Abstract
Accurate assessment of the long chain polyunsaturated fatty acid (LC-PUFA) content of human milk (HM) provides a powerful means to evaluate the FA nutrient status of breastfed infants. The conventional standard for FA composition analysis of HM is liquid extraction, trans-methylation, and analyte detection resolved by gas chromatography. This standard approach requires fresh or frozen samples, storage in deep freeze, organic solvents, and specialized equipment in processing and analysis. Further, HM collection is often impractical for many studies in the free living environment, particularly for studies in developing countries. In the present study, we compare a novel and more practical approach to sample collection and processing that involves the spotting and drying ∼50 μL of HM on a specialized paper stored and transported at ambient temperatures until analysis. Deming regression indicated the two methods aligned very well for all LC-PUFA and the abundant HM FA. Additionally, strong correlations (r>0.85) were observed for DHA, ARA, EPA, linoleic (LA), and alpha-linolenic acids (ALA), which are of particular interest to the health of the developing infant. Taken together, our data suggest this more practical and inexpensive method of collection, storage, and transport of HM milk samples could dramatically facilitate studies of HM, as well as understanding its lipid composition influences on human health and development.
Supplementary keywords: Omega-3 fatty acids, human milk, gas chromatography, mass spectrometry, flame ionization detection
Introduction
The fatty acid (FA) composition of human breast milk (HM) is receiving increased attention, as the developmental and molecular functions of individual FA present within HM are increasingly understood in infant health [1-3]. FA have a multitude of structural, energetic, and bioactive functions [4], and postnatal FA excesses and deficiencies can influence responses to the nutrient and hormone milieu during development (cell division or differentiation). As a result, HM during the breastfeeding period is known to impart persistent protective effects against the risk of chronic diseases like diabetes and obesity [2]. Particular attention has been paid to HM levels of docosahexaenoic acid (DHA) and arachidonic acid (ARA), because these PUFA are most commonly associated with improved infant development metrics, especially in premature infants [5, 6]. In industrialized countries, trans FA originating from partially hydrogenated oils occlude beneficial LC-PUFA when esterified into HM triglyceride, and are of concern primarily due to potential adverse effects on infant neural development [7, 8]. Given the importance of HM composition to human development and health, there is a great need to be able to rapidly and robustly assess the FA profile in studies of HM and breastfeeding infants.
The primary challenge facing all investigators conducting FA analyses of HM is the storage and transport of samples, especially when collection occurs outside of the clinical setting. This is particularly challenging with studies in remote locations, such as developing countries or in rural settings. Traditional approaches include the collection and allquotting of small volumes of expressed milk, freezing the samples, and then shipping the frozen samples on dry ice to the laboratory setting for analysis. Outside of the clinic, maintaining consistency of sample temperatures following collection, storage, and transport to the laboratory becomes a burden that is too impractical or too costly to overcome. Failure of this cold chain imparts a wide range of variability in sample integrity that may include PUFA oxidation and milk fat separation. Recently, a “dried milk spot” (DMS) method for the collection, storage, transport, and FA profile integrity has been developed [9], which overcomes several of these logistical concerns.
The objective of the present study was to validate the accuracy and confirm the utility of the DMS when compared to the established approach of liquid-liquid extraction for HM FA analysis. Using an analytical gas chromatography mass spectroscopy (GCMS) approach that includes lipid extraction, saponification, generation of pentafluorobenzyl (PFB) FA derivatives, and quantitative data integration, we validated this alternative DMS method that uses direct FA trans-methylation of HM and analyte detection with GC flame ionization detection (FID). Our principle interest in comparing these two methods was the integrity of bioactive LC-PUFA present in HM assessed by Deming regression and Spearman correlations. Establishing the DMS-GCFID approach for HM FA profiling would overcome a significant barrier in HM research and provide a tool that could greatly enhance our understanding of HM on health and disease.
Methods
Clinical Procedures
Participants were recruited and consented during pregnancy 5. The Colorado Multiple Institutional Review Board (COMIRB) approved all aspects of this study. The primary purpose of the parent study was to examine the relationships between maternal body weight, hormonal status and infant microbiota [10]. Briefly, mid-feed human milk samples (2mL) were collected using a hand pump at 2-weeks postpartum. All collections occurred in the morning hours and mothers were fasted. Samples were placed immediately on ice, 1 mL stocks were aliquoted, and stored at -80°C within 2 hours of collection until analysis.
Dried Milk Spot and GCFID
Milk was briefly thawed at 37°C, mixed well by inversion, and 50 μL was spotted onto absorbent paper (Ahlstrom 226, PerkinElmer, Greenville, SC), pretreated with a proprietary antioxidant solution (OmegaQuant Analytics, Sioux Falls, SD) that prevents PUFA oxidation in the sample, as determined previously for FA analysis from DMS and dried blood spots [11, 9]. Samples were allowed to dry and then mailed at ambient temperature to OmegaQuant Analytics for analysis. DMS data were collected using GCFID as previously described [9]. Briefly, a hole punched from the DMS card was added to 14% w/vol boron trifluoride in methanol (trans-methylation reagent) and other solvents (toluene and methanol), shaken, and heated at 100°C for 45 min. After cooling, hexane and distilled water were added to extract fatty acid methyl esters (FAMEs). An aliquot of the hexane layer containing the FAMEs was taken for analysis by GCFID. Data are expressed as a percent of a total of 26 identified FA (10:0 – 22:6n-3). The laboratory coefficient of variation for the DMS/GCFID method for DHA is 6.8%. FA stability was tested in DMS samples stored at different temperatures for varyied amounts of time (-20°C and -80°C for up to 36 months; 4°C and 23°C for up to 4 weeks) and all %mol DHA values were observed within 15% of the referent [9].
