Abstract
Ubiquitylation of histone H2B, associated with gene activation, leads to chromatin decompaction through an unknown mechanism. We used a hydrogen-deuterium exchange strategy coupled with nuclear magnetic resonance spectroscopy to map the ubiquitin surface responsible for its structural effects on chromatin. Our studies revealed that a previously uncharacterized acidic patch on ubiquitin comprising residues Glu16 and Glu18 is essential for decompaction. These residues mediate promiscuous electrostatic interactions with the basic histone proteins, potentially positioning the ubiquitin moiety as a dynamic “wedge” that prevents the intimate association of neighboring nucleosomes. Using two independent cross-linking strategies and an oligomerization assay, we also showed that ubiquitin-ubiquitin contacts occur in the chromatin environment and are important for the solubilization of the chromatin polymers. Our work highlights a novel, chromatin-related aspect of the “ubiquitin code”, and sheds light on how the information rich ubiquitin modification can orchestrate different biochemical outcomes using different surface features.
Genomic DNA in eukaryotic cells is tightly packaged with proteins to form a polymer structure known as chromatin. The repeating unit of chromatin is the nucleosome, which consists of 147 DNA base pairs wrapped around a protein core containing two copies of the histones H2A, H2B, H3 and H4. The histone proteins are dynamic hubs of post-translational modifications (PTMs) that control the physical access to DNA either directly, by disrupting or creating contacts within the nucleo-protein complex, or indirectly through the localized recruitment of effector proteins1. While most histone PTMs (e.g. acetylation, methylation or phosphorylation) result in the addition of a small chemical functional group, ubiquitin is unique in that it is a stable 76-residue protein and its attachment to the nucleosome provides significant steric bulk and a large surface area (~ 4800 Å2) with distinct properties2. These properties include a network of seven lysine residues that serve as anchoring points in the formation of polyubiquitin chains, as well as two hydrophobic patches centered at Ile36 and Ile44, essential for ubiquitin’s other cellular functions such as proteasomal degradation and protein localization2–4. In chromatin, depending on the attachment site, monoubiquitylation can be associated with gene repression5–7 (H2A Lys119), or transcription activation and elongation (H2B Lys120) through dynamic cross-talks that also involve methylation of histone H38–12. The latter function of ubiquitin is mediated by a surface hotspot comprising Leu71 and Leu73 that interacts with the N-terminus of histone H2A, thus orienting the ubiquitin moiety and potentially “corralling” the H3 methyltransferase into a productive binding mode with the nucleosome13,14. H2B ubiquitylation also increases access to DNA by directly disrupting local and higher order chromatin compaction15,16. While it is clear that this effect is not just due to steric bulk, as Hub1, a protein of similar size and fold, does not have the same structural effect15, the specific interactions responsible for decompaction have so far remained elusive.
Unraveling the molecular mechanisms underlying the regulation of chromatin structure by histone PTMs such as ubiquitin remains a tremendously challenging task since it requires access to post-translationally modified substrates that extend beyond the mononucleosome, and structural methods that can tackle the inherent dynamics and complexity of the chromatin polymer. Here we exploited the residue-level information afforded by hydrogen-deuterium (H/D) exchange in conjunction with nuclear magnetic resonance (NMR) spectroscopy17–19 to map the surfaces of ubiquitin responsible for its effect on chromatin. These studies were performed in the context of segmentally isotopically labeled 12-mer nucleosome arrays that reconstructed the chromatin environment and the spatial relationships between neighboring nucleosomes. Guided by the NMR data, we then performed a series of biochemical and biophysical experiments that collectively led to an electrostatic model for how ubiquitin impedes the formation of higher-order chromatin structure. We found that a small acidic patch on the ubiquitin surface is central to this model and that ubiquitin-ubiquitin contacts play an important role in the dissociation of chromatin fibers. We imagine that the structural approaches developed herein will find application in the study of other large ubiquitylated protein assemblies.
