Abstract
In the past few years, the number of autosomal DNA sequences from human fossils has grown explosively and numerous partial or complete sequences are available from our closest relatives, Neanderthal and Denisovans. I review commonly used statistical methods applied to these sequences. These methods fall into three broad classes: methods for estimating levels of contamination, descriptive methods, and methods based on population genetic models. The latter two classes are largely methods developed for the analysis of present-day genomic data. When they are applied to ancient DNA (aDNA), they usually ignore the time dimension. A few methods, particularly those concerned with inferring something about selection or ancestor-descendant relationships, take explicit account of the ages of aDNA samples.
Keywords: Population genetics, population genomics, principal components analysis, D-statistics
Although mitochondrial DNA (mtDNA) sequences have been obtained from human fossils since the 1980s and from Neanderthals since 1997, only since 2010 have nuclear DNA sequences been obtained in any abundance. A variety of statistical methods have been applied to hominin aDNA but only a few are widely used.
“Model free” methods
Some methods of analyzing aDNA data are purely descriptive. Principal components analysis (PCA) is the most commonly used. PCA assumes nothing about population genetics and is in that sense model free. Other methods (notably D- and F-statistics), make some assumptions about population genetics but do not yield estimates of population genetic parameters. They are used primarily to test for the occurrence of admixture. Other methods discussed later are based on population genetic models and estimate parameters of those models.
Principal components analysis
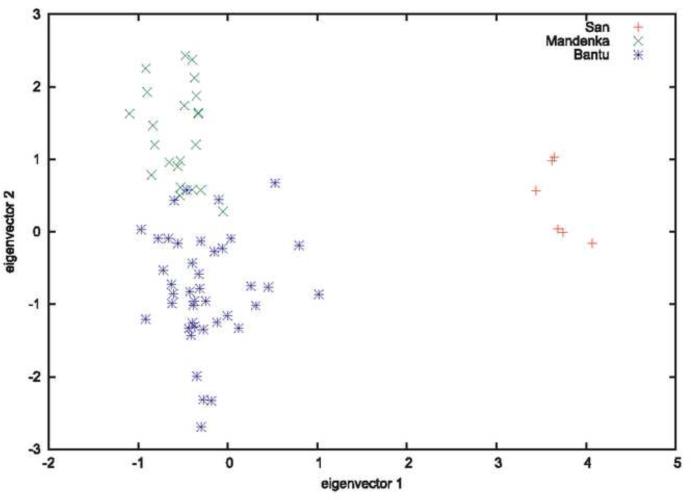
In population genetics, PCA is usually performed by finding the eigenvalues and eigenvectors of the matrix of covariances of allele frequencies between all pairs of individuals. [1] The highest order eigenvectors indicate the directions in the high dimensional allele-frequency space which account for most of the covariance. Often the individuals are plotted on the plane spanned by the first two eigenvectors. An example is shown in Figure 1. Relative distances in this space indicate their overall similarity. PCA is convenient because it is both easy to use and may provide a visually compelling result. For example, in an analysis of European populations, Novembre et al. [2] found a close correspondence between the geographic locations of individuals and their locations in principal components space.
Figure 1.
Example of principle components analysis. Plots of the first two eigenvectors for some African populations in the CEPH–HGDP dataset. (Figure 4 from reference [1]).
Ancient DNA sample are routinely combined with present-day samples in PCA. Usually the goal is to determine which present-day population an ancient sample is closest to. Drawing inferences about the relationship between the ancient and present-day samples requires additional assumptions. Skoglund et al. [3] showed that time differences in samples can be reflected in one of the principal component axes. Duforet-Frebourg and I [4] found that the sufficient migration between the time the ancient sample was taken and the present day can cause the ancient sample to not cluster with its present-day counterpart.