Liquid-liquid Extraction, TG quantification, and GCMS Method
50 μL of thawed whole milk (as above) was added to 200 μL of potassium phosphate buffer pH 7.0, then 400 μL of methanol was added and samples were vortexed vigorously. Total lipid extraction was performed as previously described [12] by adding 2 × 1 mL of isooctane/ethyl acetate 3:1 v/v and vortexed vigorously. The organic phase was collected and taken to dryness by evaporation under nitrogen gas at 40°C, and samples were resuspended in 500 μL of isooctane.
Quantification of Milk Triglyceride
25 μL of isooctane suspended total milk lipid was taken to dryness, were resuspended in 200 μL dichloromethane containing 15 μL of a 20% nonaethylene glycol monododecyl ether (Sigma Aldrich, St. Louis, MO) dissolved in dichloromethane (wt/vol), and samples were incubated for 5 minutes at 25°C. Samples were taken to dryness at 40°C for 25 minutes ensuring solvent was completely evaporated. Pellets containing triglyceride/nonionic surfactant complexes were reconstituted in 200 μL of ultrapure water without mixing and incubated at 40°C for 10 minutes, followed by a gentle vortex. A regression curve was prepared from 80 nmol of tripalmitin (Sigma Aldrich, St. Louis MO) combined with 25 μL of 20% nonaethylene glycol monododecyl ether in dichloromethane (wt/vol), processed as above, resuspended in 100 μL of ultrapure water, and a standard curve of 20, 10, 5, 2.5, 1.25, 0.625, and 0.3125 nmol tripalmitin was made. Triglyceride from the organic fraction was quantified relative to tripalmitin using a modified colorimetric assay [13]. Triglyceride Reagent and Free Glycerol Reagent were purchased from Sigma Aldrich (St. Louis, MO) and prepared according to the manufacturer's instructions.
Fatty Acid Quantification by GCMS
Individual stable isotope FA stock solutions were made in 90% methanol, a mixture containing 1.0 μg/μL FAs was made in 90% methanol that was further diluted to 50 ng/μL into 90% methanol, and stable isotope reference FA regression curves were prepared according to [14, 12]. 500 ng of stable isotope reference FA mixture was added to 5 nmol of extracted HM TAG (equal TAG loading) and taken to dryness under N2 gas. Sample saponification, derivatization, and GCMS is described elsewhere [14, 12]. Briefly, dried samples were immediately resuspended in 500 μL of 100% methanol, saponified with 500 μL of 1M NaOH at 90° C for 45 min in Teflon capped tubes, and then neutralized by addition of 525 μL pf 1M HCl. Free fatty acids were re-extracted using 1 mL of isooctane (twice) and dried under N2. Fatty acids were derivatized using 1% pentafluorobenzyl bromine and 1% N,N-Diisopropylethylamine in acetonitrile at room temperature for 30 minutes and taken to dryness under N2. The pentafluorobenzyl FA esters were resuspended in 200 μL of hexane, and diluted 1:10 into hexane for auto-sampler injection. Analyte data were acquired in NICI full scan, the FA-analyte peak area ratio to that of its stable isotope reference FA was calculated for each analyte, and ratios were converted to absolute amounts relative to regression curves for each each chain length and saturation [14, 12]. Quantitative GCMS data were mathematically converted to mol% for equivalent units during Deming regression analysis or presented as nmole/mL of HM.
Statistical Methods
Median and interquartile ranges (IQR) were calculated for all FA of interest due to non-normally distributed variables. Deming regression analysis was performed to compare the two methods for FA when expressed as mol% [15]. Spearman correlations were calculated for HM FA content expressed as mol% and when GCMS data were reported as absolute amounts (nmole/mL of HM). The FA data from both methods were evaluated for the agreement and the average discrepancy (the bias) using Bland-Altman calculations [16, 17]. Method A was lipid extract-GCMS and method B was DMS-GCFID in the calculations, and method B was subtracted from A for the Bland-Altman plots.
