Abstract
DNA repair-deficient Ercc1Δ/− mice show numerous accelerated aging features limiting lifespan to 4–6 month1–4. Simultaneously they exhibit a ‘survival response’, which suppresses growth and enhances maintenance, resembling the anti-aging response induced by dietary restriction (DR)1,5. Here we report that subjecting these progeroid, dwarf mutants to 30% DR tripled median and maximal remaining lifespan, and drastically retarded numerous aspects of accelerated aging, e.g. DR animals retained 50% more neurons and maintained full motoric function, even far beyond the lifespan of ad libitum (AL) animals. Repair-deficient, progeroid Xpg−/− mice, a Cockayne syndrome model6, responded similarly, extending this observation to other repair mutants. The DR response in Ercc1Δ/− mice closely resembled DR in wild type animals. Interestingly, AL Ercc1Δ/− liver showed preferential extinction of expression of long genes, a phenomenon we also observe in several normal aging tissues. This is consistent with accumulation of stochastic, transcription-blocking lesions, affecting long genes more than short ones. DR largely prevented declining transcriptional output and reduced γH2AX DNA damage foci, indicating that DR preserves genome function by alleviating DNA damage. Our findings establish Ercc1Δ/− mice as powerful model for interventions sustaining health, reveal untapped potential for reducing endogenous damage, provide new venues for understanding the molecular mechanism of DR, and suggest a counterintuitive DR-like therapy for human progeroid genome instability syndromes and possibly neurodegeneration in general.
DR is the best documented intervention extending lifespan in numerous species by retarding many aging symptoms7–10. Despite decade-long research its underlying mechanisms are still unresolved, although suppression of GH/IGF1 and mTOR signaling are likely implicated8,10. The molecular underpinnings of aging itself are also poorly understood, although progeroid syndromes associated with impaired genome maintenance4 point towards a connection with compromised genome stability11,12. Links between accumulating DNA damage and the GH/IGF1 axis emerged when DNA repair deficient progeroid mice were found to suppress the GH/IGF1 somatotrophic axis and upregulate anti-oxidant defenses, presumably in an attempt to extend their lifespan by redirecting resources from growth to maintenance and stress resistance1,5. Also normal mice and mammalian cells induce this so-called ‘survival response’ after induction of persisting DNA damage, indicating its general nature1,5,13.
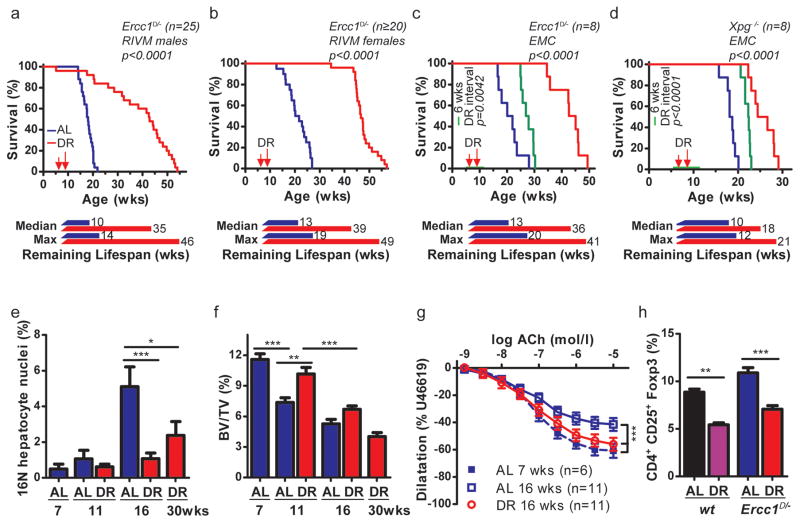
In view of spontaneous activation of DR-like responses in growth-suppressed progeroid repair mutants and other signs suggestive of DR, including reduced subcutaneous fat and paradoxical features of delayed aging14,15 we wondered whether subjecting progeroid mice to actual DR would be beneficial or -in view of their poor growth and frail appearance- detrimental. We subjected Ercc1Δ/− progeroid repair mutants with a lifespan of only 4–6 months3,16,17, to gradual food restriction, starting at week 7 with 10%, reaching 30% restriction from 9 weeks onward. Impressively, DR in both genders extended median and maximal remaining lifespan by ~200%; in males from 10 to 35 weeks (250% extension; p<0.0001) and females from 13 to 39 weeks (200% extension; p<0.0001, Fig. 1a,b).
Figure 1. Dietary restriction extends life- and healthspan of Ercc1Δ/− and Xpg−/− mouse mutants.
a–d, Survival of mice with ad libitum (AL) access to AIN93G diet or on 30% dietary restriction (DR; red, AL; blue throughout) at two separate test sites. Male (a) and female (b) Ercc1Δ/− mice, group housed at the RIVM (n=20–25 animals/group, separate experiments), and Ercc1Δ/− (c) and Xpg−/− (d) mice, solitary housed at the ErasmusMC (n=8 animals/group, 4 of each gender), under AL or DR regimens. DR was initiated at 7 weeks of age with 10%, (when the mice almost reached maximum body weight and development was completed), and increased weekly with 10%, until 30% was reached from 9 weeks onward. Remaining median and maximum lifespan are indicated (week 8 was considered the start of effective DR). Simultaneously, a cohort of Ercc1Δ/− (c) and Xpg−/− (d) mice received a temporary DR regime for a period of 6 weeks (green; n=8 animals/group, 4 of each gender). At the age of 6 weeks they received 30% DR and were switched back to AL from 12 weeks onward. p values were calculated by the log-rank test. e, Quantification of 16N nuclei in hepatocytes of AL or DR male Ercc1Δ/− mice by FACS analyses; n=5 animals/group. f, Trabecular bone volume fraction (Bone Volume/Tissue Volume of interest, BV/TV, in %) in femurs of Ercc1Δ/− male mice, measured using micro-CT. AL- and DR-treated animals were analyzed at different ages with n≥6 animals/group. g, Age-dependent decline of vasodilatation in Ercc1Δ/− aorta segments, ex vivo. DR-Ercc1Δ/− aorta segments show significantly more relaxation at age of 16 weeks than AL-Ercc1Δ/− aorta. h, Frequency of CD4+CD25+Foxp3+ T regulatory cells among all CD4+ T cells from spleen of 16 weeks old Ercc1Δ/− mice under DR or AL and aged-matched wt controls. n≥3 animals/group. Error bars denote mean ± SE. * p<0.05, ** p<0.01, ***p<0.001.
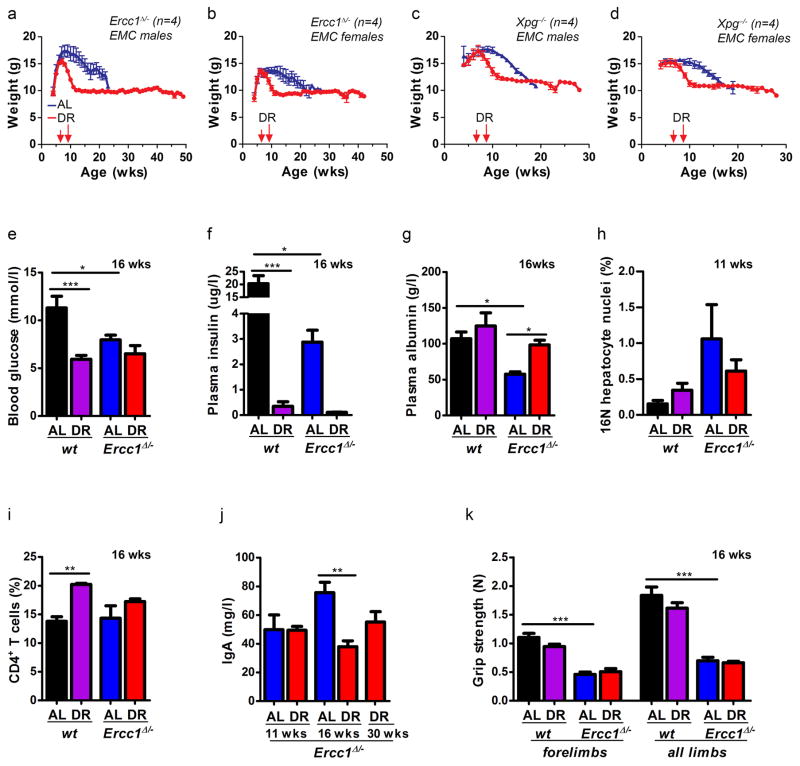
As lifespan can be influenced by factors other than food18, we repeated the study in another animal facility under different housing but similar food and DR conditions. Indeed, 30% DR extended median remaining lifespan now by 180% (p<0.0001; Fig. 1c). We decided to test another repair-deficient progeroid mutant, the Cockayne syndrome (CS)-like Xpg−/− mouse, carrying defects in partially different repair pathways2, and having an even shorter lifespan than the Ercc1Δ/− mutant (~18 versus 22–25 weeks)6. The same DR regimen induced a highly significant ~80% increase in remaining median lifespan (p<0.0001, Fig. 1d), extending this observation beyond Ercc1. Even a six week DR interval, from 6 to 12 weeks of age, yielded a striking median lifespan extension of 6 weeks for Ercc1Δ/− (p=0.0042) and 4 weeks for Xpg−/− (p<0.0001; Fig. 1c,d), indicating that the DR effect persists, consistent with reducing the short-term death risk found in Drosophila19. Comparing Ercc1Δ/− with Xpg−/− one should realize that Xpg−/− animals were already biologically older when DR started. Importantly, AL Ercc1Δ/− and Xpg−/− mice never become obese: upon DR bodyweights uniformly stabilized to gender- and genotype-specific values, persisting till the end of their extended lifespan. Paradoxically, when bodyweights of AL animals approached the weight of DR mice, they died (Extended Data Fig. 1a–d).
