Kitzman JO, Antonacci F, Alkan C, Malig M, Tsalenko A, Sampas N, Bruhn L, Shendure J, Genomes P, et al. Diversity of human copy number variation and multicopy genes. Science. 2010;330:641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Coe BP, Witherspoon K, Rosenfeld JA, van Bon BW, Vulto-van Silfhout AT, Bosco P, Friend KL, Baker C, Buono S, Vissers LE, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. A CNV morbidity map from an unprecedented 29,085 children with developmental delay compared with 19,584 healthy controls used to characterize CNVs significantly associated with neurocognitive disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaminsky EB, Kaul V, Paschall J, Church DM, Bunke B, Kunig D, Moreno-De-Luca D, Moreno-De-Luca A, Mulle JG, Warren ST, et al. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Charrier C, Joshi K, Coutinho-Budd J, Kim JE, Lambert N, de Marchena J, Jin WL, Vanderhaeghen P, Ghosh A, Sassa T, et al. Inhibition of SRGAP2 function by its human-specific paralogs induces neoteny during spine maturation. Cell. 2012;149:923–935. doi: 10.1016/j.cell.2012.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dennis MY, Nuttle X, Sudmant PH, Antonacci F, Graves TA, Nefedov M, Rosenfeld JA, Sajjadian S, Malig M, Kotkiewicz H, et al. Evolution of human-specific neural SRGAP2 genes by incomplete segmental duplication. Cell. 2012;149:912–922. doi: 10.1016/j.cell.2012.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Florio M, Albert M, Taverna E, Namba T, Brandl H, Lewitus E, Haffner C, Sykes A, Wong FK, Peters J, et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science. 2015;347:1465–1470. doi: 10.1126/science.aaa1975. The first study to show evidence that HSD ARHGAP11B may have contributed to the expansion of the neocortex in humans. It represents one of only a handful of studies that provides functional evidence of an HSD contributing to a human-specific trait. [DOI] [PubMed] [Google Scholar]
- 35•.Antonacci F, Dennis MY, Huddleston J, Sudmant PH, Steinberg KM, Rosenfeld JA, Miroballo M, Graves TA, Vives L, Malig M, et al. Palindromic GOLGA8 core duplicons promote chromosome 15q13. 3 microdeletion and evolutionary instability. Nat Genet. 2014;46:1293–1302. doi: 10.1038/ng.3120. Characterization of extraordinary structural diversity of the 15q13.3 microdeletion-associated region in human populations. Included in this study is the evolution of the ARHGAP11B-truncated SD and the role that GOLGA core duplicons played in mediating this and all other structural variant events. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Boyd JL, Skove SL, Rouanet JP, Pilaz LJ, Bepler T, Gordan R, Wray GA, Silver DL. Human-chimpanzee differences in a FZD8 enhancer alter cell-cycle dynamics in the developing neocortex. Curr Biol. 2015;25:772–779. doi: 10.1016/j.cub.2015.01.041. Not discussed in depth here, but a remarkable example of a rapidly evolving regulatory region important in brain expansion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jobling MA, Hurles M, Tyler-Smith C. Human evolutionary genetics: origins, peoples & disease. New York: Garland Science; 2004. [Google Scholar]
- 39.Sharp AJ, Mefford HC, Li K, Baker C, Skinner C, Stevenson RE, Schroer RJ, Novara F, De Gregori M, Ciccone R, et al. A recurrent 15q13. 3 microdeletion syndrome associated with mental retardation and seizures. Nat Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Locke DP, Segraves R, Carbone L, Archidiacono N, Albertson DG, Pinkel D, Eichler EE. Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization. Genome Res. 2003;13:347–357. doi: 10.1101/gr.1003303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41•.Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Hsi-Yang Fritz M, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. Comprehensive characterization of strucutural variation from the 1000 Genomes Project. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Handsaker RE, Van Doren V, Berman JR, Genovese G, Kashin S, Boettger LM, McCarroll SA. Large multiallelic copy number variations in humans. Nat Genet. 2015;47:296–303. doi: 10.1038/ng.3200. Improving on previous sequencing read-depth assays, the authors use whole-genome sequencing data to better characterize large mutiallelic CNVs (mCNVs) in 849 human genomes from the 1000 Genome Project. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell CD, Sampas N, Tsalenko A, Sudmant PH, Kidd JM, Malig M, Vu TH, Vives L, Tsang P, Bruhn L, et al. Population-genetic properties of differentiated human copy-number polymorphisms. Am J Hum Genet. 2011;88:317–332. doi: 10.1016/j.ajhg.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Population-genetic properties of differentiated human copy-number polymorphisms. Nature. 2006;444:444–454. [Google Scholar]