Results
Strong Agreement in the Analysis of Human Milk PUFA
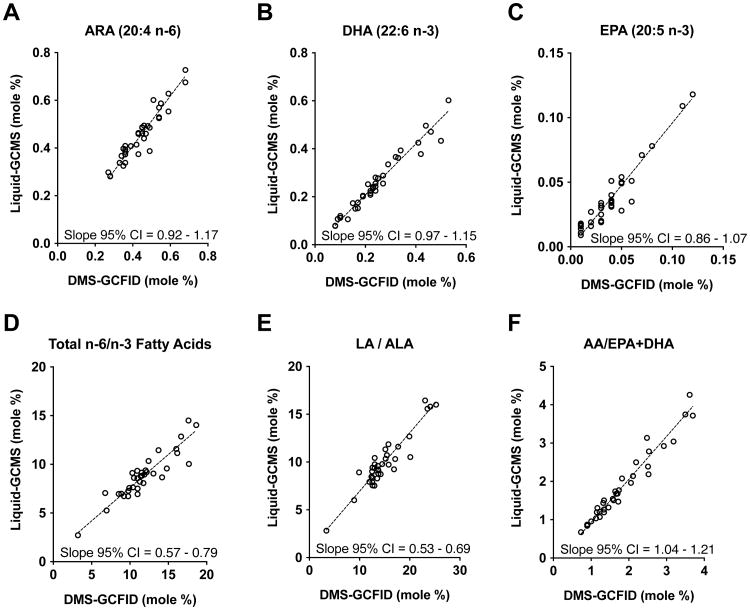
DHA (22:6n-3) and ARA (20:4n-6) were of particular interest because of their known bioactive importance for infant development as structural and signaling FA components. Of secondary interest was other PUFA including eicosapentaenoic acid (EPA, 20:5n-3), and the essential FA linoleic acid (LA, 18:2n-6) and α-linolenic acid (ALA, 18:3n-3). Other HM FA present in relatively high abundance were also evaluated, such as palmitic (16:0) and oleic (18:1 n-9) acids. In general, Deming regression analysis using the mol% measurements for LC-PUFA agreed very well between the two methods (Figure 1A, B, and C), as the mol% values are tightly nested along the slope. Each of the 95% confidence interval (CI) ranges for these LC-PUFAs contained the value 1.0, indicating the two methods are not significantly different and data are distributed equally above and below the slope. In contrast, the 95% CI ranges did not contain 1.0 for three FA ratios – Total n-6/n-3, LA/ALA, and ARA/DHA+EPA – indicating data are distributed disproportionately above or below the slope line, such that a proportional dissimilarity exists between the two methods for FA ratios (Figure 1D, E, and F). Moreover, assessment using Bland-Altman calculations supported the Deming analyses, indicating both methods had very good agreement and low bias (Online Resource Figure 1); narrow 95% limits of agreement ranges were observed for ARA, DHA, and EPA (dotted lines), values were evenly distributed along the mean line (solid line), and the overall bias between the two methods (denoted by amplitude of the mean line) was centered around zero.
Figure 1. Deming regression analysis of chief PUFA and their ratios in 2-week human milk.
Comparison between liquid-GCMS and DMS-GCFID methods showing no significant differences for the important human milk fatty acids ARA (20:4 n-6), DHA (22:6 n-3), and EPA (20:5 n-3) (A-C), as indicated by 95% CI values that contain 1.0. In contrast, the ratios of n-6 to n-3 fatty acids (D-F) are significantly different between methods (95% CI values do not contain 1.0), indicating data from the two methods distribute disproportionately above or below the slope and the mol% values are therefore not in alignment. Both methods produced measurements that were reasonably nested along the slope demonstrating tight method agreement. (n = 35)
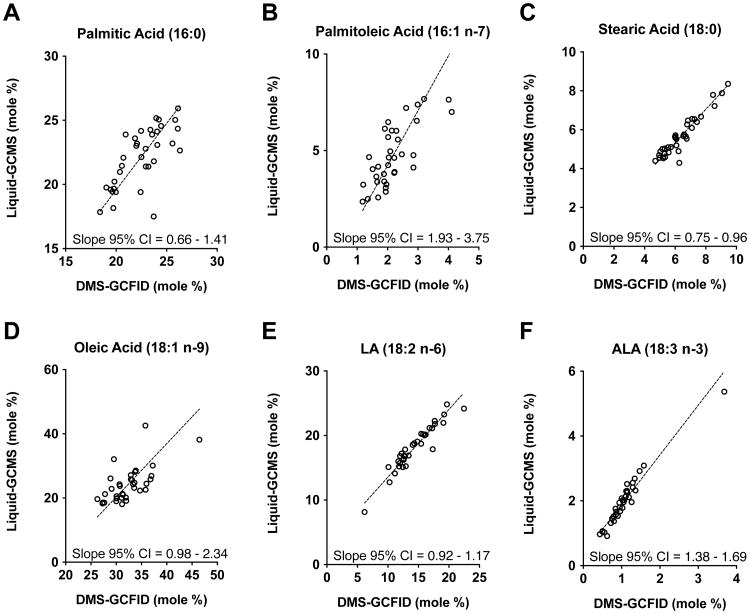
The most abundant FAs present in HM are palmitic (16:0), oleic (18:1 n-9), and LA (18:2 n-6) acids, each comprising between 17 – 45% of the total moles. Liquid-GCMS and DMS-GCFID methods were not significantly different for palmitic, stearic (18:0), LA, and oleic acids, as indicated by 95% CIs that contain 1.0 (Figure 3A, C, D, and E). Furthermore, Bland-Altman calculations supported method agreement and little bias for palmitic, stearic, and ALA (18:3 ω-3) acids, as the mean line was centered on zero (Online Resource Figure 2). Unlike those FA, the methods differed significantly for less abundant palmitoleic acid (16:1 n-7) and the essential dietary FA ALA (18:3 n-3), as indicated by 95% CI that does not contain 1.0 (Figure 3B and F). These findings were supported by Bland-Altman calculations, as the mean line was not centered on zero and the difference became more negative as the average became greater. The Deming regression equations relating relative HM FA levels (mol%) between the liquid-GCMS and DMS-GCFID are reported in Online Resource Table 1.
