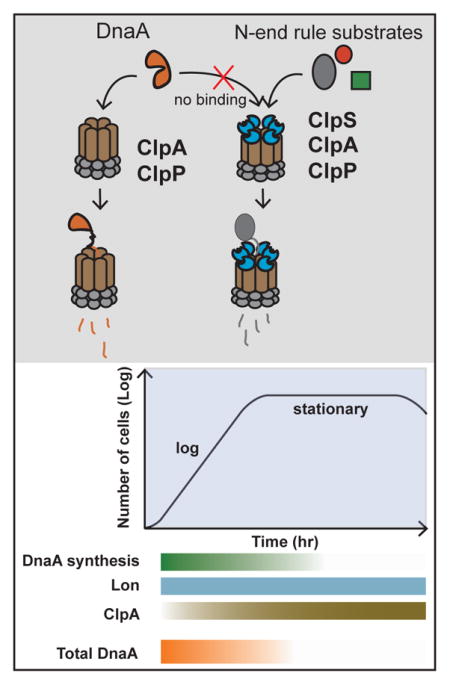
Summary
The Clp family of proteases is responsible for controlling both stress responses and normal growth. In Caulobacter crescentus, the ClpXP protease is essential and drives cell cycle progression through adaptor-mediated degradation. By contrast, the physiological role for the ClpAP protease is less well understood with only minor growth defects previously reported for ΔclpA cells. Here, we show that ClpAP plays an important role in controlling chromosome content and cell fitness during extended growth. Cells lacking ClpA accumulate aberrant numbers of chromosomes upon prolonged growth suggesting a defect in replication control. Levels of the replication initiator DnaA are elevated in ΔclpA cells and degradation of DnaA is more rapid in cells lacking the ClpA inhibitor ClpS. Consistent with this observation, ClpAP degrades DnaA in vitro while ClpS inhibits this degradation. In cells lacking Lon, the protease previously shown to degrade DnaA in Caulobacter, ClpA overexpression rescues defects in fitness and restores degradation of DnaA. Finally, we show that cells lacking ClpA are particularly sensitive to inappropriate increases in DnaA activity. Our work demonstrates an unexpected effect of ClpAP in directly regulating replication through degradation of DnaA and expands the functional role of ClpAP in Caulobacter.
Graphical Abstract
Introduction
Regulated proteolysis helps maintain correct intracellular protein levels and controls many essential biological processes. In bacteria, proteolysis of intracellular proteins is achieved by diverse members of the AAA+ (ATPases Associated with diverse cellular activities) family of oligomeric proteases (Neuwald et al., 1999; Sauer and Baker, 2011). Those proteases share similar structural composition of an ATP hydrolysis powered unfoldase coordinating with a nonspecific peptidase (Gottesman et al., 1997; Baker and Sauer, 2006). Some proteases are encoded by two polypeptides that form a fully functional protease (such as ClpXP and ClpAP) that encodes ATPase domain and peptidase domain separately, while others are defined by single polypeptides (Lon or FtsH). Unlike in eukaryotes, where one proteasome is sufficient for all ubiquitin-based degradation, bacterial proteases have more specialized substrate preferences that often rely on unique sequence motifs or additional adaptor proteins (Ciechanover, 1994; Dougan et al., 2003; Chien et al., 2007; Mukherjee et al., 2015; Lau et al., 2015; Joshi et al., 2015). This specificity allows for accurate special and temporal regulation of multiple cellular factors and coordination between protein level and cell development stages (Dougan et al., 2002; Inobe and Matouschek, 2008).
ClpA is a chaperone that forms a hexameric ring structure that unfolds and translocates substrate through its central pore to the ClpP proteolytic chamber. In many bacteria ClpA is co-transcribed from the same operon as the adaptor protein ClpS, which binds ClpA N-terminus and helps degrade N-end rule substrate (Guo et al., 2002; Zeth et al., 2002; Erbse et al., 2006; De Donatis et al., 2010). ClpS appears to tether N-end rule substrates to ClpA to form high affinity complexes where ClpA engages an unstructured ClpS N-terminal extension to transfer substrates through the same pore (Román-Hernández et al., 2011; Rivera-Rivera et al., 2014). ClpA can also recognize a number of substrates on its own, such as ssrA-tagged proteins and RepA (Gottesman, et al. 1998; Herman et al. 1998; Pak and Wickner, 1997; Hoskins et al. 2000; Hoskins et al. 2002). In many of these cases where ClpAP can act on its own, ClpS inhibits proteolytic activity, demonstrating a dual role for ClpS in ClpA mediated protein degradation (Dougan, et al. 2002; Wang, et al. 2007).
Regulated proteolysis is critical in the gram-negative bacterium Caulobacter crescentus to control cell cycle progression, DNA replication, division and stress responses (Quardokus et al., 1996; Domian et al., 1997; Jenal and Fuchs, 1998; Tsai and Alley, 2001; Abel et al., 2011). ClpAP was recently reported to degrade the cell divisome proteins FtsZ and FtsA to promote asymmetric cell division. This degradation occurs both in vivo and in vitro, with in vitro results showing no requirement for ClpS (Williams et al., 2014). Similarly, the FliF protein was shown to be cyclically degraded by ClpA in a ClpS independent manner (Grünenfelder et al., 2004). Despite these known substrates, loss of ClpA was originally reported to result in only slightly slower growth and moderate morphological defects (Grünenfelder et al., 2004). However, the need for ClpA may be more accentuated during different growth conditions, during stress, or when known ClpA substrates are misregulated (Williams et al., 2014).
In this study we show an unexpected ClpA-specific cell growth defect in Caulobacter crescentus that links ClpA with chromosome regulation. We show that the replication initiator DnaA is directly recognized by ClpAP and characterize this activity both in vivo and in vitro. ClpS inhibits DnaA degradation by ClpAP in vitro and suppresses ClpAP degradation of DnaA during normal growth. DnaA levels fall during entry into stationary phase and ClpAP activity is needed for the complete removal of DnaA during this transition. Although our previous work showed that Lon is the dominant protease for DnaA during exponential growth (Jonas, et al. 2013), we find that upregulation of ClpAP can prevent the toxic accumulation of DnaA in cells lacking Lon. Interestingly, Lon is deficient for degrading an active ATP-bound form of DnaA, while ClpAP retains similar proteolytic kinetics for this variant. Consistent with this, cells lacking ClpA are especially sensitive to aberrant increases in DnaA activity. Together, these results suggest that ClpAP mediated degradation may be controlling levels of active DnaA species in concert with Lon to regulate DNA replication during cell growth and development.