RESULTS
Ubiquitin interaction surfaces identified by NMR
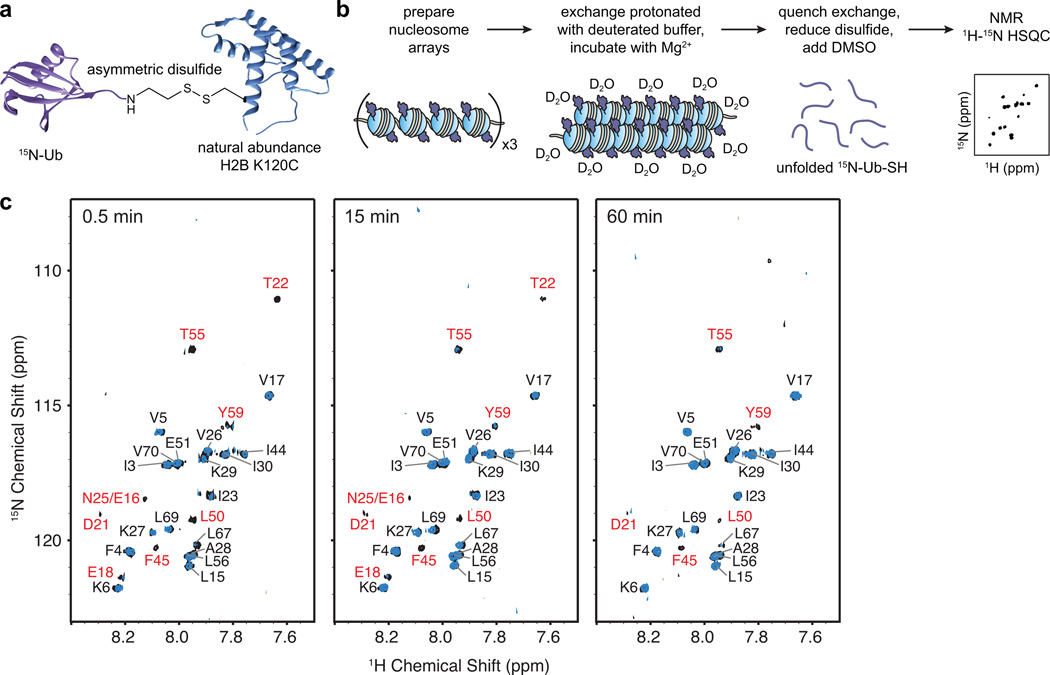
Since nucleosome arrays are a far too large a substrate for analysis by solution NMR, we adopted an approach inspired by previous studies on large biological assemblies, which involves H/D exchange in the native state, a quenching step and subsequent transfer of the exchange information to an NMR-accessible denatured protein state in a minimum exchange aprotic solvent such as dimethyl sulfoxide (DMSO)20–22. This method was enabled by a robust and efficient protocol to prepare large amounts (tens of mg) of semi-synthetic ubiquitylated H2B (H2B-Ub)23. To this end, we prepared uniformly 15N-labeled ubiquitin fused to the DnaE intein Ava24, which facilitated the addition of an aminoethanethiol moiety to the C-terminus of the protein (15N-Ub-SH). Separately, we prepared unlabeled H2B with a cysteine at position 120, and activated this residue with 2,2’-dithiobis(5-nitropyridine). Incubating the two proteins at near neutral pH ensured the selective and efficient formation of a disulfide bond between the C-terminus of ubiquitin and Cys120, thus generating a segmentally isotopically labeled version of H2B-Ub (Fig. 1a, Supplementary Results, Supplementary Fig. 1 and 2). This engineered linkage allows the synthesis of H2B-Ub on a large scale without compromising its biological activity13–15,23.
Figure 1.
Experimental strategy to detect ubiquitin interaction surfaces in nucleosome arrays. (a) Segmentally labeled H2B-15N-Ub synthesized using an asymmetric disulfide approach. (b) Schematic depicting the steps of the H/D exchange NMR experiment used to probe ubiquitin interactions in arrays. (c) 1H-15N HSQC spectra of nucleosome arrays incorporating H2B-15N-Ub incubated in deuterated buffer for 0.5, 15 and 60 min. Cross-peaks from experiments performed with H2B-Ub arrays are depicted in black, while cross-peaks from control Ub experiments are shown in blue. Resonance assignments for array-specific cross-peaks are labeled in red. Contour levels were set to 5×σnoise where σnoise is the average noise level of each spectrum.