D-statistics
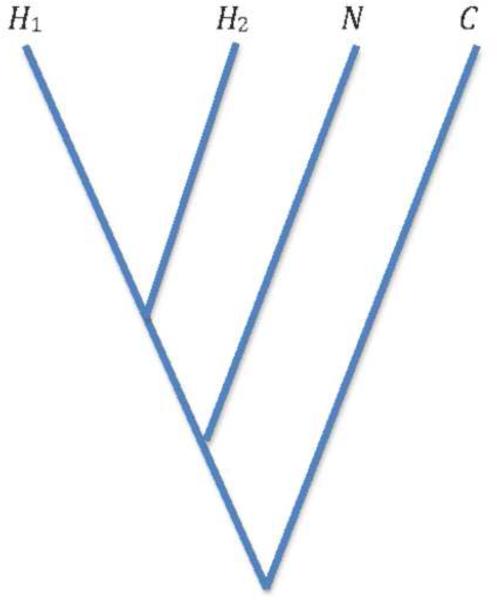
D-statistics are unusual in that they were first used with aDNA and have since been applied to present-day samples. They provided key evidence that Neanderthals and modern humans admixed. [5] D-statistics are computed for sets of four samples whose relationships are represented by a tree as shown in Figure 2. For analyzing Neanderthal admixture, H1 and H2 are two different present-day human genomes, N is the Neanderthal genome and C is the chimp genome. At each site, one nucleotide is chosen randomly from each genome and counts are made of sites at which both C and N differ and H1 and H2 differ. (H1, H2,N,C) is the percentage of sites at which N and H2 share alleles. If there had been no admixture between N and both H1 and H2, then D(H1, H2,N,C) is expected to be 0. If there is admixture between N and H2, then D will be positive.
Figure 2.
Illustration of population tree. In the application of D-statistics to Neanderthals, H1 and H2 were two different genomes from human populations (e. g. French and Yoruban), N was the Neanderthal, and C was the reference chimpanzee.
D provides a sensitive test for admixture because of the symmetry inherent in the population tree if there is no admixture. The symmetry takes account of incomplete lineage sorting provided the ancestral population has no geographic substructure. Furthermore, because sites are counted only if two copies of both alleles are seen, D is relatively insensitive to different levels of sequencing error in different genomes. [6] That is important because, if one of the genomes is from a fossil, its error rate is likely to be higher. In fact, in general it is unwise to assume the same error rate for sequences obtained in different laboratories using different sequencing platforms.. D does not directly estimate the admixture rate unless detailed assumptions are made about the branch lengths of the population tree, the time of admixture and the history of population sizes. [6]
F-statistics
F-statistics, which are closely related to D, were introduced by Patterson et al. [7]. Like D, F-statistics are used primarily to test for admixture. They are somewhat different from Wright’s F-statistics (FIS, FST etc.) [8]. Here, the term F-statistics will be used only for those defined by Patterson et al. There are three F-statistics, denoted by F2(P1, P2), F3(P1; P2, P3) and F4((P1, P2,; P3, P4) where the P’s are the populations being compared. These statistics are designed to quantify the amount of genetic drift on different parts of a population tree. Because they focus on genetic drift and not mutation they assume that the all alleles are present in an ancestral population. That constraint is satisfied if all sites analyzed were determined to be polymorphic in a population that is an outgroup to those analyzed. For human populations, that condition cannot be satisfied in most cases. In practice, F-statistics are applied to both SNP array data and whole genome data without taking account of the effect of new mutations. Patterson et al. [7] found by simulation that in some cases, mutation does not affect conclusions drawn.
F2 is defined to be the average across loci of (p1 − p2) where p1 and p2 are the frequencies of a diallelic SNP in P1 and P2. F2 is additive along branches of a tree. F2 measures the time separating P1 and P2 in units of the effective size of the intermediate population. Mutation will change this interpretation somewhat because the influx of variants by mutation depends on the history of population sizes, not merely the overall effective size.
F3 (P1;P2,P3) is the average of (p1 − p2)(p1 − p3) across sites. It will be positive for any three populations that have a tree-like history—for example H1, H2 and N in Figure 1—but it can be negative if P1 is the product of admixture between P2 and P3. This statistic is often used as a test for admixture. F4(P1,P2;P3,P4) is the average of (p1 − p2)(p3 − p4) across sites. It quantifies the covariance between the accumulated differences between P1 and P2 and between P3 and P4. If these two pairs do not share a branch of a population tree, then the covariance will be zero, a conclusion that is unaffected by incomplete lineage sorting. F4(P1,P2;P3,P4) can be regarded as the numerator of D(P1,P2,P3,P4) . Using F4 to test for admixture is equivalent to Cavalli-Sforza and Piazza’s [9] test of treeness.