Results
Loss of ClpA results in cellular defects that link to aberrant DnaA accumulation
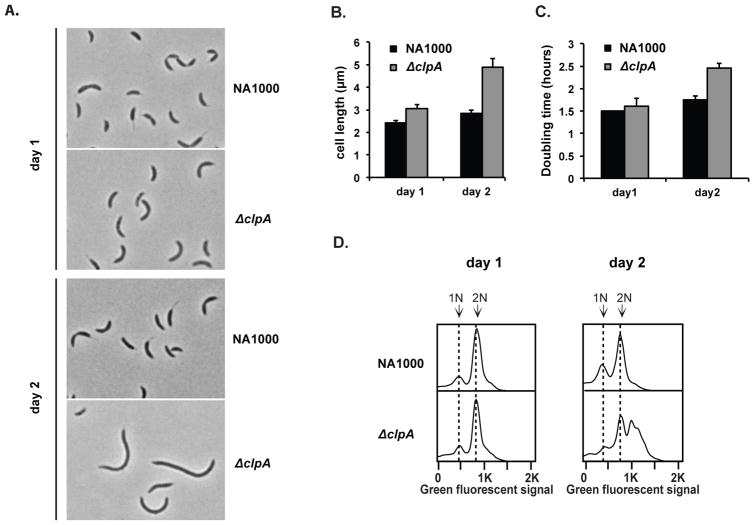
During our exploration of the roles of AAA+ proteases in Caulobacter replication and development, we found that cells lacking ClpA were defective upon extended growth in complex media. In particular, initial growth of ΔclpA cells by standard inoculation into liquid complex media from agar plates showed similar growth to wildtype cells during the initial stages (<12 hours). However, growth of this strain for another 24 hours revealed clear defects. For example, ΔclpA cells were elongated relative to wildtype cells (Fig. 1A,B), failed to grow as readily from stationary phase upon dilution into fresh media (Fig. 1C), and aberrantly accumulated chromosomes (Fig. 1D).
FIGURE 1. ClpA influences long-term growth of C. crescentus.
A. Morphology of wildtype or ΔclpA cells in nutrient rich liquid media (PYE) for short (1 day) or prolonged growth (2 days). Cells have reached stationary phase (OD~ 1.8) in these conditions. B. Quantifications of cell length after 1 or 2 days (n=200; error bars represent 95% CI). C. Doubling time of strains inoculated into fresh media from stationary phases after either 1 or 2 days of growth (n=2; error bars represent SD). D. Flow cytometry profiles showing chromosome content of strains after 1 or 2 days growth in liquid PYE media. Sytox Green fluorescence is used as a measure of DNA content (see methods for details).
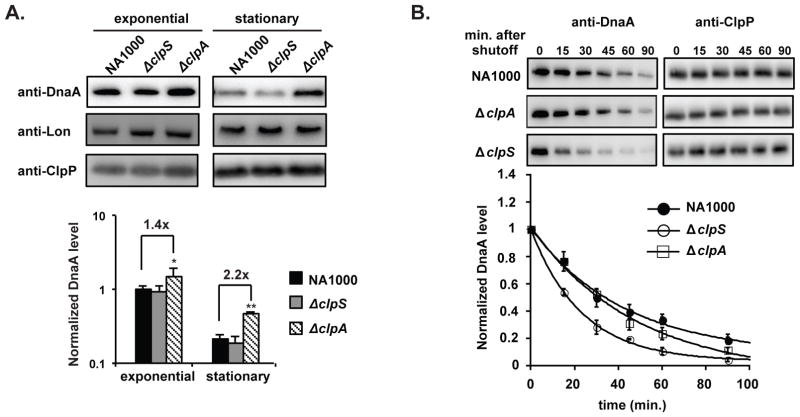
We reasoned that this chromosome accumulation defect might be linked to misregulation of the replication factors. Among these factors, the replication initiator DnaA was of particular interest, as DnaA levels are controlled partly through proteolysis and are growth phase regulated (Gorbatyuk and Marczynski, 2005; Jonas et al., 2013; Leslie et al., 2015). Interestingly, it was initially reported that the turnover of DnaA might involve ClpP related protease (Gorbatyuk and Marczynski, 2005). However, later work showed that the dominant protease for DnaA was Lon, which degrades DnaA both in exponential growth and in stationary phase (Jonas et al., 2013; Leslie et al., 2015). To determine if ClpA could be involved in DnaA regulation, we first compared levels of DnaA protein in wildtype and ΔclpA cells growing in complex media. Although DnaA levels were only slightly higher in ΔclpA cells compared to wildtype cells during exponential growth, this difference became more pronounced upon entry into stationary phase (Fig. 2A). Interestingly, we found ClpA levels increased when cells entering stationary phase, supporting an increased role for ClpAP dependent proteolysis in this growth stage (Fig. S1). We also examined other protease-regulated factors and found that CtrA (a ClpXP substrate) and FtsZ (a ClpX/AP substrate) levels were not ClpA dependent (Fig S2). Levels of the Lon substrate SciP were surprisingly reduced in cells lacking ClpA regardless of growth phase, which might contribute to the phenotype of ΔclpA cells.
FIGURE 2. ClpA reduces DnaA levels in C. crescentus.
A. Levels of DnaA, Lon and ClpP in wildtype, ΔclpS and ΔclpA strains during exponential growth (3 hrs) or entering stationary phase (12 hrs) as shown by western blotting. Quantification of DnaA levels shown below (n= 3; error bars represent SD). P<0.05 (*) or P<0.01 (**). B. DnaA degradation following translational shutoff by chloramphenicol in wildtype, ΔclpS and ΔclpA strains during exponential growth. ClpP levels shown as controls. Quantification of DnaA is shown below (n=3; error bars represent SD).