We then incorporated segmentally labeled H2B-Ub into histone octamers and nucleosome arrays using well-established protocols25 (Supplementary Fig. 3a). To initiate exchange of the amide protons, we placed the ubiquitylated nucleosome arrays into deuterated buffer, and added Mg2+ to the solution to promote a dynamic chromatin folding state (Fig. 1b). We expected that under such conditions, the chromatin compaction favored by the presence of Mg2+ would be counterbalanced by the decompaction activity of ubiquitin, and the ubiquitin residues involved in the attendant interactions would exhibit slower amide H/D exchange. After incubation, we quenched the H/D exchange and dissolved the samples in a DMSO based solution optimized for minimum exchange during the acquisition of 2D NMR HSQC spectra20. Since H2B-Ub is a 22 kDa species that tumbles relatively slowly, we increased the sensitivity of the HSQC spectra by reducing the disulfide linkage between the ubiquitin moiety and the histone (Supplementary Fig. 3b). Thus, fully reduced, isotopically labeled 15N-Ub-SH was the only species visible in the NMR experiments. Since slower amide exchange rates may result from protein-protein interactions or participation in stable structural elements in the native ubiquitin fold, we performed the same experiment with folded ubiquitin that was not incorporated into nucleosome arrays (control Ub) and superimposed the HSQC spectra of the array and control samples (Fig. 1c). To account for the influence of experimental conditions (solvent, salts, pH), we performed de novo assignments of the HSQC cross-peaks (Supplementary Fig. 4, Supplementary Table 1).
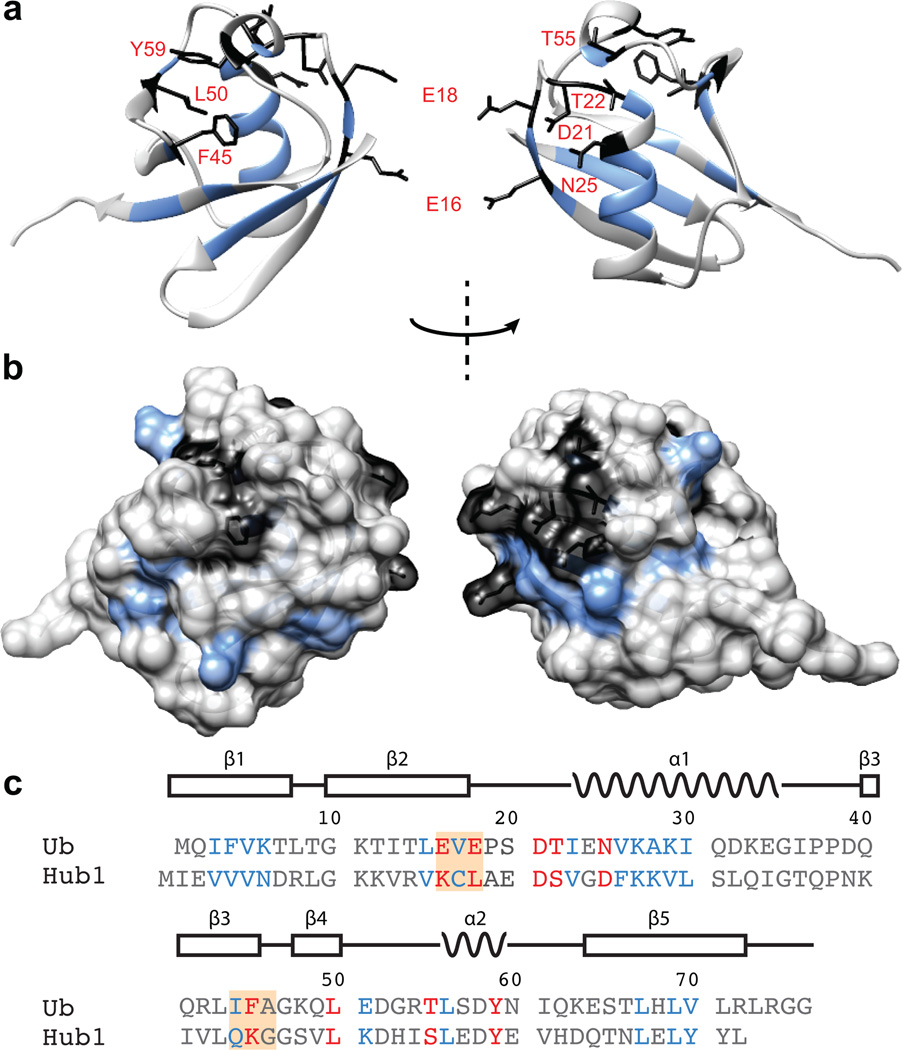
A comparison of the array and control Ub samples at the shortest exchange time (Figure 1c, Supplementary Fig. 5) revealed eighteen ubiquitin cross-peaks that appeared in both spectra, and eight cross-peaks that were unique for the array-derived sample. The common set of cross-peaks reflects residues that are part of the hydrophobic core of ubiquitin, with many previously reported to display slower exchange rate under various conditions26,27. Therefore, these cross-peaks most likely result from stable secondary and tertiary interactions inherent to the ubiquitin fold. The array-specific set of cross-peaks maps to a continuous surface on ubiquitin (Fig. 2a,b) on the side opposite to the C-terminus (i.e. the attachment site to H2B). This surface comprises a hydrophobic patch formed by Phe45, Leu50, and Tyr59, as well as a network of charged residues including Thr55, Thr22, Asp21, Asn 25, Glu18, and Glu16 that are part of loops and some secondary