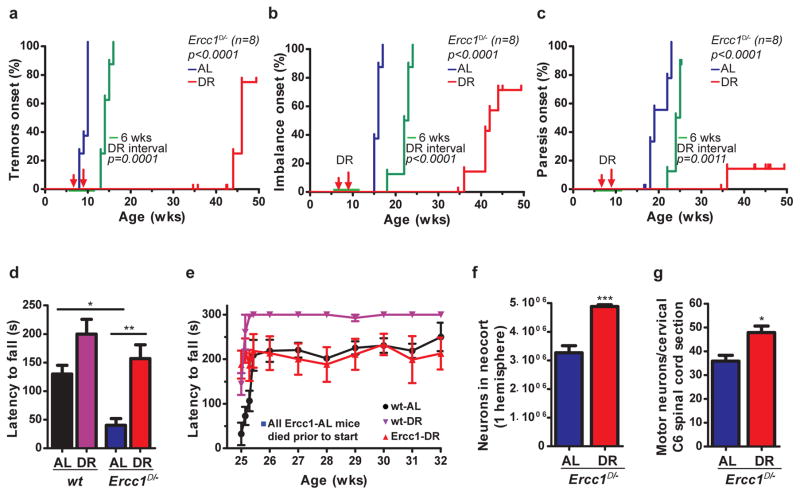
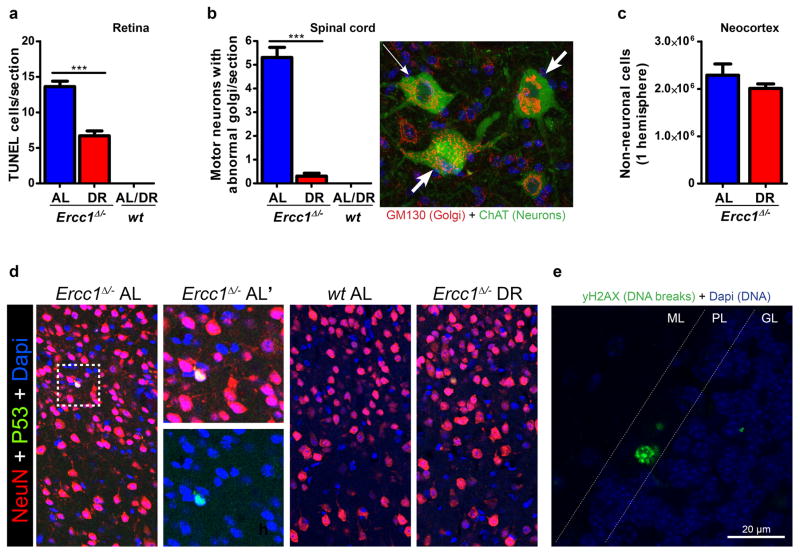
As lifespan extension can be unrelated to aging18, we examined key aging parameters. Ercc1Δ/− mutants exhibit exceptionally wide multi-morbidity, correlating with the notion that this protein is implicated in multiple DNA repair processes, including transcription-coupled and global-genome nucleotide excision repair (NER) and crosslink repair2. Aging in this mutant involves proliferative and post-mitotic organs, including the nervous system, liver, kidney, bone marrow, retina, muscle, the cardiovascular, skeletal, and gonadal systems1,2,4. Additionally, they exhibit progressively declining vision and hearing, sarcopenia, cachexia, overall frailty and an extending variety of other aging properties, many of which also noted in XpF/Ercc1 (XFE) and related syndromes1,20,21, demonstrating relevance for the human situation. Multisystem cross-sectional analyses showed that DR strongly attenuated virtually all features of premature aging investigated including anisokaryosis in liver and kidney, formation of polyploid liver nuclei, kidney tubulonephrosis, osteoporosis, disturbed vascular dilatation, B and T-cell immune parameters, testicular degeneration, etc. (Fig. 1e–h, Extended Data Fig. 1–3, Supplementary Table 1). Although the premature aging phenotype displays extreme multi-morbidity, an important cause of death of Ercc1Δ/− animals is consequential to the progressive neurodegeneration which parallels NER-deficient premature aging conditions in man22,23. Therefore, and considering its general importance, we analyzed neurofunctioning in more detail. Longitudinal examination of behavioral abnormalities showed that onset of tremors, imbalance, and paresis are dramatically postponed or even absent in Ercc1Δ/− and Xpg−/− mice under continuous and temporary DR regimes (Fig. 2a–c, Extended Data Fig. 4, Supplementary Video1). DR strongly improved motor function in Ercc1Δ/− mice; at 16 weeks of age AL mice display severe locomotor problems and frequently ‘fall’, whereas DR mice are fully capable of running (Fig. 2d, Supplementary Video2). Even well beyond the lifespan of AL mice, locomotor function is amazingly well preserved by DR (Fig. 2e). In line with behavioral data, indicating that neurodecline is virtually stopped, neurodegenerative pathology is strongly diminished, including retinal photoreceptor loss, Golgi abnormalities, axonal swellings, astrocytosis and microgliosis (Extended Data Fig. 2e, 5, 6a,b). Most importantly, stereological counting revealed that DR animals retained ~50% more neurons in the neocortex, compared to AL controls (Fig. 2f), while the number of non-neuronal cells was similar (Extended Data Fig. 6c). Likewise, significantly more motor neurons were preserved in the spinal cord upon DR (Fig. 2g). These findings indicate DR dramatically improves life- and healthspan in both Ercc1 and Xpg repair-deficient mice, most notably neurodegeneration.
Figure 2. Dietary restriction dramatically preserves neurofunctioning of Ercc1Δ/− mice.
a–c, Onset of neurological abnormalities: tremors (a), imbalance (b), paresis of the hind limbs (c) with age in AL and DR Ercc1Δ/− mice. n=8 animals/group. The onset of continuous DR is indicated by red arrows and the 6-week DR interval as green horizontal line. d–e, Average time spent on an accelerating rotarod of wt and Ercc1Δ/− mice on different diets at 16 weeks of age (d; n=8 animals/group) or weekly monitored beyond the lifespan of Ercc1-AL mice (e; Ercc1Δ/− n=4, wt n=3). A daily training period was given at 25 weeks of age. f–g, Quantitative stereological analysis of the total number of neurons (f; NeuN+; p=0.0008) in the neocortex of transverse brain sections and motor neurons (g; ChAT+; p=0.0176) in C6 cervical spinal cord sections of 16-weeks old AL and DR Ercc1Δ/− mice. Note that the selective effect on neurons is consistent with earlier observations that neurons are the primary target of Ercc1 deficiency28. n≥3 animals/group. Error bars indicate mean ± SE. * p<0.05, ** p<0.01, ***p<0.001.
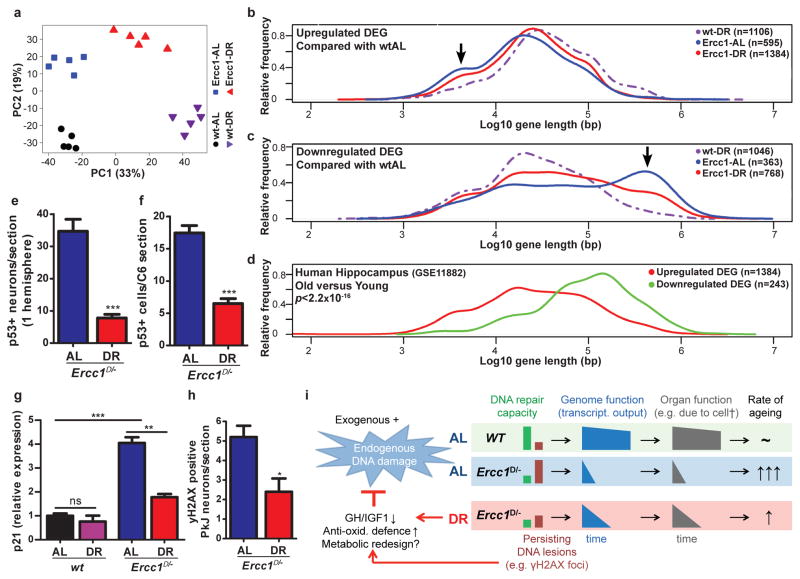
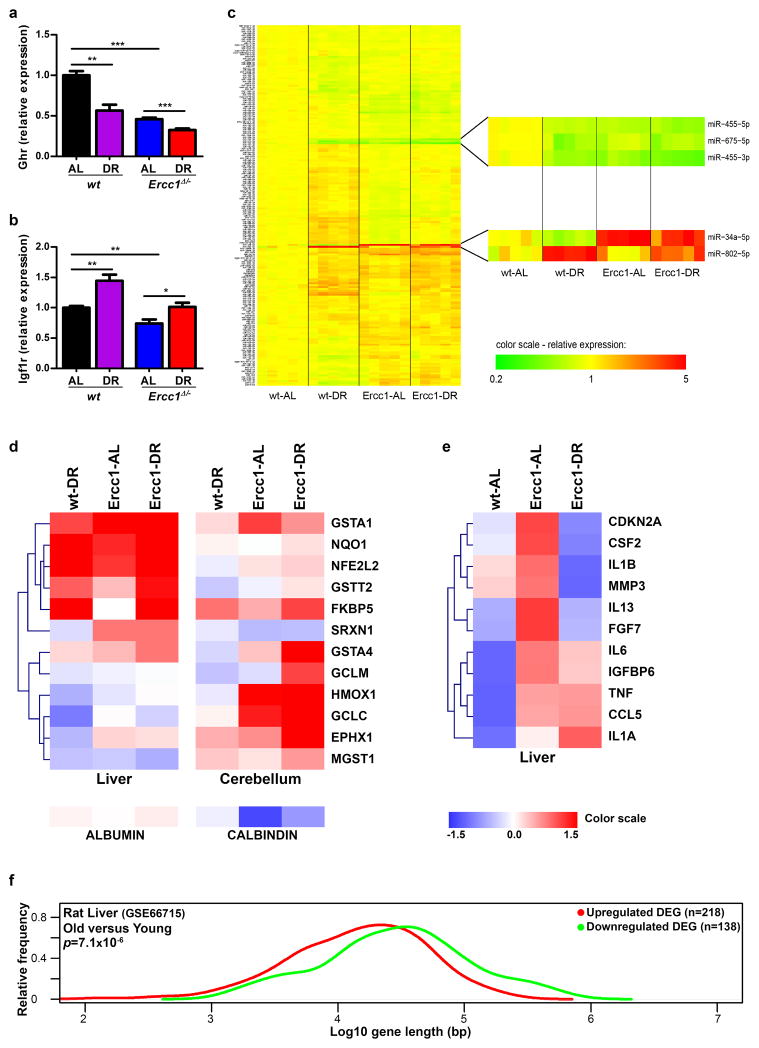
To examine whether DR in Ercc1Δ/− resembles DR in wt mice we compared full genome liver mRNA expression profiles at the age of 11 weeks. Unbiased principal component analysis revealed clear uniformity within and distinction between the 4 groups: wt and Ercc1Δ/− ad libitum (wt-AL and Ercc1-AL) and on 30% DR (wt-DR and Ercc1-DR) (Fig. 3a), primarily based on DR and genotype. Of the 1106 Ercc1 DR-induced differentially expressed genes (DEGs) ~2/3 are common with the wt-DR DEGs. Impressively, 684 of the 688 common DEGs are also changed in the same direction in wt and Ercc1Δ/− mice (Table 1, Extended Data Table 1), indicating strong mechanistic parallels between both DR responses, consistent with overlapping Gene Ontology pathways and transcription factors (Supplementary Table 2). Molecular analyses on i.a. insulin, mTOR, GH/IGF1 pathways and microRNA expression further support the parallels between wt and Ercc1Δ/−-DR (e.g. GHR gene, already suppressed in Ercc1Δ/− mice is further reduced upon DR; Extended Data Fig. 1f, 7, 8a–e). These data revealed also an unexpected link between Ercc1 DNA repair deficiency and the unfolded protein response (Extended Data Table 1, Supplementary Table 2).