Next we monitored protein turnover rate by measuring levels of DnaA following antibiotic induced arrest of translation. Loss of ClpA did not dramatically change bulk DnaA degradation during the course of the assay, but loss of ClpS yielded a modest stimulation of DnaA degradation (Fig. 2B). These results suggest that endogenous levels of ClpS inhibit most of the ClpAP dependent DnaA degradation during normal growth, similar to how ClpS inhibits ssrA-tagged protein degradation by ClpAP (Dougan, et al. 2002). This inhibitory effect may explain why the ability of ClpAP to degrade DnaA was originally overlooked (Gorbatyuk and Marczynski, 2005; Jonas et al., 2013)
Upregulation of ClpAP restores DnaA degradation to cells lacking Lon
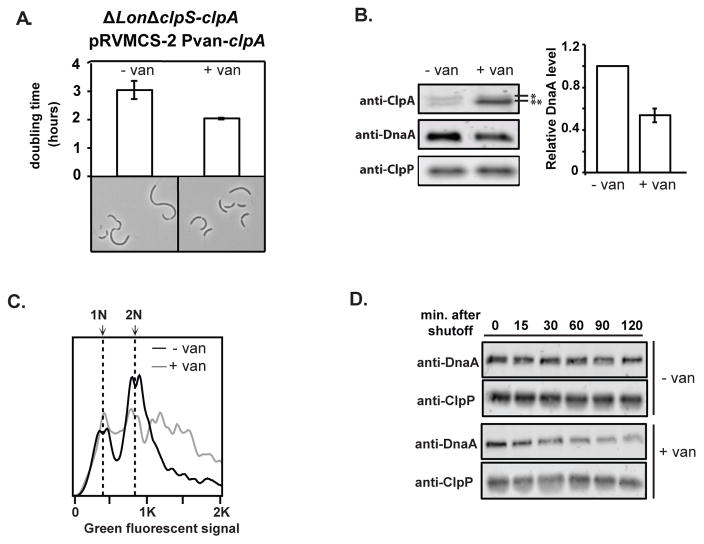
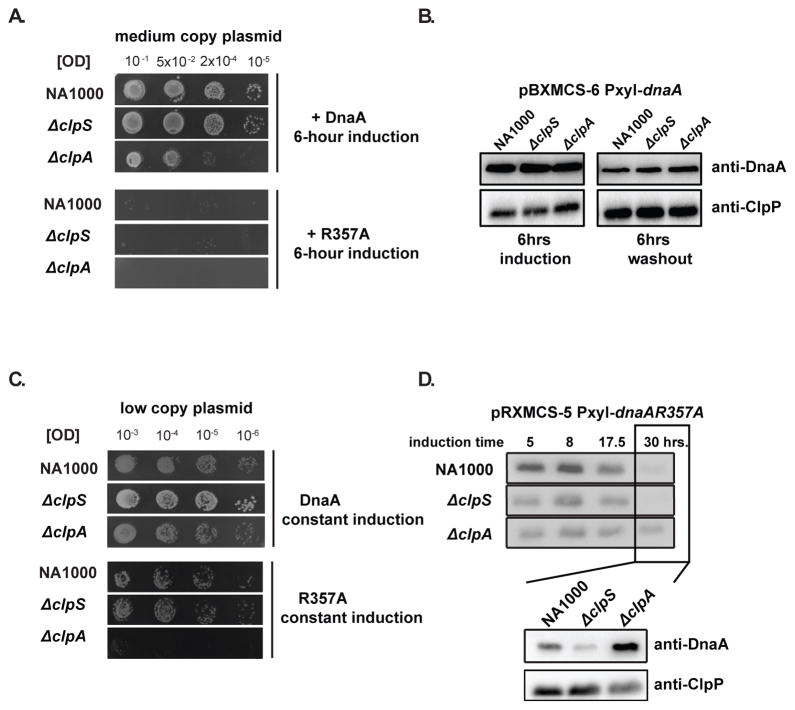
Because Lon plays a major role in DnaA degradation, we were concerned that ClpAP proteolysis of DnaA may be masked by Lon activity during normal growth conditions. Therefore, we next asked if the role of ClpAP in DnaA regulation would be accentuated in the absence of Lon. We deleted lon in a strain lacking the clpS-clpA operon to generate a triple deletion strain (ΔlonΔclpS-clpA). As expected, these cells grew poorly and showed elongated morphologies even under exponential growth conditions (Fig. 3A). All these effects were suppressed upon induction of ClpA expression, including a dramatic reversal of the abnormal chromosome accumulation (Fig. 3A–3C). Western blotting showed that increased ClpA levels were correlated with reduced steady-state levels of DnaA in this background (Fig. 3B). Most importantly, DnaA degradation was increased upon overexpression of ClpA (Fig. 3D). Taken together, these data suggest that ClpAP can degrade DnaA and shield cells from the deleterious effects of DnaA accumulation when Lon activity is compromised and/or when ClpS is absent.
FIGURE 3. ClpAP degradation of DnaA is crucial when Lon is compromised.
A. Morphology and doubling time of cells lacking ClpS, ClpA and Lon (ΔlonΔclpS-clpA), either without (−van) or with (+van) vanillate induced expression of ClpA from a low copy plasmid (pRVMCS-2 Pvan-clpA). B. Steady state levels of ClpA, DnaA and ClpP in these strains. *: cross-reacting band. **: ClpA (note leaky expression in absence of inducer) (n = 2; error bars represent SD). C. Flow cytometry profiles showing chromosome content of ΔlonΔclpS-clpA, with or without vanillate induced expression of ClpA. D. DnaA degradation upon translation shutoff in ΔlonΔclpS-clpA cells either with or without vanillate induced expression of ClpA.