structural elements but have side chains oriented towards the solvent. To verify that the H/D exchange protocol detects unique and meaningful structural interactions, we performed the same experiment with arrays containing 15N labeled ubiquitin incorporated at the other biologically relevant histone site, namely Lys119 on H2A (H2A-Ub) (Supplementary Fig. 6). The HSQC spectra from these experiments revealed a much larger set of protected amide protons, including some arising from a surface closer to the attachment site (Supplementary Fig. 7–9). Thus, H2A-Ub and H2B-Ub experience different environments in chromatin arrays, and the detected surfaces are presumably involved in specific molecular interactions.
Figure 2.
Ubiquitin residues protected from backbone H/D exchange in nucleosome arrays. (a) Residues unique for the array sample are shown in black, while residues common for both the array and ubiquitin only control samples are depicted in blue. The structure represents the ubiquitin fold42 (PDB ID: 1UBQ). (b) Surface representation of the residues depicted in (a). (c) Sequence alignment of ubiquitin and Hub1. Residues labeled in red are uniquely protected from H/D exchange in the array samples, while the residues labeled in blue are protected in both array and control samples. The orange boxes represent residues chosen for alanine mutagenesis. (a–c) Residues colored in grey undergo rapid H/D exchange and are not detectable in the NMR spectra.
Glutamate patch impairs chromatin compaction
Comparison of the ubiquitin and Hub1 sequences (Fig. 2c) revealed that three of the array specific protected residues differ substantially between the two proteins. The negative charges of the Glu16 and Glu18 side-chains have been replaced with a positively charged lysine and an aliphatic leucine side-chain respectively, thus imparting very different electrostatic properties at these positions (Supplementary Fig. 10). Additionally, Phe45 is substituted with a lysine residue, which presumably changes the structure of the hydrophobic pocket, which also includes Ile44, within this part of the protein fold. Therefore, we decided to prepare semi-synthetic chromatin with H2B-Ub containing alanine substitutions at either Glu16 and Glu18 (Ub1), or Ile44 and Phe45 (Ub2), and test whether fiber compaction is impaired (Supplementary Fig. 2). The use of double mutants was motivated by the hypothesis that a surface patch, and not a single residue, would be responsible for any potential interactions in the chromatin environment. Analysis by circular dichroism showed that both Ub1 and Ub2 preserved the global ubiquitin fold (Supplementary Fig. 11).