Correlations between concentration and mol % values
Spearman correlation was used to evaluate the mole % FA from either method, as well as to correlate known concentrations of HM FA per mL. The primary FA of interest in this study (DHA, ARA, EPA, LA, and ALA) had strong correlations (r>0.92) using the mol % values derived from liquid-GCMS and DMS-GCFID methods (Table 2, 1 versus 2). Other low abundance FA, palmitoleic (16:1 ω-7), stearic (18:0), and ALA (18:3 ω-3) acids, were strongly correlated (r = 0.75, 0.89 and 0.96, respectively). When the mol % values from either liquid-GCMS or DMS-GCFID methods are compared to the absolute concentration values (1 or 2 versus 3), the relationships became weaker. For example, for DHA the DMS-GCFID mol% to liquid-GCMS mol% correlation is 0.98 (1 versus 2), whereas the correlation between the former and absolute DHA concentration in nmol/mL is 0.70 (1 versus 3). This pattern was also observed with the other primary FA of interest, including ARA and EPA, as well as for the essential dietary FA LA and ALA (Table 2). As expected, the ratios for the n-6/n-3 FA did not exhibit this discontinuity relative to the absolute HM FA concentrations, and all correlations were r > 0.85, indicating that expressing the proportion of n-6 to n-3 FA(s) to other(s) eliminated potential method bias in comparing mol% to absolute concentration (nmol/mL).
Table 2. Spearman correlations by medians (IQR) of human milk fatty acid composition using DMS-GCFID and liquid-GCMS methods.
| Median (IQR)* | Correlation Coefficient (r) | |||||
|---|---|---|---|---|---|---|
| Method | mole % (GCFID) | mole % (GCMS) | nmol FA/mL (GCMS) | |||
| 1 | 2 | 3 | 1 vs 2 | 2 vs 3 | 1 vs 3 | |
| DHA (22:6 n-3) | 0.23% (0.17, 0.32) | 0.24% (0.17, 0.36) | 0.05 (0.03, 0.09) | 0.98 | 0.72 | 0.70 |
| ARA (20:4 n-6) | 0.45% (0.36, 0.51) | 0.46% (0.37, 0.53) | 0.10 (0.08, 0.15) | 0.92 | 0.30 | 0.31 |
| EPA (20:5 n-3) | 0.04% (0.02, 0.05) | 0.03% (0.02, 0.05) | 0.01 (0.01, 0.01) | 0.92 | 0.85 | 0.82 |
| LA (18:2 n-6) | 13.4% (12.2, 16.8) | 17.8% (16.0, 20.3) | 4.36 (3.32, 5.81) | 0.94 | 0.25 | 0.17 |
| ALA (18:3 n-3) | 1.02% (0.80, 1.19) | 1.96% (1.52, 2.31) | 0.44 (0.34, 0.56) | 0.96 | 0.44 | 0.48 |
| Total n-6/n-3 | 11.4 (9.9, 13.7) | 8.79 (7.28, 9.57) | 8.79 (7.28, 9.57) | 0.88 | 1.0 | 0.88 |
| LA/ALA | 13.8 (12.6, 16.9) | 9.41 (8.46, 10.72) | 9.41 (8.46, 10.72) | 0.85 | 1.0 | 0.85 |
| AA/EPA+DHA | 1.64 (1.22, 2.48) | 1.54 (1.19, 2.39) | 1.54 (1.19, 2.39) | 0.97 | 1.0 | 0.97 |
| Palmitic (16:0) | 22.5% (20.4, 24.1) | 22.6% (19.8, 24.1) | 5.85 (4.13, 6.61) | 0.70 | 0.23 | 0.43 |
| Oleic (18:1 n-9) | 31.9% (30.0, 33.9) | 23.2% (20.0, 26.1) | 5.29 (4.14, 7.19) | 0.63 | 0.53 | 0.19 |
| Stearic (18:0) | 6.08% (5.32, 7.09) | 5.54% (4.83, 6.40) | 1.29 (0.98, 1.72) | 0.89 | 0.30 | 0.30 |
| Palmitoleic (16:1 n-7) | 2.02% (1.70, 2.47) | 4.63 (3.40, 6.03) | 1.12 (0.75, 1.50) | 0.75 | 0.64 | 0.68 |
Median and interquartile ranges (IQR) were calculated for all FA and ratios due to non-normally distributed variables (n = 35)
Discussion
The current study compared the FA composition of identical HM samples, prepared and analyzed with two different methods: 1) liquid-liquid extraction, TAG quantification, saponification and PFB derivatization, followed by GCMS quantification, and 2) the simpler DMS direct transesterification and GCFID analyte detection. Deming regression analysis was used to analyze the strength to which two variables in each method are related by identifying method differences without ascribing superiority or “trueness” to either method [15]. The mol% measurements for principle FA of interest present in HM most associated with infant outcomes, DHA, ARA, and EPA, were highly similar between the two methods relative to the slope of the Deming regression line (Figure 2). Furthermore, the measurements for DHA and ARA, as well as the ratios of HM PUFA made by the two methods, were very tightly correlated (Table 2). These findings indicate that use of DMS methods can provide reliable FA profile data as a potential tool for sample acquisition, especially when collection/transport of liquid HM samples is not possible due to technical or financial constraints.