ClpAP degrades DnaA in vitro
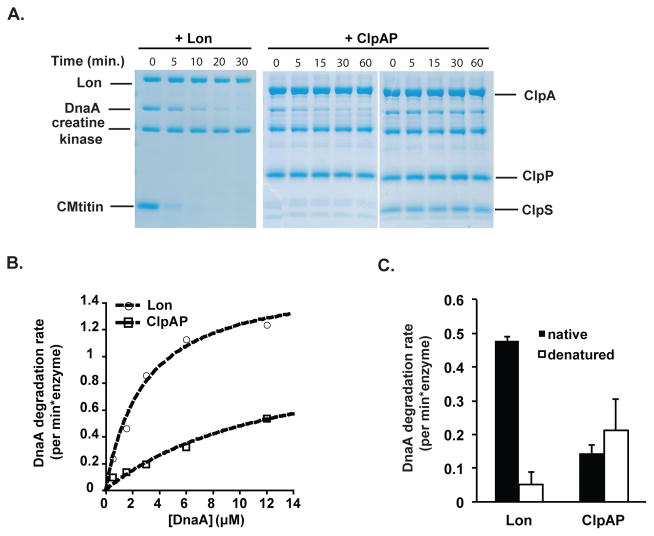
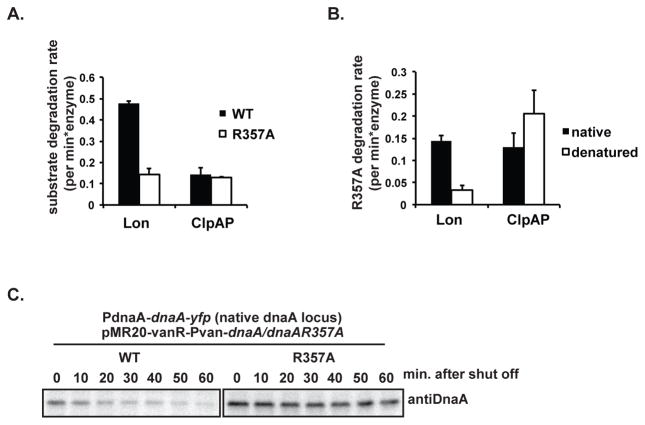
We next used purified components to reconstitute ClpAP degradation of DnaA. In these experiments we used a truncated ClpA that does not degrade itself like wildtype ClpA, but has otherwise wildtype activity (Maglica et al., 2008). Using purified proteins, we found that ClpAP alone was able to degrade DnaA and, consistent with the in vivo results, ClpS inhibits this activity (Figure 4A). A full kinetic characterization (Fig. 4B) shows that ClpAP degrades DnaA (turnover rate = 1.1 +/− 0.3 min−1; KM = 13 +/− 6 μM) with lower activity than Lon (turnover rate = 1.6 +/− 0.1 min−1; KM = 2.9 +/− 0.7 μM), supporting the stronger intracellular role for Lon in regulating DnaA levels.
FIGURE 4. ClpAP degrades DnaA in reconstituted in vitro assays.
A. DnaA degradation by 0.2 μM Lon and 1 μM ClpAP in the absence or presence of ClpS. B. Kinetics of Lon or ClpAP dependent DnaA degradation at various DnaA concentrations. Fits are to the Michaelis-Menten equation. C. Comparing degradation of native and denatured DnaA by Lon and ClpAP (n= 3; error bars represent SD). n.b. the remnant amount of urea in the final reactions do not affect global Lon activity (Figure S4).
We wondered if the differences in DnaA degradation by these proteases could be due to differences in recognition determinants dependent on the specific protease. The Lon protease is known to degrade damaged proteins upon recognizing their misfolded state, and we reasoned that Lon recognition might be more sensitive to DnaA conformational changes. We tested this hypothesis by denaturing DnaA with urea prior to the proteolysis assays. The two proteases appear to recognize DnaA in different ways: denaturation of DnaA reduced recognition by Lon, while denaturation of DnaA did not affect ClpAP degradation (Fig. 4C). Note that the small amount of urea carried over from denaturation did not abolish Lon activity (Figure S3). This result was surprising given that Lon is generally thought to recognize misfolded proteins and implies that a specific structural motif from the natively folded DnaA is recognized by Lon protease. By contrast, we speculate that ClpAP recognizes DnaA via sequence determinant(s) accessible in both folded and denatured form. These results suggest that although DnaA is degraded through redundant proteolytic pathways, these pathways may serve different purposes of DnaA regulation under different conditions
Degradation of DnaA appears linked to its activity or nucleotide bound state
Our overexpression data suggest the ClpAP can degrade DnaA, but that ClpS is normally inhibiting this activity. If there is truly so little DnaA degradation via ClpAP in vivo, why would loss of ClpA result in any aberrant chromosome accumulation? One possibility stems from the fact that DnaA regulation is highly complex and DnaA activity depends on both its levels and nucleotide bound state. ATP-bound DnaA is the active conformation with higher affinity for weak DnaA binding boxes in the replication origin than the ADP-bound state. (McGarry et al., 2004; Camara et al., 2005; Erzberger et al., 2006). Conversion between the two states is slow, requiring either exchange of nucleotide or hydrolysis of the ATP by DnaA, a normally slow process that can be accelerated by certain cellular factors. Based on our in vitro results, we hypothesized that ClpAP and Lon might recognize different DnaA conformations that might correspond to different nucleotide bound versions of DnaA. To test this hypothesis, we used a previously characterized active constitutively ATP-bound DnaA mutant, DnaAR357A, which induces replication over-initiation and aberrant chromosome accumulation in Caulobacter (Collier and Shapiro, 2009; Jonas et al., 2011).
DnaAR357A variant was degraded poorly by Lon in vitro, but ClpAP was still able to degrade this variant with kinetics similar to wildtype DnaA (Fig. 5A). The decreased Lon-specific degradation of DnaAR357A is not because it is poorly structured when purified as it retains specific DNA binding similar to wildtype DnaA (Fig. S4). Moreover, denaturation of DnaAR357A reduced its degradation by Lon to the comparable rate as denatured DnaA, indicating that once the substrate is denatured, the R357A mutation does not have an additional effect on degradation (Figure 5B). Consistent with this in vitro result and the major role of Lon in DnaA degradation in vivo, DnaAR357A was degraded more slowly than wildtype DnaA when expressed in the cell (Fig. 5C), similar to what has been recently reported (Wargachuk and Marcynski, 2015). Our working model is that while ClpAP does not dramatically affect bulk DnaA turnover, it is particularly well suited for degrading the active ATP-bound DnaA conformation. If this is true, then the inappropriate retention of active DnaA may explain the reduced viability of cells lacking ClpA upon extended growth.
FIGURE 5. Lon degrades DnaAR357A poorly.
A. Comparison of DnaA and DnaAR357A degradation by Lon and ClpAP (n= 3; error bars represent SD). B. Degradation of denatured DnaA and DnaAR357A by Lon and ClpAP (n= 3; error bars represent SD). C. In vivo degradation of DnaA or DnaAR357A following translation shutoff by chloramphenicol. DnaAR357A was expressed in a strain harboring a functional DnaA-YFP fusion to allow for resolution of wild type and mutant variants.