Figure 3. Dietary restriction in Ercc1Δ/− mice resembles wtDR and preserves genome function.
a, Principal component analysis (PCA) of full genome liver RNA expression profiles of 11-week old Ercc1Δ/− mice under AL (blue squares) and DR (red triangles) and wt controls (AL: black circles; DR: purple triangles). This analysis takes into account all the genes in the microarray platform. The two main principal components PC1 and PC2 explain 52% of the variability in the original dataset: PC1 (x-axis, 33%) differentiates on the basis of expression changes induced by DR, independent of genotype; PC2 (y-axis, 19%) reflects differences associated with genotype. b–c, Relative frequency plot of gene length (log scale) of DEGs in: wt-DR vs. wt-AL (purple interrupted lines), Ercc1Δ/− AL vs. wt-AL (blue lines) and Ercc1Δ/− DR vs. wt-AL (red lines). Plots are shown for only up-regulated genes (b), and only down-regulated genes (c). Black arrows indicate the extra peak of up-regulated short genes (b) and peak of down-regulated long genes (c) in Ercc1-AL. d, Relative frequency plot of gene length of DEGs in hippocampus of approximately 80 year old humans versus approximately 20 year young humans. Upregulated genes are depicted in red and downregulated genes in green. The DEGs from human hippocampus were selected using a log2-fold change cut-off of 0.5 and FDR <0.05. The dataset used corresponds to NCBI gene expression omnibus, number GSE11882. e–f, P53-positive cells were counted in neocortex of 3 consecutive transverse brain sections (e) at the level of the Bregma (Mouse brain atlas, Paxinos) and 3 consecutive C6 cervical spinal cord sections (f). Sections from 16-week old DR Ercc1Δ/− mice (n=4) show significantly reduced levels of P53-positive cells (p<0.0001 for neocortex and p=0.0002 for spinal cord) as compared to sections from AL mice (n=4). g, Relative expression changes in the P53 target gene p21 in 11-week old wt and Ercc1Δ/− mice by DR (n=5) h, yH2AX-positive Purkinje (PkJ) neurons were counted in cerebellum of 5 consecutive transverse brain sections in AL- and DR-treated Ercc1Δ/− mice (p=0.014). Error bars indicate mean ± SE. * p<0.05, ** p<0.01, ***p<0.001. i. Mechanistic model for the anti-aging effect of DR. With aging DNA damage from exogenous and endogenous (metabolic) sources accumulates, which is accelerated in repair-deficient Ercc1Δ/− mice. Stochastic DNA lesions reduce transcriptional output in a gene-size dependent manner leading to cell dysfunction and death, stem cell exhaustion, organ/tissue atrophy and functional decline, causing aging-related diseases. Accumulating DNA damage and DR trigger an anti-aging response, which involves suppression of growth, up-regulation of anti-oxidant defense and presumably metabolic redesign which reduces steady state levels of reactive metabolites, thereby preserving genome integrity and delaying aging-related functional decline.
Table 1.
Number of differentially expressed gene probes (DEGs) of wt and Ercc1Δ/− liver under AL and DR conditions
| Comparison | DEG | Up | Down | Ratio up:down | Concordant | Discordant |
|---|---|---|---|---|---|---|
| Wt-DR vs. Wt-AL | 2,704 | 1,522 | 1,182 | 1.29* | ||
| Ercc1-DR vs. Ercc1-AL | 1,106 | 669 | 437 | 1.53* | ||
| Common | 688 | 391 | 293 | 684 | 4 | |
| Enrichment factor | 10.3 | 17.3 | 25.6 |
P=0.019 (Chi-square test with Yates correction)
Transcription-coupled repair (TCR) deficiencies in Ercc1Δ/− and other mouse mutants and human patients, which prevent resumption of RNA synthesis after transcription-blocking DNA damage, appear uniformly associated with premature aging, particularly affecting non- or slowly proliferating organs such as the nervous system, liver and kidney2. This suggests time-dependent accumulation of transcription-blocking lesions to contribute to accelerated aging. Since DNA damage occurs stochastically, long genes would predictably accumulate more lesions and consequently become transcriptionally more crippled than short genes. Indeed, the DEGs of Ercc1-AL liver profiles at 11 weeks displayed a highly significant bias for long genes to be overrepresented in the class of down-regulated genes and underrepresented in the category of up-regulated genes (Fig. 3b,c, arrows). These and other data (Table 1, Extended Data Table 2) support genome-wide accumulation of lesions stalling transcription. Interestingly, we observed a similar, albeit milder expression bias upon normal aging in rat liver and human hippocampus (Fig. 3d, Extended Data Fig. 8f). Importantly, DR in Ercc1Δ/− mice strongly retarded this expressional shift (red curve in Fig. 3b,c, Extended Data Table 2).
Reduction of P53+-cells in brain (Fig. 3e,f and Extended Data Fig. 6d), diminished apoptosis of retinal photoreceptors (Extended Data Fig. 6a) and strong preservation of neurons (Fig. 2f,g) also support reduced DNA damage loads in Ercc1Δ/− DR animals. We also found indications for suppression of p53-transcriptional activity in Ercc1 liver upon DR (Supplementary Table 2) further supported by DR-induced downregulation of miR-34a, a target of p53 (Extended Data Fig. 8c). Additionally, key senescence parameters, elevated in Ercc1-AL conditions, are mitigated by DR including p21 (Fig. 3g), p16, IL6 (Extended Data Fig. 8e). The proportion of Purkinje cell nuclei containing γH2AX foci which reflect DNA breaks, appeared significantly reduced (Fig. 3h and Extended Data Fig. 6e). We conclude that DR concomitantly with delaying aging, attenuates accumulation of genome-wide DNA damage and preserves transcriptional output most likely contributing to improved cell viability in Ercc1 mutants (Fig. 2f,g, 3c,h).
How does DR preserve genome function? It is inconceivable that enhanced repair is implicated here, as compensatory pathways for Ercc1 deficiency are unknown. Alternatively, DR may reduce damage induction and/or alter damage responses, to which DNA repair mutants may over-respond. Indeed, DR increases stress resistance, improves antioxidant defenses, alters insulin and other hormonal signaling pathways redesigning major metabolic routes (glycolysis, oxidative phosphorylation, pentose phosphate shunt), alters mitochondrial function and apoptotic responses and induces a shift from pro- to anti-inflammatory cytokines10,24, as also apparent from our DR expression profiles (Extended Data Fig. 7,8, Supplementary Table 2). Fig. 3i presents a model integrating our findings on DR, DNA damage and (accelerated) aging.
Post-mitotic neurons have to reconcile one of the highest metabolic rates of the body with preservation of a delicate homeostatic balance for over a century in man. While everything else in cells can be turned over, the up to 105 daily DNA lesions per cell25,26 can only be repaired, requiring continuous efficient repair. The notion that TCR defects are linked with severe neurodegeneration in CS and trichothiodystrophy (TTD) indicates that cleaning the transcribed compartment of the genome from transcription-obstructing lesions is vital for neuronal survival23. Using tissue-specific repair mutants we have shown that neurodegeneration is at least in part cell-autonomous6,27,28 consistent with endogenous DNA damage as a main driver. The strong protection by DR indicates that neurons possess considerable reserves to restrict the DNA damage problem and prevent cell death. Cell-intrinsic mechanisms and systemic inflammatory and hormonal responses modulated by DR may contribute to the remarkable resilience of the neuronal system, e.g. protecting 50% more neurons from death, strongly postponing tremors, imbalance and paresis and fully preserving motoric performance (Fig. 2, Supplementary Video1,2). The striking preservation of neuronal function in the Ercc1Δ/− mutant by DR aligns with increasing evidence for beneficial effects of DR in animal models of various neurodegenerative disorders29,30 and opens perspectives for nutritional and pharmacological interventions preventing the onset of these devastating diseases.
The strong evolutionary conservation of the DR response and striking parallels between mouse and human symptoms, render it likely that the DR effect is preserved from mice to man. Therefore, an obvious future application may be the counterintuitive use of DR(mimetics) to treat DNA repair defective progeroid syndromes. Besides CS and TTD this may apply to xeroderma pigmentosum (XP), XPCS, Cerebro-oculo-facio-skeletal (COFS) syndrome, Fanconi’s anemia, the RecQ helicase family of conditions: Werner, Bloom’s and Rothmund Thomson syndrome, Ataxia Telangiectasia, and others. Finally, the repair-deficient mice might prove useful tools for understanding anti-aging interventions and finding alternatives to DR by strongly reducing labor, time, costs and number of animals and for defining long-term health effects of nutrition.
Methods
Mouse models
The generation and characterization of Ercc1Δ/+ and Ercc1+/− mice have been previously described16. Ercc1Δ/− mice were obtained by crossing Ercc1Δ/+ (in a pure C57BL6J or FVB background) with Ercc1+/− mice (in a pure FVB or C57BL6J background respectively) to yield Ercc1Δ/− offspring with a genetically uniform F1 C57BL6J/FVB hybrid background (see6 for motivation). Wild-type F1 littermates were used as controls. Xpg−/− mice were characterized previously6 and were generated by crossing Xpg+/− (in a pure C57BL6J background) with Xpg+/− mice (in a pure FVB background). Hence, all animals used in the studies described here were of the same F1 C57BL6J/FVB hybrid background. Typical unfavorable characteristics, like blindness in an FVB background or deafness in a C57BL6J background, do not occur in this hybrid background.
Mice were weighed, visually inspected weekly, and scored blindly for gross morphological and motor abnormalities. Since the Ercc1Δ/− and Xpg−/− mice were smaller, food was administered within the cages and water bottles with long nozzles were used from around two weeks of age. Animals were maintained in a controlled environment (20–22°C, 12h light:12h dark cycle) and were housed in individual ventilated cages under SPF conditions. Animals were individually housed at the EMC location and group housed at the RIVM location. Experiments were performed in accordance with the Principles of Laboratory Animal Care and with the guidelines approved by the Dutch Ethical Committee in full accordance with European legislation.
For the lifespan studies the indicated number of mice per group for ad libitum (AL) and 30% dietary restriction (DR) were generated. Additionally, several cross sectional cohorts were generated. For Ercc1Δ/− mice we generated groups which were sacrificed at 7, 11, 16 or 30 weeks of age. The 7-week group only consisted of AL-fed animals while the 30-week group consisted of only DR-treated mice. For wild type mice, AL-fed and DR-treated groups were sacrificed at 11, 16 or 20 weeks. Animals were randomly divided over all groups to prevent selection bias. All mice were clinically diagnosed daily and when moribund, euthanized after which necropsy was performed. Animals from cross sectional cohorts were euthanized when necropsy age was reached. Organs were stored at −80°C for molecular analysis or (perfusion) fixated in (para)formaldehyde for pathological examinations. Statistics was performed with survival curve analysis using the product limit method of Kaplan and Meier in GraphPad Prism.
Diets
All animals were bred and maintained on AIN93G synthetic pellets (Research Diet Services B.V., Wijk bij Duurstede, The Netherlands; gross energy content 4.9 kcal/g dry mass, digestible energy 3.97 kcal/g). The initial lifespan cohort, shown in Figure 1a, was performed on standard AIN93G pellets containing 2.5 g/kg choline bitartrate. To avoid potential formation of bladder and kidney stones, we replaced choline bitartrate for choline chloride in all subsequent experiments. The amount of DR was determined in a prior pilot study but as control food intake of the AL-fed mice was continuously monitored. On average, Ercc1Δ/− and Xpg−/− mice ate 2.3 g food per day. DR was initiated at 7 weeks of age with 10% food reduction (2.1 g/day), when animals almost reached the maximum bodyweight and development was completed. DR was gradually increased weekly with 10%, until 30% DR (1.6 g/day) from 9 weeks of age onward. Temporary DR was initiated directly with 30% food reduction at 6 weeks of age. These mice received ad libitum food again from 12 weeks onward. Wild type mice ate on average 3.0 g food per day, resulting in 2.1 g/day for 30% DR. Food was given to the animals just before the start of the dark (active) period to avoid alterations in the biological clock.