Linkage disequilibrium and introgressed fragments
Admixture from one population to another results in the production of hybrid offspring that carry one parental chromosome from each population. Subsequent interbreeding and recombination break down introgressed chromosomes into shorter fragments. Several methods have used this process both to provide evidence of admixture and to estimate the time when admixture took place. There are two classes of methods. One uses the extent of linkage disequilibrium between pairs of sites. The method rolloff [7] is of this type. Sankararaman et al. [10] used a similar method to estimate the date of admixture from Neanderthals into human populations.
The other class of methods uses the lengths of fragments that derive from another population. These methods have been particularly useful for characterizing admixture from Neanderthals into humans because Neanderthal and humans differ sufficiently that identifying introgressed Neanderthal fragments is relatively easy. Several groups have taken this approach. [11-15]
Population genetic models
George Box’s dictum "All models are wrong but some are useful"[16] applies with special force in population genetics. It is hopeless to think that the complexity of human history can be represented by any model. Yet, the right model can capture the essential events that account for broad patterns in genomic data. Many computer-intensive methods have been developed to estimate model parameters, among others dadi [17], fastsimcoal2 [18], and G-PhoCS [19]. None of these widely used programs is tailored to aDNA. When they are used with ancient samples, the effective size in the branch leading to the ancient sample is reduced to reflect the shorter time available for genetic drift to act.
TreeMix
Pickrell and Pritchard [20] developed the program TreeMix to both test for whether a set of samples fits a population tree and, if they do not, which admixture events were mostly likely to have occurred. TreeMix has been widely applied to aDNA samples combined with present-day samples because, although it is model-based, it does not require an assumption-rich model. It begins by assuming that populations sampled have a history represented by a population tree. It then tests the tree hypothesis against alternative models that allow for admixture between different branches of the tree. It adds admixture events until the data are adequately explained. TreeMix achieves its computational speed by approximating the process of genetic drift by a Gaussian random walk.
Inferring the history of population sizes
The demographic history of a population determines the distribution of pairwise coalescence times in a population. This fact has been exploited by several programs that reconstruct the history of population size by estimating the coalescent time distribution. If a genome is sequenced to sufficient depth that heterozygotes can be identified with high confidence, even a single genomic sequence can be used. Li and Durbin [21] developed PSMC (for pairwise sequentially Markovian coalescent), a model that efficiently estimates past population sizes of a single population. Refinements of this type of method have been developed by Sheehan et al. [22] and Schiffels and Durbin [23]. PSMC is popular for the same reasons that TreeMix is. Although PSMC is based on a model of genetic drift, it makes few assumptions because it needs to model only the population ancestral to the population sampled.
Testing for direct ancestry
Rasmussen et al. [24] developed a test for direct ancestry of an aDNA sequence that is essentially model-free, even though it is presented in the context of coalescent theory. The test is especially convenient because it is applicable to pairs of sequences, one present-day and the other ancient. It is a likelihood ratio test of whether the length of the branch leading to the aDNA sequence is zero. If the branch is non-zero in length, then a maximum likelihood estimate of the relative branch lengths scaled by effective population sizes is obtained.
If aDNA samples are of different age are available, the method of projections [25] can be used to test whether those samples come from a lineage directly ancestral to a present-day population. [26]
Contamination
A major problem with analyzing hominin aDNA is contamination by present-day humans. Based only on the DNA sequences themselves, contaminating reads are difficult to distinguish from those endogenous to the source of the aDNA. For Neanderthals and Denisovans, the mtDNA sequences are sufficiently different that contaminating human mtDNA is easily detected. It is possible, however, that levels of mtDNA and nuclear DNA contamination differ. If the archaic individual was a female, the extent of contamination is indicated by the amount of Y-chromosome DNA detected. It is also possible to directly estimate the level of nuclear contamination from the presence of heterozygous aDNA sites [5,27].
Skoglund et al. [28] developed a method that uses the tendency of aDNA fragments to carry distinctive miscoding lesions to estimate the probability that each fragment is endogenous. Racimo et al. [29] took a different approach and used an explicit model of the archaic and present-day populations to jointly estimate the level of contamination of the archaic sample and the time of divergence of archaic and contaminating populations. Applying this method to different potentially contaminating populations allows identification of the source of contamination with some confidence.
Time sequence of samples
When aDNA is available from samples of different age, it is possible to approximate the changes in allele frequency of individual loci [30,31]. Several method have been developed both to test for selection and to estimate selection intensity. [32-35] As increasing numbers of aDNA is sequenced, more data will be available for the application of these methods. These methods all assume that the ancient and present-day DNA all come from the same population lineage, something that may be difficult to test for.