Loss of ClpA sensitizes cells to increased DnaA activity
Our results so far indicate a role for ClpAP in regulating functional levels of DnaA in the cell. Because an excessive amount of DnaA is toxic (Jonas et al., 2011), we hypothesized that ΔclpA cells would be even more sensitive to increased DnaA activity. Transient overexpression of wildtype DnaA from a medium copy plasmid was more toxic to strains that lacked ClpA (Fig. 6A). However, no substantial changes in DnaA levels were observed between strains using this overexpression system (Fig. 6B). We speculated that excessive overexpression of DnaA may result in so much substrate that the role of ClpA is masked in these circumstances and hypothesized that milder chronic upregulation of DnaA activity may better reveal the regulation of ClpA. Mild upregulation of wildtype DnaA from a low copy plasmid did not affect growth of either wildtype or ΔclpA strains (Fig. 6C). By contrast, this level of expression of the active ATP-bound DnaAR357A variant resulted in poor growth for ΔclpA strains (Fig. 6C) and increased levels of DnaA in extended growth conditions (Fig 6D). Taken together with the increased accumulation of DnaA seen with ΔclpA strains (Figure 2), these data support a model where ClpA works in concert with the Lon protease to protect cells from the toxic consequences of excessive DnaA activity.
FIGURE 6. ClpA protects cells from over-activation of DnaA.
A. Wildtype or mutant strains carrying inducible dnaA on a medium copy plasmid were induced for 6 hours (0.2% xylose). Cells were serially diluted, then plated on PYE agar without inducer. B. Protein levels in wildtype, ΔclpS and ΔclpA strains after 6 hours of DnaA induction and after an additional 6 hours following removal of the inducer. C. Wildtype or mutant strains carrying dnaA or dnaAR357A on a low copy plasmid were serially diluted and plated on PYE agar with 0.2% xylose. D. Levels of DnaA in wildtype, ΔclpS and ΔclpA strains during growth where DnaAR357A is continuously induced from a low copy plasmid. Inset compares DnaA levels directly across all three strains and shows ClpP as a control.
Discussion
Our study identifies DnaA as a ClpAP substrate in Caulobacter crescentus. Degradation of DnaA by ClpAP in vitro is inhibited by the ClpS regulator and loss of ClpS accelerates loss of DnaA in vivo, supporting a role for ClpS in inhibiting ClpAP activity under normal conditions. Our triple deletion results suggest that ClpA plays an important role in regulating DnaA when Lon is absent or Lon activity is compromised. One possible scenario is that under conditions where the Lon protease is occupied by high levels of other substrates, ClpAP can serve to support DnaA degradation to prevent unwanted accumulation. This type of saturation has been seen in E. coli, where RpoS degradation by ClpXP is reduced upon upregulation of other ClpXP substrates (Fredriksson et al., 2007; Cookson et al., 2011). Identifying conditions that result in reduction in Lon activity is clearly an interesting direction for future work.
ClpAP appears to play a particularly useful role in eliminating residual DnaA during stationary phase, where DnaA levels fall and ClpA levels rise. Interestingly, prior studies showed that ClpA levels also increase upon entry into stationary phase in Escherichia coli (Farrell et al., 2005). Perhaps increased ClpAP activity in stationary phase is a universal feature of bacteria entering this nutrient limiting, potentially stressful condition. In addition, the ClpS adaptor can both stimulate and repress substrate degradation, allowing for switches in ClpAP activity. A recent study in Agrobacterium tumefaciens found that levels of a ClpS paralog increased during entry into stationary phase, which suggests the potential for altering degradation by ClpAP in different growth stages (Stein et al., 2016). It will be interesting for future studies to explore in Caulobacter crescentus how relative ClpS and ClpA activity may vary under different growth conditions to understand the biological range of this inhibition. Finally, although we focus on the degradation of DnaA in this current work, we note that ClpAP also degrades the cytoskeletal proteins FtsZ and FtsA in Caulobacter (Williams, et al. 2014) and this activity may contribute to or even drive the cellular defects associated with ClpA loss in stationary phase cells, even though steady-state levels of FtsZ are unchanged (Figure S2). In addition, the loss of ClpA may have indirect effects, such as the loss of SciP in ΔclpA cells (Figure S2), that might also contribute to the over-replication and cellular defects described here.
Our identification of ClpAP as a redundant protease for DnaA may help integrate prior observations showing a possible role for the ClpP proteases in DnaA turnover (Gorbatyuk and Marczynski, 2005) with recent results illustrating the need for Lon in degrading bulk DnaA (Jonas, et al. 2013). Our working model is that during normal growth, Lon is the main protease responsible for DnaA degradation in part because ClpAP is inhibited by ClpS. When cells enter stationary phase, ClpAP activity increases either due to increased protease levels or altering the role of ClpS inhibition. Consistent with this interpretation, DnaA degradation seems increased in cells lacking clpS and is reduced in cells lacking lon. Despite its secondary role during normal exponential growth, ClpAP limits the toxic consequences of DnaA overexpression even during this stage. This is especially clear when overexpressing the active ATP-bound DnaA variant, which is resistant to Lon degradation. Together, our work suggests that ClpAP may act as a backup proteolysis pathway during stress conditions that works in concert with Lon or becomes dominant when the Lon protease is incapable of responding to increased DnaA activity.
Experimental Procedures
Strains and growth conditions
Strains and their genotypes in this study are listed in Table S1. Caulobacter crescentus cells were grown at 30°C in peptone yeast extract (PYE) media. For protein induction, stock solution 20% xylose (in H2O) or 50 mM vanillic acid/NaOH (pH7.5 in H2O) were added to liquid culture or agar plate to reach final concentration of 0.2% xylose, or 0.5 mM vanillic acid/NaOH. The final concentrations of antibiotics used in this study were: for Escherichia coli liquid and solid culture, 30μg/ml chloramphenicol, 100 μg/ml ampicillin, 50μg/ml kanamycin, 15μg/ml oxytetracycline or 50 μg/ml spectinomycin; for C. crescentus, 1 μg/ml (liquid) or 2 μg/ml (solid) chloramphenicol, 5 μg/ml (liquid) or 25 μg/ml (solid) kanamycin, 1 μg/ml (liquid) or 2 μg/ml (solid) oxytetracycline, or 25μg/ml (liquid) or 100 μg/ml (solid) spectinomycin.