Pathology assessment of aging characteristics
Representative sections from the liver, kidneys, sciatic nerve, testis and femur were processed, stained with hematoxylin and eosin, and microscopically examined in a blind manner by 2 board certified pathologists (SAY, AdB) for the presence of histopathologic lesions. Severity score of lesions was semi-quantitatively assessed. Scores were given as absent (0), subtle (1), mild (2), moderate (3), severe (4), and massive (5). Digital images from the kidneys and femur cortical bone at mid-shaft area were taken for morphometric analysis using Labsense image analysis software (Olympus). Aging characteristics were assessed in >5 animals per group per sex. Groups were compared with nonparametric Mann-Whitney U and Kruskal-Wallis tests.
FACS analysis of nuclear DNA content
Polyploidy levels were assessed based on propidium iodide (PI) fluorescence using FACS analysis31,32. A small part of the left lobe (approximately 5 mm3) was dissected from AL- and DR-treated Ercc1Δ/− mice (7, 11, 16 and 30 weeks, n=5) and wild type mice (11 weeks, n=5), cut into small fragments and suspended in 800 μL PBS using a syringe (21G). 300 μL homogenate was added to 300 μL 100% ethanol for fixation. Samples were stored at least 24 hours before further processing. After fixation the liver homogenate was washed with ice cold PBS and subsequently 20 minutes incubated with a pepsin solution. After washing in PBS/Tween-20 cells were collected in 500μL PBS supplemented with 5 μg/ml PI and 250 μg/ml RNAse and samples were measured using the FACS (FACSCalibur, Becton Dickinson, Franklin Lakes, NJ, USA). Differences between groups were assessed with a two-way ANOVA with age and diet as fixed factors.
Micro-computed tomographic (micro-CT) quantification of bone thickness
AL- and DR-treated mice were sacrificed by cervical dislocation at scheduled ages, femora were excised and non-osseous tissue was removed. Two days post-fixation (4% formalin), the right femora were scanned using the Skyscan 1076 in vivo X-Ray computed tomography (Bruker microCT, Kontich, Belgium) with a voxel size of 8.88 μm. Osseous tissue was distinguished from non-osseous tissue by segmenting the reconstructed grayscale images with an automated algorithm using local thresholds33. The region of interest (ROI), i.e. distal metaphysis of the femora, was selected by using 3D data analysis software. To compensate for bone length differences, the length of each ROI was determined relative to the largest specimen femur of the cohort. Cortex and trabeculae of the metaphysis were separated using in-house developed automated software. Thickness of the trabeculae and cortices were assessed using 3D analysis software as described34 using the CT analyzer software package (Bruker microCT). A bone specimen with known bone morphometrics was included within each scan as a quantitative control. Statistical significance was calculated using one-way Anova with Bonferroni’s multiple comparison test.
Ex vivo vascular function
The responses of isolated aortic tissue were ex vivo measured in small wire myograph organ baths containing oxygenated Krebs-Henseleit buffer at 37°C. After preconstriction with 30 nmol/L U46619, relaxation concentration-response curves to acetylcholine were constructed35.
Immunological analyses
Single cell suspensions were prepared from spleen by passing the cells in a cell strainer with Hepes-buffered saline solution (HBSS) supplemented with 2% FBS and washed. Erythrocytes were eliminated with ACK buffer. For CD4+CD25+Foxp3+ staining, cells were first stained for the expression of cell surface markers and then fixed, permeabilized, and stained using the Foxp3 kit (eBiosciences, San Diego, CA) according to the manufacturer’s instructions. FACS analysis was performed using FACSCanto (Becton Dickinson, Palo Alto, CA) and analyzed with FlowJo Software (TreeStar, Ashland, OR).
Blood glucose, insulin and albumin levels
Mice were euthanized by CO2 asphyxiation and blood was immediately collected from the heart. Glucose levels were measured using a Freestyle mini blood glucose meter. Insulin and albumin levels were measured in blood plasma using an ultrasensitive mouse insulin Elisa (Mercodia AB, Uppsala, Sweden) or mouse albumin ELISA kit (Immunology Consultants Laboratory, Inc., Portland, OR), respectively. Insulin levels were determined after overnight fasting. Glucose levels were determined after feeding, at the beginning of the dark period.
IgA measurements
Euthanasia of moribund or cross-sectional animals was performed by intramuscular injection of a Ketamine–Rompun mixture, followed by exsanguination3. IgA Immunoglobulin was measured in blood serum using the commercially available bead-based multiplexed panel Mouse Immunoglobulin Isotyping (Millipore Corporation, Billerica, MA, USA). Standard analysis protocols were followed and all samples were analyzed at least in duplo.
Phenotype scoring
The mice were weighed and visually inspected weekly, and were scored in a blind manner by 2 experienced research technicians (RMCB, SB) for the onset of various phenotypical parameters. Clasping was measured by suspending mice from their tails for 20 seconds.
A clasping event was scored when retraction of both hind limbs towards the body was observed for at least 5 seconds. Whole body tremor was scored if mice were trembling for a combined total of at least 10 seconds, when put on a flat surface for 20 seconds. Impaired balance was determined by observing the mice for 20 seconds, while walking on a flat surface. Mice that had difficulties in maintaining an upright orientation during this period were scored as having imbalance. If mice showed a partial loss of function of the hind limbs, it was indicated as having paresis. Statistics was performed with survival curve analysis using the product limit method of Kaplan and Meier in GraphPad Prism.
Behavioral analyses
Rotarod performance was assessed by measuring the average time spent on an accelerating rotarod (Ugo Basile). All animals were given four consecutive trials of maximally 5 minutes with inter-trial intervals of 1 hour. For weekly monitoring the motor coordination performance two consecutive trials of maximally 5 minutes were measured. Grip strength was determined by placing mice with forelimbs or all limbs on a grid attached to a force gauge, and steadily pulling the mice by their tail. Grip strength is defined as the maximum strength produced by the mouse before releasing the grid. For each value the test is performed in triplicate.
TUNEL staining
To quantify apoptotic cells in the retina, eyes were fixed overnight in 10% buffered formalin, paraffin-embedded, sectioned at 5 mm, and mounted on Superfrost Plus slides. Paraffin sections were employed for TdT-mediated dUTP Nick-End Labeling (TUNEL) assay using a commercial kit (Apoptag Plus Peroxidase in situ apoptosis detection kit, Millipore). Sections were deparaffinized and incubated as described by the manufacturer. Statistical differences were calculated with a t-test.
Antibodies
Primary antibodies (supplier; catalog number; dilutions) used in this study were as follows: rabbit anti-ATF3 (Santa Cruz; sc-188; 1:2,000), goat anti-ChAT (Millipore; AB144P; 1:500); rabbit anti-GFAP (DAKO; Z0334; 1:8,000); mouse anti-GM130 (BD Transduction; 610823; 1:100); rabbit anti-Iba-1 (Wako; 019-19741; 1:5,000); rat anti-Mac2 (Cedarlane; CL8942AP; 1:2,000); mouse anti-NeuN (Millipore; MAB377; 1:1,000); rabbit anti-p53 (Leica; NCL-p53-CM5p; 1:1,000); mouse anti-yH2AX (Millipore; 05-636; 1:4,000). For avidin– biotin–peroxidase immunocytochemistry biotinylated secondary antibodies from Vector Laboratories, diluted 1:200 were used. Alexa488-, Cy3-, and Cy5-conjugated secondary antibodies raised in donkey (Jackson ImmunoResearch) diluted at 1:200 were used for confocal immunofluorescence.
Histological procedures
Mice were anaesthetized with pentobarbital and perfused transcardially with 4% paraformaldehyde. Brain and spinal cord were carefully dissected out, post-fixed for 1 h in 4% paraformaldehyde, cryoprotected, embedded in 12% gelatin, rapidly frozen, and sectioned at 40 μm using a freezing microtome or stored at −80°C until use. Frozen sections were processed free floating using the ABC method (ABC, Vector Laboratories, USA) or single-, double-, and triple-labelling immunofluorescence. Immunoperoxidase-stained sections were analyzed and photographed using an Olympus BX40 microscope. Immunofluorescence sections were analyzed using a Zeiss LSM700 confocal. Mean intensities were quantified using Fiji. Statistical differences were calculated with a t-test.
RNA isolation
Total RNA was extracted using QIAzol lysis Reagent from mouse tissue specimens. For increased purity miRNAeasy Mini Kits (QIAGEN, Hilden, Germany) were used. Addition of wash buffers RPE and RWT (QIAGEN) was done mechanically by using the QIAcube (QIAGEN, Hilden, Germany) via the miRNeasy program and stored at −80°C. The concentration of RNA was measured by Nanodrop (Thermo Fisher Scientific).
Real-time PCR
Gene expression analyses were performed with gene-specific real-time PCR primers (see below) using SYBR Green (Sigma-Aldrich) and Platinum Taq polymerase (Life Technologies) on a Bio-Rad CFX96 thermocycler or with pre-designed TaqMan Gene Expression Assays (given below) with a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). Relative gene expressions were calculated as previously described6. For SYBR Green method the following primers were used (Forward primer 5′ to 3′; Reverse primer 5′ to 3′): Gsta1 (CTTCTGACCCCTTTCCCTCT; ATCCATGGGAGGCTTTCTCT), Nqo1 (GGTAGCGGCTCCATGTACTC; GAGTGTGGCCAATGCTGTAA), Nfe2l2 (AGGACATGGAGCAAGTTTGG; TCTGTCAGTGTGGCTTCTGG), Gstt2 (CGAGCAATTCTCCCAGGTGA; TATTCGTGGACTTGGGCACG), Fkbp5 (TGTTCAAGAAGTTCGCAGAGC; CCTTCTTGCTCCCAGCTTT), Srxn1 (TGAGCAGCTCCTCTGATGTG; GCTGAGGTGACAATTGACTATGG), Gsta4 (TCGATGGGATGATGCTGAC; CATCTGCATACATGTCAATCCTG), Gclm (TGGAGCAGCTGTATCAGTGG; CAAAGGCAGTCAAATCTGGTG), Hmox1 (CAGGTGATGCTGACAGAGGA; ATGGCATAAATTCCCACTGC), Gclc (AGATGATAGAACACGGGAGGAG; TGATCCTAAAGCGATTGTTCTTC), Ephx1 (GAGTGGAGGAACTGCACACC; AGCACAGAAGCCAGGATGA), Mgst1 (CTCGGCAGGACAACTTGC; CCATGCTTCCAATCTTGGTC), TubG2 (CAGACCAACCACTGCTACAT; AGGGAATGAAGTTGGCCAGT), Hprt (TGATAGATCCATTCCTATGACTGTAGA; AAGACATTCTTTCCAGTTAAAGTTGAG), Rps9 (ATCCGCCAACGTCACATTA; TCTTCACTCGGCCTGGAC). As pre-designed TaqMan assays we used (Order number; Sequence 5′ to 3′): Ghr (Mm00439093_m1; GACAAGCTGCAAGAATTGCTCATGA), Igf1r (Mm00802831_m1; GGCCAGAAGTGGAGCAGAATAATCT), HPRT-E2_3 (HPRT-E2_3_F; GCCGAGGATTTGGAAAAAGTGTTTA, HPRT-E2_3_R; TTCATGACATCTCGAGCAAGTCTTT, HPRT-E2_3_M; CAGTCCTGTCCATAATCA), POLR2A-E2_3 (POLR2A-E2_3F; GCAGTTCGGAGTCCTGAGT, POLR2A-E2_3R; CCCTCTGTTGTTTCTGGGTATTTGA, POLR2A-E2_3M2; CATCCGCTTCAATTCAT).