- 45••.Sudmant PH, Mallick S, Nelson BJ, Hormozdiari F, Krumm N, Huddleston J, Coe BP, Baker C, Nordenfelt S, Bamshad M, et al. Global diversity, population stratification, and selection of human copy-number variation. Science. 2015;349:aab3761. doi: 10.1126/science.aab3761. Using whole-genome sequencing data from 236 individuals from 125 distinct human populations, this paper systematically compares the burden and contribution of SNPs, indels, and structural variants in the same genomes. It shows that almost half of the variant base pairs between two individuals are due to copy number variation in SDs. It also produces a catalog of over 1,000 differentiated CNVs between populations, including the discovery of the first introgressed SD from an archaic hominin back into human. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Locke DP, Sharp AJ, McCarroll SA, McGrath SD, Newman TL, Cheng Z, Schwartz S, Albertson DG, Pinkel D, Altshuler DM, et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am J Hum Genet. 2006;79:275–290. doi: 10.1086/505653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Groot PC, Bleeker MJ, Pronk JC, Arwert F, Mager WH, Planta RJ, Eriksson AW, Frants RR. The human alpha-amylase multigene family consists of haplotypes with variable numbers of genes. Genomics. 1989;5:29–42. doi: 10.1016/0888-7543(89)90083-9. [DOI] [PubMed] [Google Scholar]
- 49.Carpenter D, Dhar S, Mitchell LM, Fu B, Tyson J, Shwan NA, Yang F, Thomas MG, Armour JA. Obesity, starch digestion and amylase association between copy number variants at human salivary (AMY1) and pancreatic (AMY2) amylase genes. Hum Mol Genet. 2015;24:3472–3480. doi: 10.1093/hmg/ddv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50••.Usher CL, Handsaker RE, Esko T, Tuke MA, Weedon MN, Hastie AR, Cao H, Moon JE, Kashin S, Fuchsberger C, et al. Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity. Nat Genet. 2015;47:921–925. doi: 10.1038/ng.3340. Using whole-genome sequencing, droplet digital PCR, and optical mapping, the authors characterized eight structural haplotypes at the amylase locus. Though previous studies have shown evidence for diverse haplotypes, this was the first to provide a detailed reconstruction of all predicted haplotypes published to date. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fledel-Alon A, Leffler EM, Guan Y, Stephens M, Coop G, Przeworski M. Variation in human recombination rates and its genetic determinants. PLoS One. 2011;6:e20321. doi: 10.1371/journal.pone.0020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boettger LM, Handsaker RE, Zody MC, McCarroll SA. Structural haplotypes and recent evolution of the human 17q21. 31 region. Nat Genet. 2012;44:881–885. doi: 10.1038/ng.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinberg KM, Antonacci F, Sudmant PH, Kidd JM, Campbell CD, Vives L, Malig M, Scheinfeldt L, Beggs W, Ibrahim M, et al. Structural diversity and African origin of the 17q21. 31 inversion polymorphism. Nat Genet. 2012;44:872–880. doi: 10.1038/ng.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koolen DA, Vissers LE, Pfundt R, de Leeuw N, Knight SJ, Regan R, Kooy RF, Reyniers E, Romano C, Fichera M, et al. A new chromosome 17q21. 31 microdeletion syndrome associated with a common inversion polymorphism. Nat Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 56.Sharp AJ, Hansen S, Selzer RR, Cheng Z, Regan R, Hurst JA, Stewart H, Price SM, Blair E, Hennekam RC, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 57.Itsara A, Vissers LE, Steinberg KM, Meyer KJ, Zody MC, Koolen DA, de Ligt J, Cuppen E, Baker C, Lee C, et al. Resolving the breakpoints of the 17q21. 31 microdeletion syndrome with next-generation sequencing. Am J Hum Genet. 2012;90:599–613. doi: 10.1016/j.ajhg.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koolen DA, Kramer JM, Neveling K, Nillesen WM, Moore-Barton HL, Elmslie FV, Toutain A, Amiel J, Malan V, Tsai AC, et al. Mutations in the chromatin modifier gene KANSL1 cause the 17q21. 31 microdeletion syndrome. Nat Genet. 2012;44:639–641. doi: 10.1038/ng.2262. [DOI] [PubMed] [Google Scholar]
- 59.Ikram MA, Fornage M, Smith AV, Seshadri S, Schmidt R, Debette S, Vrooman HA, Sigurdsson S, Ropele S, Taal HR, et al. Common variants at 6q22 and 17q21 are associated with intracranial volume. Nat Genet. 2012;44:539–544. doi: 10.1038/ng.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaisson MJ, Wilson RK, Eichler EE. Genetic variation and the de novo assembly of human genomes. Nat Rev Genet. 2015;16:627–640. doi: 10.1038/nrg3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, Antonacci F, Surti U, Sandstrom R, Boitano M, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517:608–611. doi: 10.1038/nature13907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gordon D, Huddleston J, Chaisson MJ, Hill CM, Kronenberg ZN, Munson KM, Malig M, Raja A, Fiddes I, Hillier LW, et al. Long-read sequence assembly of the gorilla genome. Science. 2016;352:aae0344. doi: 10.1126/science.aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63•.Huddleston J, Ranade S, Malig M, Antonacci F, Chaisson M, Hon L, Sudmant PH, Graves TA, Alkan C, Dennis MY, et al. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res. 2014;24:688–696. doi: 10.1101/gr.168450.113. This paper presents a strategy to perform targeted sequencing of BAC clones using single-molecule, real-time (SMRT) sequencing to resolve complex regions associated with SDs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64•.Tai DJ, Ragavendran A, Manavalan P, Stortchevoi A, Seabra CM, Erdin S, Collins RL, Blumenthal I, Chen X, Shen Y, et al. Engineering microdeletions and microduplications by targeting segmental duplications with CRISPR. Nat Neurosci. 2016;19:517–522. doi: 10.1038/nn.4235. The authors present a method to use CRISPR/Cas9 genome editing to engineer deletions and duplications by targeting flanking SDs mediating CNVs at regions associated with neurocognitive disorders in iPSCs. This presents a potential strategy to model large CNVs associated with disease and target HSDs for future functional studies. [DOI] [PMC free article] [PubMed] [Google Scholar]