Cloning and strain constructions
ClpA or DnaA overexpression strains carrying plasmids were generated by Gibson assembly of PCR product and double digested plasmids at NdeI and NheI/SpeI sites. ΔlonΔclpS-clpA strains were generated by two-step recombination cloning as described before (Jonas et al., 2013) with small revisions. The plasmid pNPTS138 with lon gene flanking region was transformed into ΔclpA strain or ΔclpS-clpA strains (Grünenfelder et al., 2004), and integration was selected by kanamycin resistance. Cells were grown in PYE media with 5 μg/ml kanamycin overnight, then back-diluted 1:100 into fresh PYE with 3% sucrose in the absence of kanamycin, grown for four hours, and plated on PYE + 3% sucrose agar to select for the loss of the sacB gene at the integration locus. The recombination was screened by colony PCR with primers outside of integration locus, and sequencing the insertion locus validated candidate clones with correct insertion size.
Protein purification and modification
Untagged ClpA, untagged Lon, and his-tagged ClpP were purified as before (Levchenko et al., 2000; Chien et al., 2007; Micevski and Dougan, 2013; Gora et al., 2013; Williams et al., 2014), with additional ion-exchange polishing for Lon if necessary. The ClpA* construct used for in vitro assays is a stable variant where the c-terminal 9-residue degron has been removed, but otherwise retains the same proteolytic activity as wildtype ClpA (Maglica et al., 2008). Titin-I27-β20 was purified as described (Gur and Sauer, 2008) using a GE superdex 75 size exclusion chromatography column with H-buffer (25mM HEPES PH7.5, 100mM KCl, 10mM MgCl2, 10% Glycerol (v/v) and 1mM DTT). CMtitin was generated by carboxymethylating the two cysteines in Titin-I27-β20 with iodoacetamide under urea denaturation condition as described (Jonas et al., 2013). Modified protein was stored at 4°C in TK buffer (25mM Tris PH8.0, 100mM KCl, 10mM MgCl2 and 1mM DTT). ClpS, DnaA and R357A mutant were purified as his6SUMO tagged protein, followed by tag cleavage as described (Wang et al., 2007). Exceptions to this protocol were that DnaA purification was carried in S-buffer (20% Sucrose, 25mM HEPES PH7.5, 200mM L-Glutamic acid potassium, 10mM MgCl2 and 1mM DTT), and further purified with an additional ion-exchange column (GE healthcare, MonoS G5/50) using a KCl gradient from 0.1M to 1M in MS-S buffer (20% Sucrose, 25mM Tris PH8.5, 2mM DTT). DnaAR357A mutant was further purified with a size exclusion chromatography column (GE healthcare, Superdex 200 10/300 GL) after tag cleavage.
In vitro Degradation assay
Degradation for all constructs were performed at 30°C with the following protease concentration except elsewhere indicated: 0.2μM Lon6, 0.2 μM ClpA*6, 0.4 μM ClpP14 and 1μM ClpS, with 4mM ATP, 15mM creatine phosphate (Sigma) and 75ug/ml creatine kinase (Roche) as ATP regeneration components. 10μl aliquots were taken at each time point and quenched with SDS loading dye (2% SDS, 6% Glycerol, 50mM Tris PH8.0 and 2mM DTT), and examined by SDS-PAGE. Using creatine kinase or ClpP as an internal loading control, the degradation rate was determined by protein band intensity change at different time points analyzed with ImageJ 1.47(NIH) software. To perform the degradation on denatured protein, urea was added to DnaA to reach 6M final concentrations, and the denaturation was carried overnight at room temperature. Denatured proteins were then run through a desalting column (Thermo Scientific) to remove excess urea and immediately followed by degradation components addition (protease and ATP regeneration system) to initiate the assay.
Sensitivity of strains to elevated DnaA activity
Parent strains containing low copy or medium copy DnaA or DnaAR357A overexpression plasmid were grown overnight in PYE with appropriate antibiotics and inoculated into fresh media the next day (1:100 dilution). Inoculated cultures were grown for 3 hours to allow cells to exit stationary phase, then 0.2% xylose was added to the culture. For low copy plasmid expression, cells were diluted to desire OD, and 3 μl of diluted culture were directly spotted on PYE + tetracycline agar media containing 0.2% xylose. For medium copy plasmid expression, since long-term overexpression of DnaA kills all strains, cells were induced for 6 hours at which point 1 ml of cells were taken, pelleted and washed twice with fresh PYE without xylose, then resuspended in PYE to reach OD 0.1. Cells were then serially diluted to desire OD, and 3 μl culture were spotted on PYE + chloramphenicol agar plates without xylose. Plates were incubated at 30 °C for 3 days and imaged under white light (G-box; Syngene).
In vivo protein level determination, in vivo degradation assay and flow cytometry
The degradation of in vivo protein was monitored by inhibiting protein synthesis upon addition of 30 μg/ml chloramphenicol into cells growing in exponential phase (OD 0.2–0.6). At each time point, 1ml of culture was taken, centrifuged at 15k rpm for 2 minutes and supernatant was removed. 100μl 2x SDS loading dye per 0.2 OD was added to the pellet, and the sample was frozen in liquid nitrogen. Samples in 2x SDS dye were then thawed, resuspended and boiled for 5 minutes for complete cell lysing. Following centrifugation to remove insoluble material, extracts were then resolved on 10% Bis-Tris gels by running at 150 V for 1 hour at room temperature to be transferred to PVDF membrane. Membrane was blocked with 3% milk in 1x TBST (Tris-based-saline with 0.05% Tween-20) for 15 minutes, then probed with primary antibody in 1x TBST at 4°C overnight with following dilution factors: 1:5,000 dilution of DnaA antibody, 1:2,000 dilution of ClpA antibody, 1:2,000 dilution of SciP antiserum, 1:2,000 dilution of Lon antibody, 1:2,000 dilution of CtrA antiserum, 1:5,000 antiFtsZ antibody or 1:20,000 ClpP antiserum. Membranes were washed with 1x TBST for 10 minutes twice, and then probed with goat-anti-rabbit HRP conjugated secondary antibody (Millipore) with 1:50,000 dilution in 1x TBST at room temperature for 2 hours and excess secondary antibody was washed away. The protein was visualized by the luminescence from HRP substrate (Millipore) on G-box (Syngene). Flow cytometry to measure chromosomal content in rifampicin-treated cells was performed as described (Chen et al., 2009) and analyzed by FlowJo v. 10.1 software.