Microarray Hybridizations
RNA quality was assessed using the 2100 Bio-Analyzer (Agilent Technologies, Amstelveen, the Netherlands) following manufacturer’s instructions. The quality of the RNA is expressed as the RNA integrity number (RIN, range 0–10). Samples with a RIN below 8 were excluded from analysis. Hybridization to Affymetrix HT MG-430 PM Array Plates was performed at the Microarray Department of the University of Amsterdam, the Netherlands according to Affymetrix protocols. Quality control and normalization were performed using the pipeline at the www.arrayanalysis.org website (Maastricht University, the Netherlands).
miRNA expression
The same total RNA extracts were used as extracted for mRNA analysis (above). miRNA expression levels were assessed using a miRNA micro-array (miRCURY LNA™ microRNA Array (7th Gen), Exiqon, Vedbæk, Denmark). All probes with more than three calls were selected for assessing differential expression between groups. Differences in mean expression were compared using a One-Way ANOVA. Probes with a FDR of 5% were considered as significantly differentially expressed.
Total RNAseq
RNA expression analysis was also performed by next generation sequencing approach on one animal per treatment as described in36.
Data Pre-analysis
Raw data (CEL files) were normalized by robust multichip average (RMA) in the oligo BioConductor package, which summarizes the perfect matches through median polish and collapses probes into core transcripts based on .CDF annotation file provided by Affymetrix using the R open statistical package (http://www.r-project.org/). All data files have been submitted to the NCBI gene expression omnibus under accession number GSE77495.
Principal component analysis (PCA)
PCA was performed using all the probe sets in the array. A graphical representation was generated to show the relation among the different samples. PCA is a linear projection method that defines a new dimensional space that captures the maximum information present in the initial data set. It is an unsupervised exploratory technique used to remove noise, reduce dimensionality and identify common/dominant signals oriented to try to find biological meaning37. The two principal components with the higher amount of variance were plotted. PCA was performed using the prcomp package and the plot was drawn with gplots, both from the Bioconductor project.
Detection of differentially expressed genes (DEG)
The linear model from Limma38 implemented in R was used to identify the DEGs. Pairwise comparisons for each genotype between ad libitum and DR samples were applied to calculate the fold change (FC), p value and false discovery rate (FDR) for each probe in the microarray. Cut-off values for a DEG were put at FDR <5% with fold change ≥ |1.5|. For all mouse analyses differentially expressed probes were considered as DEGs.
Determination of enrichment factor and p values of overlap
Overlap between lists of DEGs was identified looking by the intersection between pair of lists. To determine if the overlap was higher than expected by chance the hypergeometric distribution was used as is implemented in phyper function in R. Additionally the factor of enrichment was calculated with the formula:
Where:
nA = Number of DEG in experimental group A
nB= Number of DEG in experimental group B
nC= Number of total genes in the microarray
nAB = Number of common DEG between A and B
Pathway analysis
Pathway enrichment analysis was conducted via overrepresentation analysis (ORA). ORA was performed in the Interactive pathway analysis (IPA) of complex genomics data software (Ingenuity Systems, Qiagen) by employing a pre-filtered list of differentially expressed genes. Genes were selected as differentially expressed if they had a fold change ≥ |1.5| and a FDR lower than 0.05. The over-represented canonical pathways were generated based on information in the Ingenuity Pathways Knowledge Base. A pathway was selected as deregulated when the p value in the Fisher test was lower than 0.01.
Additionally, IPA transcription factor (TF) analysis was performed to identify the cascade of upstream transcriptional regulators that can explain the observed gene expression changes in the different lists of DEGs. To do that the data base with prior knowledge of expected effects between TF and their target genes stored in the Ingenuity Knowledge Base was used. The analysis examines how many known targets of each TF are present in the list of DEGs, and also compares their direction of change to what is expected from the literature in order to predict likely relevant transcriptional regulators. If the observed direction of change is mostly consistent with a particular activation state of the transcriptional regulator (“activated” or “inhibited”), then a prediction is made about that activation state. For each TF two statistical measures are computed (overlap p-value and activation z- score). The overlap p-value labels upstream regulators based on significant overlap between dataset genes and known targets regulated by a TF. The activation z-score is used to infer likely activation states of upstream regulators based on comparison with a model that assigns random regulation directions. Overlap p value lower than 0.05 and z-score higher than |2| were selected as thresholds to identify a TF as relevant.
Gene length analysis
Limma was used to identify the DEGs among wt AL samples compared with the others experimental conditions (wtDR, Ercc1Δ/− AL and Ercc1Δ/− DR). Next, probe-sets in the Affymetrix array with multiple gene annotation were filtered out. BiomaRt39 was used to retrieve the gene length for the remaining probe sets (32,930 probe-sets from 45,142 probe-sets in the original microarray). Differentially expressed genes were selected using FDR <0.05 and linear fold change ±1.5. Shapiro–Wilk test was applied to contrast the normality of the distribution of gene length in the different lists of DEGs. Because most of the distributions were not normal a Mann-Whitney for non-paired samples test was used to evaluate whether the distributions of DEGs were different between the different comparisons. Finally, a Relative Frequency (Kernel density) plot of gene length and probability density for DEG in each comparison was drawn using the density function implemented in R. Kernel density estimates are related to histograms, but with the possibility to smooth and continuity by using a kernel function. The y axis represents the density probability for a specific range of values in the x axis.
Immuno-blotting
Liver extracts from AL- and DR-treated Ercc1Δ/− and wild type mice (n=6, 11 weeks) were prepared by mechanical disruption in lysis buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris), which was supplemented with Mini cOmplete protease inhibitor (Roche Diagnostics, Indianapolis, IN, USA) and phosphates inhibitors (5 mM NaF, 1mM Na-Orthovanadate). After mechanical disruption, lysates were incubated on ice for one hour and subsequently centrifuged at 4 degrees Celsius for 20 minutes. 25 – 50 μg was loaded on a 10% SDS-PAGE gel (Life Technologies LTD, Paisley, UK) and transferred to a PVDF transfer membrane (GE-Healthcare Life Sciences, Uppsala, Sweden). Levels of S6 (#2217S Lot5; 1:2,000), S6(Ser240/244; #2215 Lot14; 1:500), Akt (#9272 Lot25; 1:500), Akt(Ser473; #9271S Lot13; 1:250) and Akt(Thr308; #9275S Lot19; 1:500) were detected (Cell Signaling Technology, Danvers, MA, USA), semi-quantified using the ImageJ software package (http://rsb.info.nih.gov/ij/index.html) and phospho/total ratios relative to AL were calculated and differences between groups were assessed with a t-test. B-Actin was used as loading control (Sigma; A5441 Lot064M4789V; 1:25,000).
Extended Data
Extended Data Figure 1. Effect of dietary restriction on body weight and various healthspan parameters of Ercc1Δ/− mice primarily related to glucose metabolism and liver pathology.
a–d, Body weights curves of Ercc1Δ/− (a–b) and Xpg−/− (c–d) male (a, c) and female (b, d) mice with ad libitum (AL; blue) access to AIN93G diet or on 30% dietary restriction (DR; red) shown as mean±SE at weekly intervals; n=4 animals/group solitary housed at the ErasmusMC. DR was initiated at 7 weeks of age with 10%, when the mice almost reached the maximum body weight avoiding disruption of development of young animals, and increased weekly with 10%, until 30% was reached from 9 weeks of age onwards. e–g, Blood glucose after feeding (e), plasma fasting insulin (f), and plasma albumin levels (g), indicative of liver functioning, in AL and DR wt and Ercc1Δ/− mice at 16 weeks. n≥3 animals/group. h, Quantification of 16N nuclei in hepatocytes32 of 11 week old male wt and Ercc1Δ/− mice under AL or DR regimens by FACS analyses; n=5 animals/group. i, Total numbers of splenic CD4+ T cell levels from spleen of 16 weeks old Ercc1Δ/− mice under DR or AL and aged-matched wt controls. n≥3 animals/group. j, IgA blood levels in male Ercc1Δ/− mice at different ages under DR or AL regimes. n=5 animals/group. k, Average grip strength of the forelimbs and all limbs of 16-week old Ercc1Δ/− and wt mice is similar under AL and DR conditions; n= 4 animals/group. Error bars denote mean ± SE. * p<0.05, ** p<0.01, ***p<0.001.
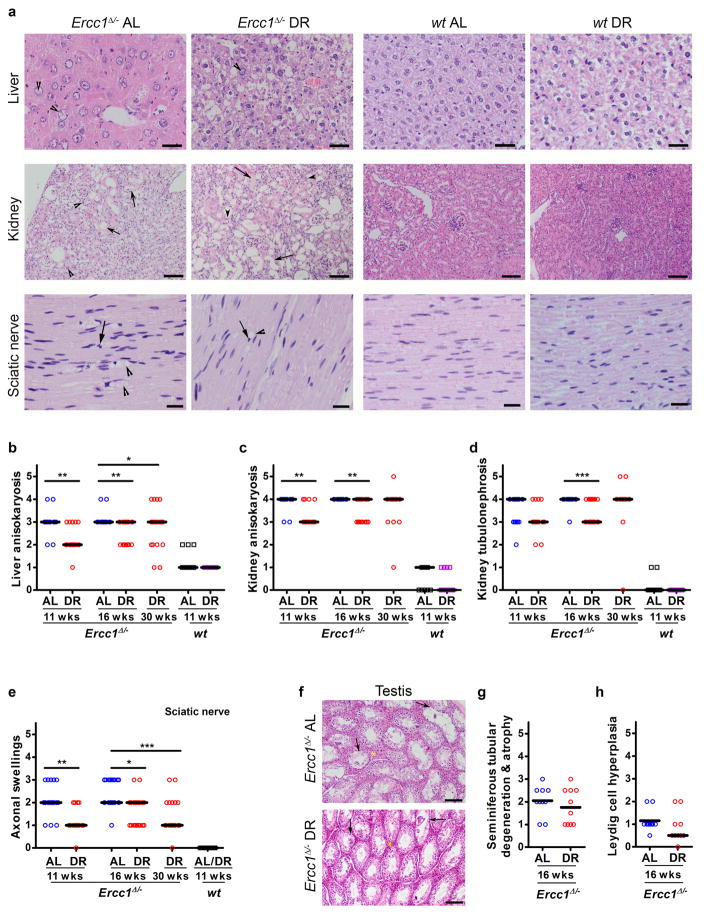
Extended Data Figure 2. Dietary restriction improves aging-related histopathological phenotypes in different tissues of Ercc1Δ/− mice.