Figure 2. Deming regression analysis of abundant and essential dietary fatty acids in 2-week human milk.
Comparison between liquid-GCMS and DMS-GCFID methods showed no significant differences for the abundant human milk fatty acids (A, C, D, E), including palmitic (16:0), stearic (18:0), oleic (18:1 n-9), and LA (18:2 n-6), indicated by 95% CI values that contain 1.0. For the comparatively less abundant fatty acids palmitoleic (16:7 n-7) and ALA (18:3 n-3) (B and F), significant differences between methods were observed (95% CI values do not contain 1.0), indicating data from the two methods distribute disproportionately above or below the slope and the mol% values had method bias for these FA. The data for palmitic (16:0), palmitoleic (16:7 n-7), oleic (18:1 n-9) acids were dispersed away from the slope, despite significant similarities, indicating moderate agreement between methods for mol% values. Both methods produced mol% measurements for stearic (18:0), LA (18:2 n-6), and ALA (18:3 n-3) that were tightly nestled along the slope demonstrating outstanding method agreement. (n = 35)
The n-6/n-3 ratios for PUFA were in reasonable agreement with the Deming regression line, even though the 95% CI values did not contain the value 1.0. This observation indicates data points fell disproportionately above or below the slope. Accordingly, a proportional dissimilarity exists between the two methods for n-6/n-3 ratios that was not observed for regressions of the individual DHA, ARA, and EPA, as well as the highly abundant LA, oleic, and palmitic acids. The implications from this observation are that the numeric values for n-6/n-3 PUFA ratios may be relatively different between the two methods, but the DMS approach provides an adequate assessment of these ratios as indicated by a reasonably tight dispersion of points along the Deming slope. This observation is supported by Bland-Altman calculations indicating low overall bias and strong agreement for DHA, ARA, EPA and the ratio of ARA/EPA+DHA, which was not observed for the total n-6/n-3 and LA/ALA ratios (Online Resource Figure 1). Altogether, DMS-GCFID performs equally well compared to liquid-GCMS in measuring the relative amounts (mol%) of DHA, ARA, EPA, LA, and ALA present in HM, which provide critical information about bioactive FA commonly associated with health outcomes of infant development [5, 6].
Data from both methods expressed in mol % were compared to the absolute FA concentrations quantified using GCMS (nmol FA/mL HM) in order to relate mol% data to absolute FA concentration (Table 2). Here, the correlation coefficients between mol% and concentration data were, not unexpectedly, considerably weaker. The exceptions to this were the ratios of n-6 to n-3 PUFA present in HM, which preserved their same strong correlations because ratios eliminate units of measurement by definition. This observation points out numerical differences that can occur when comparing FA measurements as mol% with FA data as concentration (nmol/mL), and as such, conclusions that compare HM FA data in this way should be made with this in mind.
The liquid-liquid and the DMS methods have advantages and disadvantages depending on the research question, design, and setting. For research investigations that aim to determine the DHA and/or ARA “status” of HM (as mol% of total FA), either method can be used effectively. Only the liquid-GCMS based method provided quantitative concentrations of individual FA present in the milk. Importantly, FA data expressed as mol% (generated by either method) can be directly compared across a large body of existing research on HM FA composition, whereas HM FA concentrations are less frequently reported. Strengths of the DMS-GCFID method include the ability to assess the FA status in large cohorts or clinical studies, low patient and researcher burden for milk collection, and simplified storage/transport procedures; relatively inexpensive instrumentation (vs the GCMS) and relatively lower technical skill necessary to run analyses in the laboratory. Even though the TAG concentration HM varies throughout the day or within a feeding session [18, 19], milk can be collected at any time of the day as the mol% FA composition of HM does not change [20]. In fact, milk TAG amounts in this study were highly variable, ranging from 43-184 mM presumably due to the 8-hour maternal fast, demonstrating the importance of TAG quantification for equal loading into saponification when using the liquid-GCMS method. Thus, DMS-GCFID methods provide a simpler, practical, and potentially less expensive approach to assessing HM FA status. For researchers wishing to use the DMS collection method, we have identified best practices as: 1) use filter paper cards pre-treated with the proprietary antioxidant cocktail (we do not recommend DMS without antioxidant), and 2) store DMS cards in the presence of provided desiccant pouch at room temperature (23°C) for up to 1 month, frozen at -20°C for up to 36 months, or at -80°C indefinitely [9]. Limitations of this, or any method that generates FA proportions, are that mol% of low abundance FA could be affected by changes in levels of the highly abundant FA present [21-23], and the absolute concentrations (at present) cannot be ascertained.