Branch shortening
An aDNA sequence is from a population lineage that is shorter than the lineage leading to any present-day sample. As a consequence, the aDNA sequence has had somewhat less time to accumulate mutations, a phenomenon called branch shortening. If the error rate in the aDNA sequence is low enough, branch shortening can be detected. In fact, detecting branch shortening is another way to verify that the aDNA sequence is endogenous. If the age of the fossil yielding the aDNA sequence is known, then the extent of branch shortening provides an estimate of mutation rates. Fu et al. [12] were able to estimate nuclear, Y-chromosome and mtDNA mutation rates using this method.
Discussion
Methods for analyzing aDNA data will flourish as more aDNA data become available. Methods will focus increasingly on whole-genome sequences. Although many methods for analyzing present-day sequences have been applied to aDNA, they do not take account of the additional complexity that aDNA creates. Sequence quality may be lower both because only low coverage may be obtained and because aDNA is subject to post-mortem modification. Furthermore, the age difference between ancient and present-day sequences may not be adequately taken account of. Analyzing aDNA gives access to a time dimension which had been previously been unseen. New methods will have to take better advantage of the ages of aDNA sequences.
One shortcoming of most analysis aDNA samples is shared by most current analysis of present-day samples. The starting point is a population tree, possibly with some admixture represented by connections among branches. This is convenient for modelers because samples are usually identified with named populations and most population genetics theory is developed in terms of randomly mating populations. Yet human populations are dispersed widely. True population distinctness may be rare even for island populations. The assumption of a population tree with or without admixture may be a reasonable starting point but little effort has been expended on testing the adequacy of that modeling framework. Unfortunately, accounting for the real geographic structure of gene flow and isolation-by-distance is difficult and taking account of spatial variation in population density adds to the difficulty. At present, we do not know what effect complexities in population structure might have on conclusions drawn from tree-based models. Future modelers will have to take up this challenge sooner or later.
Acknowledgements
This work was supported in part by NIH grant R01-GM40282.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1**.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genetics. 2006;2:2074–2093. doi: 10.1371/journal.pgen.0020190. This paper introduced principal components analysis to the population genetics community and presented a widely used program for carrying out that analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, Auton A, Indap A, King KS, Bergmann S, Nelson MR, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skoglund P, Sjödin P, Skoglund T, Lascoux M, Jakobsson M. Investigating Population History Using Temporal Genetic Differentiation. Molecular Biology and Evolution. 2014;31:2516–2527. doi: 10.1093/molbev/msu192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duforet-Frebourg N, Slatkin M. Isolation-by-distance-and-time in a stepping-stone model. Theoretical Population Biology. 2016;108:24–35. doi: 10.1016/j.tpb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5**.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. Several methods including D-statistics were used to provide evidence of Neanderthal admixture in the to ancestors of non-African human populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durand EY, Patterson N, Reich D, Slatkin M. Testing for ancient admixture between closely related populations. Molecular Biology and Evolution. 2011;28:2231–2237. doi: 10.1093/molbev/msr048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7**.Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human history. Genetics. 2012;192:1065–1093. doi: 10.1534/genetics.112.145037. This paper introduces several statistics that have been widely applied to ancient and present-day DNA and relates those statistics to population genetic processes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright S. The genetical structure of populations. Annals of Eugenics. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- 9.Cavalli-Sforza LL, Piazza A. Analysis of evolution: evolutionary rates, independence and treeness. Theoretical Population Biology. 1975;8:127–165. doi: 10.1016/0040-5809(75)90029-5. [DOI] [PubMed] [Google Scholar]