a, Representative pictures of Hematoxylin-Eosin stained slides from liver, kidney, and sciatic nerve. Left panel represents Ercc1Δ/− ad libitum (Ercc1Δ/− AL), left middle panel represents DR-treated Ercc1Δ/− group (Ercc1Δ/− DR), right middle panel is from wt-AL group, and right panel represents DR-treated wt. Lesions were semi-quantitatively assessed in scores ranging from absent (0) through massive (5). Liver of female Ercc1Δ/− AL mouse with moderate anisokaryosis (score=3) and intranuclear inclusions (score =3, arrowheads), liver of female Ercc1Δ/− DR mouse with moderate hydropic degeneration with mild anisokaryosis (score=1) and presence of few hepatocellular intranuclear inclusions (score=1, arrowhead), and histologically normal liver from wt-AL and wt-DR. Kidney of female Ercc1Δ/− AL and Ercc1Δ/− DR mouse with severe tubular attenuation and degeneration (score=5, arrows) with marked anisokaryosis (score=4, arrowheads) next to histologically normal kidneys from female wt-AL and wt-DR mice. Sciatic nerve of a female Ercc1Δ/− AL mouse with severe axonal swellings (score=3, arrowheads). These axonal swellings most likely represent vacuoles containing myelin debris and/or fragmented axons. The Schwann cell nuclei around vacuolated areas are pyknotic (arrow). The sciatic nerve of a female Ercc1Δ/− DR mouse displays mild vacuole-like structures (score=1, arrowhead) with pyknosis of Schwann cell nuclei (arrow), while the histologically normal sciatic nerves of female wt-AL and wt-DR mice display no axonal swellings. Scale bar in liver = 50 μm, kidney = 100 μm, and in sciatic nerve = 20 μm. b, Pathology assessment of anisokaryosis in liver of Ercc1Δ/− mice at different ages under AL (blue) or DR (red) regimen and young AL (black) and DR (purple) wt controls. Scores range from absent (0) through massive (5); n≥10 animals/group; bars indicate group medians. c–d, Pathology assessment of anisokaryosis (c) and tubulonephrosis (d) in kidney of Ercc1Δ/− and wt mice at different ages under AL and DR regimes. Scores range from absent (0) through massive (5); n≥10 animals/group. e, Pathology assessment of axonal swellings in sciatic nerves of Ercc1Δ/− mice at different ages under AL or DR regimen. Scores range from absent (0) through massive (5); n≥10 animals/group. f, Representative pictures to the testicular lesions observed in Ercc1Δ/− males. The AL testes (upper panel) exhibited moderate testicular degeneration and atrophy (arrows). Also, the Leydig cells (yellow asterisk) appeared more prominent likely secondary to the tubular loss/attenuation or may be due to true Leydig cell hyperplasia (a common aging lesion in rodent testes). These phenotypes were slightly rescued in the DR testes (lower panel). g–h, Pathology assessment of seminiferous tubular degeneration and atrophy (g) and Leydig cell hyperplasia (h) in testis of Ercc1Δ/− mice at 16 weeks of age under AL (blue) or DR (red) regimen. Scores were given as absent (0), subtle (1), mild (2), moderate (3), severe (4), and massive (5) for each criteria with 0.5 interval; n=10 animals/group; bars indicate group medians. Note that testicular development is mostly completed at the start of DR. * p<0.05, ** p<0.01, ***p<0.001. As for wt controls the values do not change significantly in the timeframes used here (see40). Pathological scores, including those of other liver and kidney aging-related histopathological phenotypes are given in Supplementary Table 1.
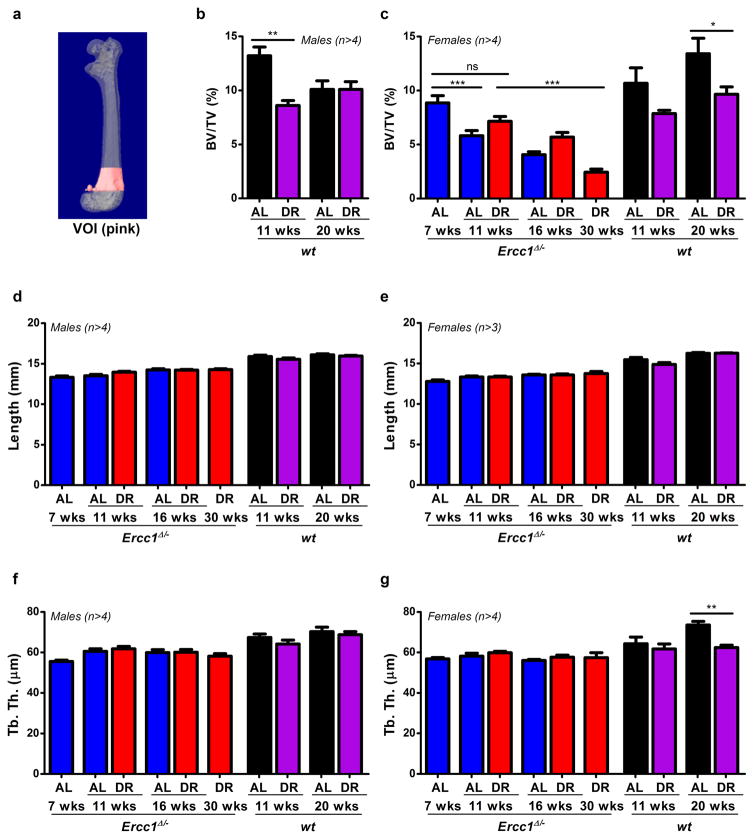
Extended Data Figure 3. Dietary restriction preserves skeletal structure in Ercc1Δ/− mice.
a, Illustration depicting the femural volume of interest (VOI) for microCT analyses. b–c, Trabecular bone volume fraction (BV/TV) representing the amount of trabecular bone in the femur VOI of (b) wt male mice as well as (c) Ercc1Δ/− and wt female mice expressed as percentage measured using micro-CT. d–e, Femur length of Ercc1Δ/− and wt (d) male and (e) female mice. f–g, Trabecular thickness in the femur VOI of Ercc1Δ/− and wt (f) male and (g) female mice. AL- and DR-treated animals were measured at different ages with n≥3 animals/group. Values of Ercc1Δ/− mice are depicted in blue (AL) and red (DR). Young wt controls are depicted in black (AL) and purple (DR). Error bars denote mean ± SE. * p<0.05, ** p<0.01, ***p<0.001.
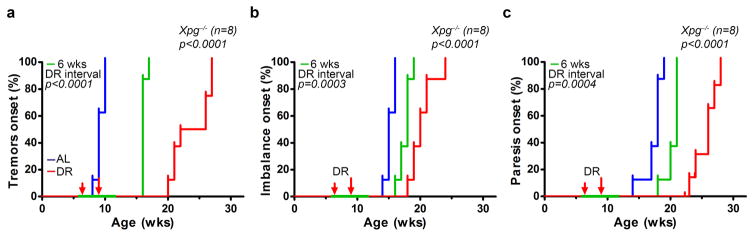
Extended Data Figure 4. Dietary restriction preserves neurofunctional behavior of Xpg−/− mice.
a–c, Onset of neurological abnormalities as tremors (a), imbalance (b), and paresis of the hind limbs (c) with age in Xpg−/− mice under AL and DR regimens. n=8 animals/group. The onset of continuous DR is indicated by the red arrows. Average age in the onset of tremors is delayed from 9 to 24 weeks of age, imbalance from 15 to 20, and paresis from 18 to 26 weeks of age. Temporary DR was given between 6 and 12 weeks of age and is indicated in green. This short period of DR yielded in a median delay in onset of tremors of 7 weeks while the median age of onset of both imbalance and paresis was 3 weeks delayed. p values were calculated against Xpg−/−-AL using the log-rank test.
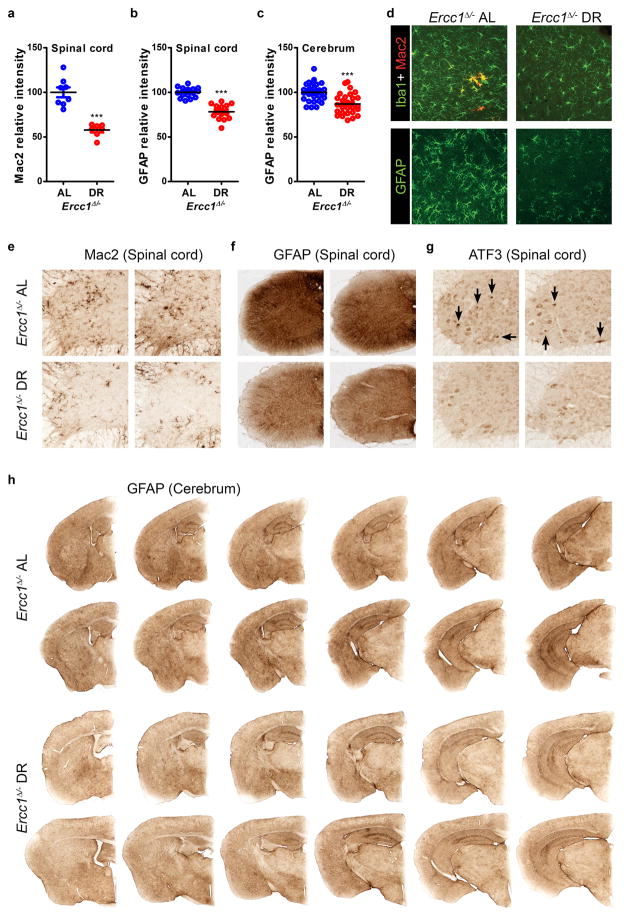
Extended Data Figure 5. Dietary restriction improves microgliosis and astrocytosis in brain and spinal cord of Ercc1Δ/− mice.
a–c, Quantification of the relative intensity of consecutive transverse brain and spinal cord sections immunoperoxidase-stained for Mac2 in spinal cord (a), and GFAP in spinal cord (b) and cerebrum (c). n>3 animals/group; bars indicate group medians. d, Iba1, Mac2, and GFAP immunofluorescent confocal images showing that reduced astrocytosis (GFAP) in cortex is paralleled by reduced staining for microglia (Iba1). Also Mac2-immunoreactivity, which outlines a subset of phagocytosing microglia cells, is reduced in 16 week old DR (n=4) as compared to AL (n=3) cortex in Ercc1Δ/− mice. e–g, Representative pictures of spinal cord sections of 16 weeks old AL and DR Ercc1Δ/− mice immunoperoxidase-stained for Mac2 (e) GFAP (f) reflecting reduced microgliosis and astrocytosis respectively, in the DR nervous system. Immunoperoxidase-stained spinal cord sections for ATF3 (g) showed that activation of the stress-inducible transcription factor ATF3 (which is induced following genotoxic stress via p53-dependent and -independent pathways) is less pronounced in DR nervous system. Per condition, sections from two different animals are presented next to each other. Black arrows indicate cells with high nuclear ATF3 staining. h, Representative pictures of consecutive transverse brain sections of 16 weeks old AL and DR Ercc1Δ/− mice immunoperoxidase-stained for GFAP, showing reduced GFAP staining, in the DR nervous system. Per animal, represented in one row, six 40μm slices are shown with 360μm cerebrum thickness in between each slice. Error bars denote mean ± SE. ***p<0.001.