Strengths of the liquid-GC/MS method include its utility in a variety of basic and clinical research questions (including quantification of stable isotopes in pulse/chase experiments in vivo), greater sensitivity to measure FA present in very low concentrations, quantification of non-esterified FA, and the ability to express FA data either quantitatively or as mol%. Limitations of liquid-GCMS method include the extensive training and expertise required to prepare and run the lipid mass spectrometry analyses; the greater difficulty in collecting, storing and transferring samples; the large amount of organic solvents required and their corresponding hazardous waste; the higher cost to purchase and maintain a GCMS vs. a GCFID; and, the more complex data output requiring expertise for interpretation. In sum, the greater precision and quantitative nature of the liquid-based GCMS method must be balanced against the superior efficiency and lower cost of the DMS-GCFID method in light of specific needs of the research project.
Conclusions
HM FA status can be measured equally well using a DMS collection system coupled with direct trans-methylation and GCFID analysis as it can with a liquid-based collection system coupled with equal TAG loading for saponification, PFB derivatization, and GCMS quantification. Each method can play an important role in the evolving field of HM and FA research depending on the experimental needs.
Supplementary Material
Online Resource Figure 1. Bland-Altman Plots for principle human milk LC-PUFA and n-6/n-3 fatty acid ratios. The fatty acid data from both methods was evaluated for the agreement and the average discrepancy (the bias) between the two methods. (A-C) The methods have narrow 95% limit of agreement ranges for ARA, DHA, and EPA (dotted lines), and the values are evenly distributed along the mean line (solid line). Overall bias between the two methods, denoted by amplitude of the mean line, was centered on zero and indicates tight method agreement. (D-F) Method agreement was good for HM n-6/n-3 fatty acid ratios with very little proportional bias for ARA/EPA+DHA (F), however, a positive trend exists for the total ω-6/ω-3 and LA/ALA ratios (D and E) that is proportional to the magnitude of measures (x-axis), indicating the method bias becomes greater as these ratios increase.
Online Resource Figure 2. Bland-Altman Plots for highly and moderately abundant human milk fatty acids. The fatty acid data from both methods was evaluated for the agreement and the average discrepancy (the bias) between the two methods. Calculations for palmitic (16:0) and stearic (18:0) acids (A and C) had tight method agreement and little bias, however, palmitic acid had a broader dispersion for the 95% limit of agreement range. In contrast, palmitoleic (16:1 n-7), oleic (18:1 n-9), LA (18:2 n-6), and ALA (18:3 n-3) acids had reasonable agreement between the two methods (B, D, E, and F), however, their means deviated from zero indicating proportional bias. Further, palmitoleic, oleic, and ALA acids had negative trends proportional to increasing amounts of FAs measured, indicating the method bias becomes less as the amounts of these fatty acids increase.
Online Resource Table 1. Deming regression equations relating human milk fatty acids using the liquid-GCMS and DMS-GCFID methods (n = 35).
Table 1. Methodological Comparison between liquid-GCMS and DMS-GCFID workflows.
| Liquid-GCMS Method | DMS-GCFID Method | |
|---|---|---|
| Sample Collection | Mid-feed breast milk collected with hand pump (20mL); placed on ice | Place 50uL (roughly 1 drop) of whole milk onto treated filter paper and let dry for ≥ 15 min at room temperature |
| Sample Transport and Storage | Transfer on ice, aliquot, freeze and store at -80 °C until analysis | Send dried milk spot on filter paper through regular mail to lab |
| Sample Preparation |
|
|
| Analysis |
|
Analyze FAMEs by GCFID relative to a reference standards mixture |
| Results | moles of FA/mole TAG; nmol FA/mL | Percent of total identified FA |
Acknowledgments
grant support: The authors would like to express our sincere appreciation to all of our study participants and those who provided critical reading. 6 MCR is supported by Building Interdisciplinary Careers in Women's Health NIH K-12 HD057022, NICHD T32-HD007186 training grant, and Nutrition and Obesity Research Center pilot funding P30-DK048520. BEY by NICHD F32-HD0978068, T32-DK007658-21, Thrasher Research Fund Early Career Award, and Center for Women's Health Research. NFK by K24-DK083772. PSM by P01-HD038129, P50 HD073063, R01 CA164166, and R01-HD075285. Lipid mass spectrometry supported by Dr. Robert Murphy and NIH/NCATS Colorado CTSA Grant UL1 TR001082. OmegaQuant internally funded the dried milk spot analysis. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- ALA
alpha-linolenic acid
- ARA
arachidonic acid (20:4 n-6)
- DHA
docosahexaenoic acid (22:6 n-3)
- DMS
dried milk spot
- EPA
eicosapentaenoic acid (20:5 n-3)
- GC
gas chromatography
- FA
fatty acid
- FID
flame ionization detection
- HM
human milk
- LC-PUFA, LA
linoleic acid; long chain polyunsaturated fatty acid
- MS
mass spectrometry
- PFB
pentafluorobenzyl
- TG
triglyceride
Footnotes
The parent study is registered at clinicaltrials.gov under the following: NCT01693406(https://clinicaltrials.gov/ct2/show/NCT01693406?term=12-0629&rank=1)
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Contributor Information
Michael C. Rudolph, University of Colorado Denver, Center for Human Nutrition | Division of Endocrinology, Metabolism & Diabetes, Mail Stop F-8305; RC1 North, 12800 E. 19th Avenue P18-5402M, Aurora, CO 80045-2537, phone: 303.724.6878, fax: 303.724.3031
Bridget E. Young, Department of Pediatrics, Section of Nutrition, 12700 East 19th Ave, Box C-225, University of Colorado School of Medicine, Aurora, CO, 80045
Kristina Harris Jackson, OmegaQuant Analytics, LLC. 5009 W. 12th St, Ste 8, 57106, Sioux Falls, South Dakota. Phone: 913-302-0456.