- 10.Sankararaman S, Patterson N, Li H, Pääbo S, Reich D. The Date of interbreeding between Neandertals and Modern Humans. PLoS Genetics. 2012;8:e1002947. doi: 10.1371/journal.pgen.1002947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wall JD, Yang MA, Jay F, Kim SK, Durand EY, Stevison LS, Gignoux C, Woerner A, Hammer MF, Slatkin M. Higher levels of Neanderthal ancestry in East Asians than in Europeans. Genetics. 2013;194:199–209. doi: 10.1534/genetics.112.148213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Q, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, Johnson PLF, Aximu-Petri A, Prufer K, de Filippo C, et al. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature. 2014;514:445–449. doi: 10.1038/nature13810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Q, Hajdinjak M, Moldovan OT, Constantin S, Mallick S, Skoglund P, Patterson N, Rohland N, Lazaridis I, Nickel B, et al. An early modern human from Romania with a recent Neanderthal ancestor. Nature. 2015;524:216–219. doi: 10.1038/nature14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vernot B, Akey JM. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 15.Sankararaman S, Mallick S, Dannemann M, Prufer K, Kelso J, Paabo S, Patterson N, Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Box GEP . Robustness in the strategy of scientific model building. In: Launer RL, Wilkinson GN, editors. Workshop sponsored by the Mathematics Division, Army Research Office, held at Army Research Office, Weiss Building. Vol. 1979. Apr 11-12, 1978. pp. 201–236. [Google Scholar]
- 17.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genetics. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Excoffier L, Dupanloup I, Huerta-Sanchez E, Sousa VC, Foll M. Robust demographic dnference from genomic and SNP data. PLoS Genetics. 2013;9:e1003905. doi: 10.1371/journal.pgen.1003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gronau I, Hubisz MJ, Gulko B, Danko CG, Siepel A. Bayesian inference of ancient human demography from individual genome sequences. Nature Genetics. 2011;43:1031–1034. doi: 10.1038/ng.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genetics. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. This paper introduced the program TreeMix which has become widely applied to aDNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. The program PSMC described in this paper has widely used to infer the history of population sizes from single high-coverage genomic sequences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheehan S, Harris K, Song YS. Estimating Variable Effective Population Sizes from Multiple Genomes: A Sequentially Markov Conditional Sampling Distribution Approach. Genetics. 2013;194:647–662. doi: 10.1534/genetics.112.149096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nature Genetics. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rasmussen M, Anzick SL, Waters MR, Skoglund P, DeGiorgio M, Stafford TW, Jr, Rasmussen S, Moltke I, Albrechtsen A, Doyle SM, et al. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature. 2014;506:225–229. doi: 10.1038/nature13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang MA, Harris K, Slatkin M. The projection of a test genome onto a reference population and applications to humans and archaic hominins. Genetics. 2014;198:1655–1670. doi: 10.1534/genetics.112.145359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang MA, Slatkin M. Using ancient samples in projection analysis. G3: Genes|Genomes|Genetics. 2016;6:99–105. doi: 10.1534/g3.115.023788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28**.Skoglund P, Northoff BH, Shunkov MV, Derevianko AP, Pääbo S, Krause J, Jakobsson M. Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proceedings of the National Academy of Sciences. 2014;111:2229–2234. doi: 10.1073/pnas.1318934111. This paper presents a useful method for enriching a sample for endogenous sequences. It will be especially useful when only low coverage or partial sequences can be obtained. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Racimo F, Renaud G, Slatkin M. Joint estimation of contamination, error and demography for nuclear DNA from ancient humans. PLoS Genetics. 2016;12:e1005972. doi: 10.1371/journal.pgen.1005972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig A, Pruvost M, Reissmann M, Benecke N, Brockmann GA, Castanos P, Cieslak M, Lippold S, Llorente L, Malaspinas A-S, et al. Coat Color Variation at the Beginning of Horse Domestication. Science. 2009;324:485. doi: 10.1126/science.1172750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathieson I, Lazaridis I, Rohland N, Mallick S, Llamas B, Pickrell J, Meller H, Rojo Guerra MA, Krause J, Anthony D, et al. Genome-wide patterns of selection in 230 ancient Eurasians. Nature. 2015;528:499–503. doi: 10.1038/nature16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bollback JP, York TL, Nielsen R. Estimation of 2Nes from temporal allele frequency data. Genetics. 2008;179:497–502. doi: 10.1534/genetics.107.085019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malaspinas A-S, Malaspinas O, Evans SN, Slatkin M. Estimating Allele Age and Selection Coefficient from Time-Serial Data. Genetics. 2012;192:599–607. doi: 10.1534/genetics.112.140939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Terhorst J, Schlötterer C, Song YS. Multi-locus analysis of genomic time Series data from experimental evolution. PLoS Genetics. 2015;11:e1005069. doi: 10.1371/journal.pgen.1005069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schraiber JG, Evans SN, Slatkin M. Bayesian inference of natural selection from allele frequency time series. Genetics. 2016;203:493–511. doi: 10.1534/genetics.116.187278. [DOI] [PMC free article] [PubMed] [Google Scholar]