Extended Data Figure 6. Dietary restriction dramatically preserves neurofunctioning of Ercc1Δ/− mice.
a, Quantification of TUNEL-positive cells in the outer nuclear layer of retinal sections of 16 week old AL (blue) or DR (red) Ercc1Δ/− mice; n=4 animals/group. b, Analysis of the total number of motor neurons with an abnormal Golgi apparatus (indicative of cells with ill health; see thick arrows in representative image; neuron with normal Golgi is indicated by a thin arrow) in C6 cervical spinal cord sections from 16 week old DR and AL Ercc1Δ/− mice. n=4 animals/group. TUNEL-positive cells (a) and neurons with abnormal Golgi morphology (b) were absent in both AL-17 and DR-treated young wt mice. c, Quantitative stereological analysis of the total number of non-neuronal cells (Dapi+/NeuN-; p=0.2744) in the neocortex of transverse brain sections of 16 weeks old AL and DR Ercc1Δ/− mice. n≥3 animals/group. Error bars indicate mean ± SE. ***p<0.001. d, Representative images of neocortex stained for NeuN (neurons), P53 and Dapi (for staining DNA) used for quantitative stereological analysis of the total number of neurons (NeuN+) and non-neuronal cells (Dapi+/NeuN−) in 16 weeks old AL- (n=3) and DR-(n=4) treated Ercc1Δ/− mice. Quantification of the number of P53-positive neurons is shown in Figure 3f. The analysis was performed using the optical dissector probe from StereoInvestigator on a Zeiss LSM700 laser scanning microscope. e, Representative image of cerebellum stained for yH2AX (green; double stranded DNA breaks) and Dapi (blue; for staining DNA) in 16 weeks old AL (n=3) and DR (n=4) treated Ercc1Δ/− mice. The Purkinje (PkJ) neurons are present in a single layer (PL; purkinje layer) in between the molecular layer (ML) and granular layer (GL)41. Quantification of the number of yH2AX-positive PkJ-neurons is shown in Figure 3i. The analysis was performed using a Zeiss LSM700 laser scanning microscope.
Extended Data Figure 7. Effect of DR on mTorc1, mTorc2 and Ins/PDK1 signaling using immunoblot analysis of wt and Ercc1Δ/− liver extracts.
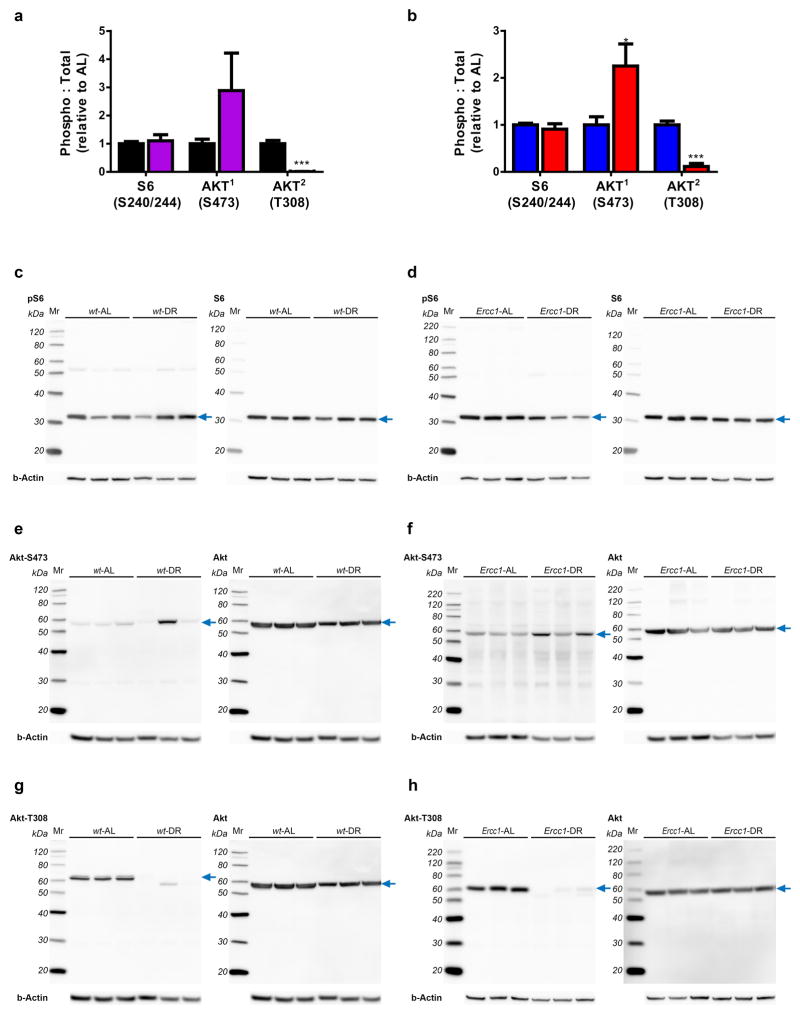
a–b, Quantified relative S6 and AKT phosphorylation by DR in both wt (a) and Ercc1Δ/− (b) liver extracts. 6 Animals per group were used; 11 weeks-of-age. c–h, Representative images used for the quantification of the ratio of S6 and AKT phosphorylation versus total S6 and AKT respectively. Phosphorylation of AKT at position S473 seems increased by DR in liver homogenates of 11 week old wt (e) and Ercc1Δ/− (f) mice but is suppressed at position T308 (g, h). Phosphorylation of S6 at S240/244 is unaffected by DR (c, d). For immunoblots, three animals per group are shown. For graphs and statistics, six animals per group were used. The blue arrow indicates signals used for quantification. Below each blot b-actin is presented as a loading control.
Extended Data Figure 8. Molecular analysis of expression changes by diet, DNA damage, or aging.
a–b, Ghr and Igf1r gene expression changes measured by quantitative real-time PCR in liver samples of 11 week old wt and Ercc1Δ/− mice by DR (n=5). Gene-specific real-time PCR primers as described in Methods. c, MicroRNA expression profile comparison of wt and Ercc1Δ/− liver under AL and DR conditions. 188 significant regulated miRNAs (FRD ≤ 5%) between groups are shown. Five of the most significantly changed microRNAs are zoomed in. miR-34a, a downstream target of p53 and involved in cell cycle and apoptosis induced by DNA-damage42,43, showed a differential effect between liver homogenates of 11 week old wt and Ercc1Δ/− mice. It was downregulated by DR in liver of wt mice (1.62 fold, p=0.02), but strongly upregulated in liver of AL-fed Ercc1Δ/− mice compared to AL-fed wt mice (4.7 fold, p=0.0001) and seems suppressed in DR-treated Ercc1Δ/− profiles. These chances were confirmed by qPCR (data not shown). d, Heatmap of key antioxidant defense genes in liver and brain of wt and Ercc1Δ/− mice. Fold changes were calculated for wt-DR, Ercc1-AL, and Ercc1-DR against wt-AL using microarray expression profiles of liver at 11 weeks of age (n=5) or quantitative real-time PCR for cerebellum at 16 weeks of age (n=4). DR induced an antioxidant response in liver, which is less pronounced in brain specimens, consistent with earlier findings of Swindell44, likely due to the already high antioxidant defense levels in the nervous system. The difference in antioxidant response between liver and brain by genotype is conform previous results6. Interestingly, the Purkinje-neuron marker Calbindin is clearly reduced in cerebella of Ercc1-AL mice and is less reduced in Ercc1-DR mice, confirming the drastic reduction in DNA damage induced neuronal (Purkinje cell) loss by DR. Blue color represents decreased expression, red represents increased expression. Hierarchical clustering on liver and cerebellum genes was performed using Pearson correlation. e, DR reduces the p16-RB branch of senescence and the senescence-associated secretary phenotype as assessed by next generation sequencing expression analysis of liver RNA of Ercc1Δ/− mice. To assess the p16-RB branch of the senescence phenotype45, we followed a next generation sequencing approach as described in36 using 16 week old livers from AL- and DR-fed wt and AL- and DR-fed Ercc1Δ/− mice. By sequencing >150M sequence reads per sample, we detected the p16-ink4a (Cdkn2a) transcript at sufficient levels. P16-ink4 (Cdkn2a) is considered a key marker for cellular senescence, but difficult to quantitatively analyze using other methods due to the high ratio normal cells compared to senescent cells. Datasets were normalized by calculating reads per kilobase million (RPKM). Subsequently, z-scores were calculated and plotted in a heatmap: red refers to increased expression, blue to decreased expression. In AL-fed Ercc1Δ/− liver RNA p16-ink4a (Cdkn2a) is upregulated compared to wt AL animals, but down-regulated after DR, indicating that Ercc1Δ/− mice have increased cellular senescence that is reduced upon DR. Secondly, we monitored the transcriptionally-induced senescence-associated secretory phenotype (SASP) as described in46. Many, if not all, SASP factors are not exclusively specific for cellular senescence. To reduce the probability that observed SASP factor expression changes are contributed by other cells, we only selected those SASP factors that have an absolute expression (RPKM) in the same range as p16-ink4a in across these datasets, since these are most likely the result of cellular senescence. The figure shows that most SASP factors are down-regulated after DR such as IL-6, the most prominent SASP cytokine supporting the idea that cellular senescence and associated SASP is increased in AL-fed Ercc1Δ/− liver and is reduced by DR. Hierarchical clustering was performed using Pearson correlation. f, Suppression of long genes in normal aging of rat liver. Relative frequency plot of gene length of DEGs in livers of 24 month old rat versus 6 month young rat. Upregulated genes are depicted in red and downregulated genes in green. The DEGs from rat liver were selected using a fold change cut-off of 1.5 and FDR < 0.05. The dataset is publicly available in the NCBI gene expression omnibus under number GSE66715.
Extended Data Table 1. Discordant DEG in DR response for wt and Ercc1Δ/−.
Genes DEG in common in DR response in liver of wt and Ercc1Δ/− mice but with discordant direction of change.
| ProbeID | Gene Symbol | logFC wt-DR vs wt-AL | P.Val Wt-DR vs wt-AL | logFC Ercc1-DR vs Ercc1-AL | P.Val Ercc1-DR vs Ercc1-AL |
|---|---|---|---|---|---|
| 1438583_PM_at | Ern1 | 0.77 | 0.003 | −0.74 | 0.010 |
| 1438997_PM_at | Ern1 | 0.69 | 0.005 | −0.70 | 0.010 |
| 1429295_PM_s_at | Trip13 | −0.72 | 0.005 | 0.77 | 0.007 |
| 1441098_PM_at | Pnldc1 | −0.66 | 0.030 | 1.10 | 0.002 |
Extended Data Table 2. Average gene length of up- and down-regulated genes of wt and Ercc1Δ/− liver expression profiles under AL and DR conditions.