Nancy F. Krebs, Department of Pediatrics, Section of Nutrition, University of Colorado School of Medicine, Aurora, CO, 80045
William S. Harris, Department of Internal Medicine, University of South Dakota School of Medicine; and OmegaQuant Analytics, LLC. 5009 W. 12th St, Ste 8, 57106, Sioux Falls, South Dakota. Phone: 605-271-6917
Paul S. MacLean, Division of Endocrinology, Metabolism, and Diabetes, University of Colorado School of Medicine, RC1 North Rm 5108, Campus Box F-8301, 12800 East 19th Avenue, Aurora, CO 80045
References
- 1.Novak EM, Innis SM. Impact of maternal dietary n-3 and n-6 fatty acids on milk medium-chain fatty acids and the implications for neonatal liver metabolism. Am J Physiol Endocrinol Metab. 2011;301(5):E807–17. doi: 10.1152/ajpendo.00225.2011. [DOI] [PubMed] [Google Scholar]
- 2.Innis SM. Metabolic programming of long-term outcomes due to fatty acid nutrition in early life. Maternal & child nutrition. 2011;7(2):112–23. doi: 10.1111/j.1740-8709.2011.00318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novak EM, Keller BO, Innis SM. Metabolic development in the liver and the implications of the n-3 fatty acid supply. American journal of physiology Gastrointestinal and liver physiology. 2012;302(2):G250–9. doi: 10.1152/ajpgi.00189.2011. [DOI] [PubMed] [Google Scholar]
- 4.German JB, Dillard CJ. Composition, structure and absorption of milk lipids: a source of energy, fat-soluble nutrients and bioactive molecules. Critical reviews in food science and nutrition. 2006;46(1):57–92. doi: 10.1080/10408690590957098. [DOI] [PubMed] [Google Scholar]
- 5.Hadley KB, Ryan AS, Forsyth S, Gautier S, Salem N. The Essentiality of Arachidonic Acid in Infant Development. Nutrients. 2016;8(4) doi: 10.3390/nu8040216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koletzko B, Boey CC, Campoy C, Carlson SE, Chang N, Guillermo-Tuazon MA, et al. Current information and Asian perspectives on long-chain polyunsaturated fatty acids in pregnancy, lactation, and infancy: systematic review and practice recommendations from an early nutrition academy workshop. Ann Nutr Metab. 2014;65(1):49–80. doi: 10.1159/000365767. [DOI] [PubMed] [Google Scholar]
- 7.Decsi T, Boehm G. trans Isomeric fatty acids are inversely related to the availability of long-chain PUFAs in the perinatal period. Am J Clin Nutr. 2013;98(2):543S–8S. doi: 10.3945/ajcn.112.039156. [DOI] [PubMed] [Google Scholar]
- 8.Szabo E, Boehm G, Beermann C, Weyermann M, Brenner H, Rothenbacher D, et al. trans Octadecenoic acid and trans octadecadienoic acid are inversely related to long-chain polyunsaturates in human milk: results of a large birth cohort study. Am J Clin Nutr. 2007;85(5):1320–6. doi: 10.1093/ajcn/85.5.1320. [DOI] [PubMed] [Google Scholar]
- 9.Jackson KH, Polreis J, Sanborn L, Chaima D, Harris WS. Analysis of breast milk fatty acid composition using dried milk samples. Int Breastfeed J. 2016;11:1. doi: 10.1186/s13006-016-0060-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemas DJ, Young BE, Baker PR, 2nd, Tomczik AC, Soderborg TK, Hernandez TL, et al. Alterations in human milk leptin and insulin are associated with early changes in the infant intestinal microbiome. The American journal of clinical nutrition. 2016;103(5):1291–300. doi: 10.3945/ajcn.115.126375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baack ML, Puumala SE, Messier SE, Pritchett DK, Harris WS. What is the relationship between gestational age and docosahexaenoic acid (DHA) and arachidonic acid (ARA) levels? Prostaglandins Leukot Essent Fatty Acids. 2015 doi: 10.1016/j.plefa.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudolph MC, Wellberg EA, Lewis AS, Terrell KL, Merz AL, Maluf NK, et al. Thyroid hormone responsive protein Spot14 enhances catalysis of fatty acid synthase in lactating mammary epithelium. J Lipid Res. 2014;55(6):1052–65. doi: 10.1194/jlr.M044487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Van Veldhoven PP, Swinnen JV, Esquenet M, Verhoeven G. Lipase-based quantitation of triacylglycerols in cellular lipid extracts: requirement for presence of detergent and prior separation by thin-layer chromatography. Lipids. 1997;32(12):1297–300. doi: 10.1007/s11745-006-0166-1. [DOI] [PubMed] [Google Scholar]