Limited proteolysis and electrophoretic mobility shift assay
To perform limited trypsin digestion, a serial titrations of trypsin with concentration from 10μg/ml were added to 10μM DnaA in S-buffer, and the reactions were kept at 25°C for half an hour. To stop the reaction, 5mM protease inhibitor phenylmethylsulfonyl fluoride (PMSF) was added to the reactions, and the resulting fragments were separated by SDS-PAGE.
The electrophoretic mobility shift assays were performed with 5′ 6-FAM fluorophore labeled oligos. G1 DnaA box has the sequence AACGGATGATCCA CAGGAGAG (underline highlights G1 box) from Caulobacter crescentus origin and annealed with its reverse complement strand by heating at 95 °C and slow cooling (TK buffer). 0.5 μM annealed G1 DnaA box containing dsDNA or dT25 and various concentration of DnaA or DnaAR357A (2-fold dilution from 8μM) were incubated in TK buffer containing 1mM ATP for 10 minutes, and run on 0.8 % agarose gel in TAMK buffer (40mM Tris, 20mM acetic acid, 10mM MgCl2 and 100mM KCl, PH 8.5) at 80 volts for 30 minutes. Gels were scanned by a Typhoon 7000 scanner (GE health Life Sciences) and analyzed by Image J software.
Supplementary Material
Abbreviated Summary.
ClpA affects chromosomal content in Caulobacter crescentus.
The replication initiator DnaA is recognized by the ClpAP protease.
ClpAP degradation of DnaA is inhibited by the ClpS adaptor.
ClpA has a stronger effect on regulating DnaA levels in stationary phase.
Loss of ClpAP sensitizes cells to excess DnaA activity.
ClpAP is not the dominant protease for bulk DnaA degradation, but is particularly useful for degrading active DnaA.
Acknowledgments
We thank Urs Jenal (Biozentrum) for the ΔclpS and ΔclpS-clpA strains, and Tania Baker (MIT) for anti-ClpA antibodies. We thank members of the Chien lab for helpful feedback. Funding for KJ was provided by the Excellence Program LOEWE of the State Hessen, Germany. This work was sponsored by NIH R01GM111706 to P.C. and in part by funding from a Chemistry Biology Interface Program Training Grant (NIH T32GM08515) to J.L.
Footnotes
Author Contributions
P.C. and J.L. conceived the idea for the project. J.L., L.F., and K.J. performed experiments. J.L., K.J. and P.C. analyzed the results. J.L. and P.C. wrote the paper. All authors read the paper.
Conflict of interests
The authors declare that they have no conflicts of interest with the contents of this article.
References
- Abel S, Chien P, Wassmann P, Schirmer T, Kaever V, Laub MT, et al. Regulatory cohesion of cell cycle and cell differentiation through interlinked phosphorylation and second messenger networks. Mol Cell. 2011;43:550–560. doi: 10.1016/j.molcel.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara JE, Breier AM, Brendler T, Austin S, Cozzarelli NR, Crooke E. Hda inactivation of DnaA is the predominant mechanism preventing hyperinitiation of Escherichia coli DNA replication. EMBO Rep. 2005;6:736–741. doi: 10.1038/sj.embor.7400467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YE, Tsokos CG, Biondi EG, Perchuk BS, Laub MT. Dynamics of two Phosphorelays controlling cell cycle progression in Caulobacter crescentus. J Bacteriol. 2009;191:7417–7429. doi: 10.1128/JB.00992-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien P, Perchuk BS, Laub MT, Sauer RT, Baker TA. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc Natl Acad Sci USA. 2007;104:6590–6595. doi: 10.1073/pnas.0701776104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- Collier J, Shapiro L. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J Bacteriol. 2009;191:5706–5716. doi: 10.1128/JB.00525-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson NA, Mather WH, Danino T, Mondragón-Palomino O, Williams RJ, Tsimring LS, Hasty J. Queueing up for enzymatic processing: correlated signaling through coupled degradation. Mol Syst Biol. 2011;7:561–561. doi: 10.1038/msb.2011.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Donatis GM, Singh SK, Viswanathan S, Maurizi MR. A single ClpS monomer is sufficient to direct the activity of the ClpA hexamer. J Biol Chem. 2010;285:8771–8781. doi: 10.1074/jbc.M109.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Mogk A, Zeth K, Turgay K, Bukau B. AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett. 2002;529:6–10. doi: 10.1016/s0014-5793(02)03179-4. [DOI] [PubMed] [Google Scholar]
- Dougan DA, Weber-Ban E, Bukau B. Targeted delivery of an ssrA-tagged substrate by the adaptor protein SspB to its cognate AAA+ protein ClpX. Mol Cell. 2003;12:373–380. doi: 10.1016/j.molcel.2003.08.012. [DOI] [PubMed] [Google Scholar]
- Erbse A, Schmidt R, Bornemann T, Schneider-Mergener J, Mogk A, Zahn R, et al. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature. 2006;439:753–756. doi: 10.1038/nature04412. [DOI] [PubMed] [Google Scholar]
- Erzberger JP, Mott ML, Berger JM. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat Struct Mol Biol. 2006;13:676–683. doi: 10.1038/nsmb1115. [DOI] [PubMed] [Google Scholar]
- Farrell CM, Grossman AD, Sauer RT. Cytoplasmic degradation of ssrA-tagged proteins. Mol Microbiol. 2005;57:1750–1761. doi: 10.1111/j.1365-2958.2005.04798.x. [DOI] [PubMed] [Google Scholar]
- Fredriksson A, Ballesteros M, Peterson CN, Persson O, Silhavy TJ, Nyström T. Decline in ribosomal fidelity contributes to the accumulation and stabilization of the master stress response regulator sigmaS upon carbon starvation. Genes Dev. 2007;21:862–874. doi: 10.1101/gad.409407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gora KG, Cantin A, Wohlever M, Joshi KK, Perchuk BS, Chien P, Laub MT. Regulated proteolysis of a transcription factor complex is critical to cell cycle progression in Caulobacter crescentus. Mol Microbiol. 2013;87:1277–1289. doi: 10.1111/mmi.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbatyuk B, Marczynski GT. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol Microbiol. 2005;55:1233–1245. doi: 10.1111/j.1365-2958.2004.04459.x. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Maurizi MR, Wickner S. Regulatory subunits of energy-dependent proteases. Cell. 1997;91:435–438. doi: 10.1016/s0092-8674(00)80428-6. [DOI] [PubMed] [Google Scholar]