Comparisons of the mean gene size distributions of DEGs (up- and down-regulated) among wt-DR vs. wt-AL, Ercc1-AL vs. wt-AL and Ercc1-DR vs. wt-AL. wt-DR Up, means the up-regulated DEGs in wt-DR compared with wt-AL (n = 1106); wt-DR Down, means the down-regulated genes in wt-DR, compared with wt-AL (n = 1046); Ercc1-AL Up means up-regulated DEGs in Ercc1-AL compared with wt-AL (n = 595), Ercc1-AL Down means the down-regulated DEGs in Ercc1-AL compared with wt-AL (n = 363); Ercc1-DR UP means up-regulated genes in Ercc1-DR compared with wt-AL (n = 1384): Ercc1-DR Down means down-regulated genes in Ercc1-DR compared with wt-AL (n = 768). Because the distributions of the gene lengths do not have a normal distribution the means should be taken as a reference value.
| Comparison | Length (bp) means | wilcox.test |
|---|---|---|
| Wt-DR Up vs. Ercc1-AL Up | 66,364 vs. 40,021 | <2.2e-16 |
| Wt-DR Down vs. Ercc1-AL Down | 62,352 vs. 212,957 | <2.2e-16 |
| Wt-DR Up vs. Ercc1-DR Up | 66,364 vs. 41,284 | <2.2E-16 |
| Wt-DR Down vs. Ercc1-DR Down | 62,352 vs. 141,391 | 3.24E-14 |
| Ercc1-AL Up vs. Ercc1-DR Up | 40,021 vs. 41,284 | 0.001 |
| Ercc1-AL Down vs. Ercc1-DR Down | 212,957 vs. 141,391 | 0.0002 |
Supplementary Material
Acknowledgments
We thank Piet de With, Jolanda Rigters, Elize Haasdijk, Sylvia Gabriels, Marjolein Steinenbosch, Nicole van Vliet, Yvette van Loon, Jery Baan and the animal caretakers for general assistance with mouse experiments and prof. A.H.J. Danser and prof. J.P. van Leeuwen for support. We acknowledge financial support of the National Institute of Health (NIH)/National Institute of Ageing (NIA) (1PO1 AG-17242-02), the National Institute for Public Health and the Environment and the Ministry of Health, Welfare and Sport of The Netherlands (S/340005), European Research Council Advanced Grant DamAge and Proof of Concept Grant Dementia to JHJH, the European commission FP7 Markage (FP7-Health-2008-200880), DNA Repair (LSHG-CT-2005-512113), EU ITN Address (GA-316390), the KWO Dutch Cancer Society (5030), the Dutch CAA Foundation and the Royal Academy of Arts and Sciences of the Netherlands (academia professorship to JHJH). The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement No. HEALTH-F2-2010-259893. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions W.P.V., M.E.T.D., E.R., J.V., H.S., and J.H.J.H. designed the research and wrote the manuscript. D.J., C.P.G., A.J.M.R., S.M.B., B.C.E., A.B., A.G., and J.P. contributed to editing the manuscript. W.P.V., M.E.T.D., E.R., R.M.C.B., and S.B. performed and analyzed the mouse lifespan cohorts. E.R., B.N., C.T.O., R.M.C.B., S.B., and S.I. performed genotyping and coordinated animal sectioning. S.A.Y., R.V.K., and A.B. assessed the aging pathology characteristics. R.V.K. and S.I. performed FACS analysis of nuclei. S.M.B. and B.C.E. quantified bone changes. H.W. and A.J.M.R. quantified vascular function. C.R.B. performed the immunological analyses. W.P.V., D.J., R.M.C.B., and S.B. performed and analyzed phenotypical scoring and behavioral analysis. W.P.V. and D.J. characterized neuropathological changes. W.P.V., E.R., C.P.G., C.T.O., J.L.A.P., A.G., and J.P. performed transcriptomic analyses and analyzed the data. W.P.V., M.E.T.D., E.R., C.T.O. R.M.C.B., S.B., and S.I. performed the molecular studies.
The expression data have been deposited to the Gene Expression Omnibus database under accession number GSE77495.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of the paper.
References
- 1.Niedernhofer LJ, et al. A new progeroid syndrome reveals that genotoxic stress suppresses the somatotroph axis. Nature. 2006;444:1038–43. doi: 10.1038/nature05456. [DOI] [PubMed] [Google Scholar]
- 2.Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–81. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 3.Dolle ME, et al. Broad segmental progeroid changes in short-lived Ercc1(−/Delta7) mice. Pathobiol Aging Age Relat Dis. 2011;1 doi: 10.3402/pba.v1i0.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vermeij WP, Hoeijmakers JH, Pothof J. Genome Integrity in Aging: Human Syndromes, Mouse Models, and Therapeutic Options. Annual Review of Pharmacology and Toxicology. 2016;56:427–45. doi: 10.1146/annurev-pharmtox-010814-124316. [DOI] [PubMed] [Google Scholar]
- 5.Garinis GA, et al. Persistent transcription-blocking DNA lesions trigger somatic growth attenuation associated with longevity. Nat Cell Biol. 2009;11:604–15. doi: 10.1038/ncb1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnhoorn S, et al. Cell-autonomous progeroid changes in conditional mouse models for repair endonuclease XPG deficiency. PLoS Genet. 2014;10:e1004686. doi: 10.1371/journal.pgen.1004686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colman RJ, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–4. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–6. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barzilai N, Huffman DM, Muzumdar RH, Bartke A. The critical role of metabolic pathways in aging. Diabetes. 2012;61:1315–22. doi: 10.2337/db11-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Speakman JR, Mitchell SE. Caloric restriction. Mol Aspects Med. 2011;32:159–221. doi: 10.1016/j.mam.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Hoeijmakers JH. DNA damage, aging, and cancer. N Engl J Med. 2009;361:1475–85. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 12.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Pluijm I, et al. Impaired genome maintenance suppresses the growth hormone--insulin-like growth factor 1 axis in mice with Cockayne syndrome. PLoS Biol. 2007;5:e2. doi: 10.1371/journal.pbio.0050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wijnhoven SW, et al. Accelerated aging pathology in ad libitum fed Xpd(TTD) mice is accompanied by features suggestive of caloric restriction. DNA Repair (Amst) 2005;4:1314–24. doi: 10.1016/j.dnarep.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Schumacher B, et al. Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 2008;4:e1000161. doi: 10.1371/journal.pgen.1000161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weeda G, et al. Disruption of mouse ERCC1 results in a novel repair syndrome with growth failure, nuclear abnormalities and senescence. Curr Biol. 1997;7:427–39. doi: 10.1016/s0960-9822(06)00190-4. [DOI] [PubMed] [Google Scholar]
- 17.de Waard MC, et al. Age-related motor neuron degeneration in DNA repair-deficient Ercc1 mice. Acta Neuropathol. 2010;120:461–75. doi: 10.1007/s00401-010-0715-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Partridge L, Gems D. Benchmarks for ageing studies. Nature. 2007;450:165–7. doi: 10.1038/450165a. [DOI] [PubMed] [Google Scholar]
- 19.Mair W, Goymer P, Pletcher SD, Partridge L. Demography of dietary restriction and death in Drosophila. Science. 2003;301:1731–3. doi: 10.1126/science.1086016. [DOI] [PubMed] [Google Scholar]
- 20.Bogliolo M, et al. Mutations in ERCC4, encoding the DNA-repair endonuclease XPF, cause Fanconi anemia. Am J Hum Genet. 2013;92:800–6. doi: 10.1016/j.ajhg.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kashiyama K, et al. Malfunction of nuclease ERCC1-XPF results in diverse clinical manifestations and causes Cockayne syndrome, xeroderma pigmentosum, and Fanconi anemia. Am J Hum Genet. 2013;92:807–19. doi: 10.1016/j.ajhg.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaarsma D, van der Pluijm I, van der Horst GT, Hoeijmakers JH. Cockayne syndrome pathogenesis: lessons from mouse models. Mech Ageing Dev. 2013;134:180–95. doi: 10.1016/j.mad.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Wilson BT, et al. The Cockayne Syndrome Natural History (CoSyNH) study: clinical findings in 102 individuals and recommendations for care. Genet Med. 2016;18:483–93. doi: 10.1038/gim.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartke A, Sun LY, Longo V. Somatotropic signaling: trade-offs between growth, reproductive development, and longevity. Physiol Rev. 2013;93:571–98. doi: 10.1152/physrev.00006.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gates KS. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol. 2009;22:1747–60. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swenberg JA, et al. Endogenous versus exogenous DNA adducts: their role in carcinogenesis, epidemiology, and risk assessment. Toxicol Sci. 2011;120(Suppl 1):S130–45. doi: 10.1093/toxsci/kfq371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Borgesius NZ, et al. Accelerated age-related cognitive decline and neurodegeneration, caused by deficient DNA repair. J Neurosci. 2011;31:12543–53. doi: 10.1523/JNEUROSCI.1589-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raj DD, et al. Priming of microglia in a DNA-repair deficient model of accelerated aging. Neurobiol Aging. 2014;35:2147–60. doi: 10.1016/j.neurobiolaging.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 29.Stranahan AM, Mattson MP. Metabolic reserve as a determinant of cognitive aging. J Alzheimers Dis. 2012;30(Suppl 2):S5–13. doi: 10.3233/JAD-2011-110899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halagappa VK, et al. Intermittent fasting and caloric restriction ameliorate age-related behavioral deficits in the triple-transgenic mouse model of Alzheimer’s disease. Neurobiol Dis. 2007;26:212–20. doi: 10.1016/j.nbd.2006.12.019. [DOI] [PubMed] [Google Scholar]
- 31.Duncan AW, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–10. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McWhir J, Selfridge J, Harrison DJ, Squires S, Melton DW. Mice with DNA repair gene (ERCC-1) deficiency have elevated levels of p53, liver nuclear abnormalities and die before weaning. Nat Genet. 1993;5:217–24. doi: 10.1038/ng1193-217. [DOI] [PubMed] [Google Scholar]
- 33.Waarsing JH, Day JS, Weinans H. An improved segmentation method for in vivo microCT imaging. J Bone Miner Res. 2004;19:1640–50. doi: 10.1359/JBMR.040705. [DOI] [PubMed] [Google Scholar]
- 34.Botter SM, et al. ADAMTS5−/− mice have less subchondral bone changes after induction of osteoarthritis through surgical instability: implications for a link between cartilage and subchondral bone changes. Osteoarthritis Cartilage. 2009;17:636–45. doi: 10.1016/j.joca.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 35.Durik M, et al. Nucleotide excision DNA repair is associated with age-related vascular dysfunction. Circulation. 2012;126:468–78. doi: 10.1161/CIRCULATIONAHA.112.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Derks KW, et al. Deciphering the RNA landscape by RNAome sequencing. RNA biol. 2015;12:30–42. doi: 10.1080/15476286.2015.1017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ringner M. What is principal component analysis? Nat Biotechnol. 2008;26:303–4. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 38.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 39.Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc. 2009;4:1184–91. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonker MJ, et al. Life spanning murine gene expression profiles in relation to chronological and pathological aging in multiple organs. Aging Cell. 2013;12:901–9. doi: 10.1111/acel.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Graaf EL, et al. Spatio-temporal analysis of molecular determinants of neuronal degeneration in the aging mouse cerebellum. Mol Cell Proteomics. 2013;12:1350–62. doi: 10.1074/mcp.M112.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Navarro F, Lieberman J. miR-34 and p53: New Insights into a Complex Functional Relationship. PLoS One. 2015;10:e0132767. doi: 10.1371/journal.pone.0132767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swindell WR. Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics. 2009;10:585. doi: 10.1186/1471-2164-10-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Campisi J, Robert L. Cell senescence: role in aging and age-related diseases. Interdiscip Top Gerontol. 2014;39:45–61. doi: 10.1159/000358899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Coppe JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.