- 14.Rudolph MC, Karl Maluf N, Wellberg EA, Johnson CA, Murphy RC, Anderson SM. Mammalian fatty acid synthase activity from crude tissue lysates tracing 13C-labeled substrates using gas chromatography mass spectrometry. Analytical Biochemistry. 2012;428(2):158–66. doi: 10.1016/j.ab.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin RF. General deming regression for estimating systematic bias and its confidence interval in method-comparison studies. Clin Chem. 2000;46(1):100–4. [PubMed] [Google Scholar]
- 16.Altman D, Bland J. Measurement in Medicine: The Analysis of Method Comparison Studies. Journal of the Royal Statistical Society. 1983;32(3):307–17. doi: 10.2307/2987937. [DOI] [Google Scholar]
- 17.Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999;8(2):135–60. doi: 10.1177/096228029900800204. [DOI] [PubMed] [Google Scholar]
- 18.Jensen RG. The lipids in human milk. Prog Lipid Res. 1996;35(1):53–92. doi: 10.1016/0163-7827(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 19.Hromadova M, Ponec J, Mackova A. Cholesterol and triacylglycerols in human breast milk before and after nursing. Endocr Regul. 1991;25(1-2):70–3. [PubMed] [Google Scholar]
- 20.Gibson RA, Kneebone GM. Effect of sampling on fatty acid composition of human colostrum. J Nutr. 1980;110(8):1671–5. doi: 10.1093/jn/110.8.1671. [DOI] [PubMed] [Google Scholar]
- 21.Lagerstedt SA, Hinrichs DR, Batt SM, Magera MJ, Rinaldo P, McConnell JP. Quantitative determination of plasma c8-c26 total fatty acids for the biochemical diagnosis of nutritional and metabolic disorders. Mol Genet Metab. 2001;73(1):38–45. doi: 10.1006/mgme.2001.3170. [DOI] [PubMed] [Google Scholar]
- 22.Carnielli VP, Pederzini F, Vittorangeli R, Luijendijk IH, Boomaars WE, Pedrotti D, et al. Plasma and red blood cell fatty acid of very low birth weight infants fed exclusively with expressed preterm human milk. Pediatric research. 1996;39(4 Pt 1):671–9. doi: 10.1203/00006450-199604000-00019. [DOI] [PubMed] [Google Scholar]
- 23.Dodds ED, McCoy MR, Rea LD, Kennish JM. Gas chromatographic quantification of fatty acid methyl esters: flame ionization detection vs. electron impact mass spectrometry. Lipids. 2005;40(4):419–28. doi: 10.1007/s11745-006-1399-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Resource Figure 1. Bland-Altman Plots for principle human milk LC-PUFA and n-6/n-3 fatty acid ratios. The fatty acid data from both methods was evaluated for the agreement and the average discrepancy (the bias) between the two methods. (A-C) The methods have narrow 95% limit of agreement ranges for ARA, DHA, and EPA (dotted lines), and the values are evenly distributed along the mean line (solid line). Overall bias between the two methods, denoted by amplitude of the mean line, was centered on zero and indicates tight method agreement. (D-F) Method agreement was good for HM n-6/n-3 fatty acid ratios with very little proportional bias for ARA/EPA+DHA (F), however, a positive trend exists for the total ω-6/ω-3 and LA/ALA ratios (D and E) that is proportional to the magnitude of measures (x-axis), indicating the method bias becomes greater as these ratios increase.
Online Resource Figure 2. Bland-Altman Plots for highly and moderately abundant human milk fatty acids. The fatty acid data from both methods was evaluated for the agreement and the average discrepancy (the bias) between the two methods. Calculations for palmitic (16:0) and stearic (18:0) acids (A and C) had tight method agreement and little bias, however, palmitic acid had a broader dispersion for the 95% limit of agreement range. In contrast, palmitoleic (16:1 n-7), oleic (18:1 n-9), LA (18:2 n-6), and ALA (18:3 n-3) acids had reasonable agreement between the two methods (B, D, E, and F), however, their means deviated from zero indicating proportional bias. Further, palmitoleic, oleic, and ALA acids had negative trends proportional to increasing amounts of FAs measured, indicating the method bias becomes less as the amounts of these fatty acids increase.
Online Resource Table 1. Deming regression equations relating human milk fatty acids using the liquid-GCMS and DMS-GCFID methods (n = 35).