- Grünenfelder B, Tawfilis S, Gehrig S, ØSterås M, Eglin D, Jenal U. Identification of the protease and the turnover signal responsible for cell cycle-dependent degradation of the Caulobacter FliF motor protein. J Bacteriol. 2004;186:4960–4971. doi: 10.1128/JB.186.15.4960-4971.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Esser L, Singh SK, Maurizi MR, Xia D. Crystal structure of the heterodimeric complex of the adaptor, ClpS, with the N-domain of the AAA+ chaperone, ClpA. J Biol Chem. 2002;277:46753–46762. doi: 10.1074/jbc.M208104200. [DOI] [PubMed] [Google Scholar]
- Gur E, Sauer RT. Recognition of misfolded proteins by Lon, a AAA(+) protease. Genes Dev. 2008;22:2267–2277. doi: 10.1101/gad.1670908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inobe T, Matouschek A. Protein targeting to ATP-dependent proteases. Curr Opin Struct Biol. 2008;18:43–51. doi: 10.1016/j.sbi.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U, Fuchs T. An essential protease involved in bacterial cell-cycle control. EMBO J. 1998;17:5658–5669. doi: 10.1093/emboj/17.19.5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Chen YE, Laub MT. Modularity of the bacterial cell cycle enables independent spatial and temporal control of DNA replication. Curr Biol. 2011;21:1092–1101. doi: 10.1016/j.cub.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Jing Jing, Chien P, Laub MT. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell. 2013;154:623–636. doi: 10.1016/j.cell.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi KK, Bergé M, Radhakrishnan SK, Viollier PH, Chien P. An Adaptor Hierarchy Regulates Proteolysis during a Bacterial Cell Cycle. Cell. 2015;163:419–431. doi: 10.1016/j.cell.2015.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J, Hernandez-Alicea L, Vass RH, Chien P. A Phosphosignaling Adaptor Primes the AAA+ Protease ClpXP to Drive Cell Cycle-Regulated Proteolysis. Mol Cell. 2015;59:104–116. doi: 10.1016/j.molcel.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie DJ, Heinen C, Schramm FD, Thüring M, Aakre CD, Murray SM, et al. Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA. PLoS Genet. 2015;11:e1005342. doi: 10.1371/journal.pgen.1005342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levchenko I, Seidel M, Sauer RT, Baker TA. A specificity-enhancing factor for the ClpXP degradation machine. Science. 2000;289:2354–2356. doi: 10.1126/science.289.5488.2354. [DOI] [PubMed] [Google Scholar]
- Maglica Z, Striebel F, Weber-Ban E. An intrinsic degradation tag on the ClpA C-terminus regulates the balance of ClpAP complexes with different substrate specificity. J Mol Biol. 2008;384:503–511. doi: 10.1016/j.jmb.2008.09.046. [DOI] [PubMed] [Google Scholar]
- McGarry KC, Ryan VT, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc Natl Acad Sci USA. 2004;101:2811–2816. doi: 10.1073/pnas.0400340101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micevski D, Dougan DA. Proteolytic regulation of stress response pathways in Escherichia coli. Subcell Biochem. 2013;66:105–128. doi: 10.1007/978-94-007-5940-4_5. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Bree AC, Jing Jing, Patrick JE, Chien P, Kearns DB. Adaptor-mediated Lon proteolysis restricts Bacillus subtilis hyperflagellation. Proc Natl Acad Sci USA. 2015;112:250–255. doi: 10.1073/pnas.1417419112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- Quardokus E, Din N, Brun YV. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc Natl Acad Sci USA. 1996;93:6314–6319. doi: 10.1073/pnas.93.13.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Rivera I, Román-Hernández G, Sauer RT, Baker TA. Remodeling of a delivery complex allows ClpS-mediated degradation of N-degron substrates. Proc Natl Acad Sci USA. 2014;111:E3853–9. doi: 10.1073/pnas.1414933111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Román-Hernández G, Hou JY, Grant RA, Sauer RT, Baker TA. The ClpS adaptor mediates staged delivery of N-end rule substrates to the AAA+ ClpAP protease. Mol Cell. 2011;43:217–228. doi: 10.1016/j.molcel.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer RT, Baker TA. AAA+ proteases: ATP-fueled machines of protein destruction. Annu Rev Biochem. 2011;80:587–612. doi: 10.1146/annurev-biochem-060408-172623. [DOI] [PubMed] [Google Scholar]
- Stein BJ, Grant RA, Sauer RT, Baker TA. Structural Basis of an N-Degron Adaptor with More Stringent Specificity. Structure. 2016 doi: 10.1016/j.str.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Alley MR. Proteolysis of the Caulobacter McpA chemoreceptor is cell cycle regulated by a ClpX-dependent pathway. J Bacteriol. 2001;183:5001–5007. doi: 10.1128/JB.183.17.5001-5007.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Sauer RT, Baker TA. ClpS modulates but is not essential for bacterial N-end rule degradation. Genes Dev. 2007;21:403–408. doi: 10.1101/gad.1511907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams B, Bhat N, Chien P, Shapiro L. ClpXP and ClpAP proteolytic activity on divisome substrates is differentially regulated following the Caulobacter asymmetric cell division. Mol Microbiol. 2014;93:853–866. doi: 10.1111/mmi.12698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargachuk R, Marczynski GT. The Caulobacter crescentus Homolog of DnaA (HdaA) Also Regulates the Proteolysis of the Replication Initiator Protein DnaA. J Bacteriol. 2015 Nov;197(22):3521–32. doi: 10.1128/JB.00460-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeth K, Ravelli RB, Paal K, Cusack S, Bukau B, Dougan DA. Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA. Nat Struct Biol. 2002;9:906–911. doi: 10.1038/nsb869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.