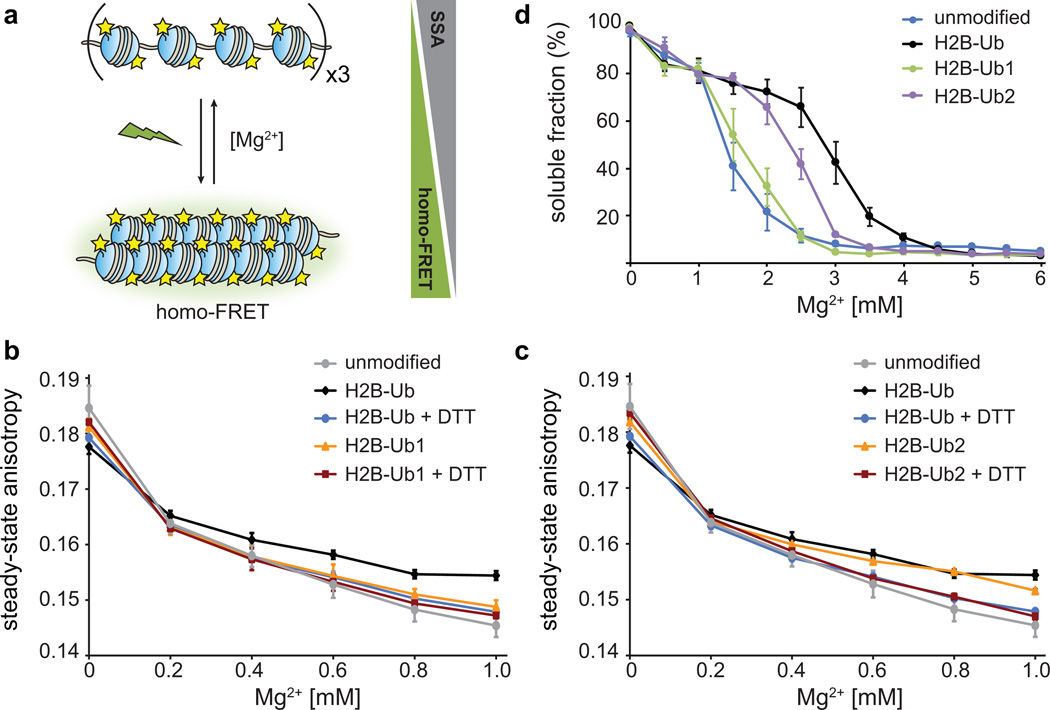
To decouple intra-fiber folding from fiber oligomerization, we took advantage of a previously published fluorescence based experiment that reports on the distances between fluorescent probes placed on each nucleosome in a chromatin array15 (Fig. 3a). Upon fiber compaction in low Mg2+ concentrations and excitation with polarized light, the neighboring fluorescent probes undergo homo-FRET, detected as a decrease in the steady-state anisotropy (SSA). The data presented in Fig. 3b,c compare the SSA curves for unmodified and ubiquitylated arrays, where the presence of H2B-Ub leads to a smaller change in SSA, and thus to a less compact ensemble of fiber conformations, as described previously15. Upon treatment with a reducing agent such as dithiothreitol (DTT), ubiquitin was detached from H2B, and the resulting arrays compacted similarly to the unmodified arrays, as expected. Performing this experiment with Ub1 and Ub2 revealed that the substitution of the two negatively charged residues in Ub1 with alanine abolished the ability of ubiquitin to decompact chromatin fibers, while the substitutions in Ub2 had no effect.
Figure 3.
Effect of ubiquitin mutations on intra- and inter-fiber compaction. (a) Homo-FRET based assay to detect intra-fiber compaction as a function of Mg2+. Each nucleosome contains fluorescein-labeled H2A, and upon compaction the fluorophores undergo homo-FRET detected as a decrease in the steady-state anisotropy. (b) Homo-FRET compaction assay of arrays containing H2B-Ub (black) and H2B-Ub1 (orange). Upon reduction of the asymmetric disulfide with dithiothreitol (DTT), the ubiquitin moiety is removed and the arrays compact similarly to unmodified nucleosome arrays (grey, blue and red). Error bars, s.d. (n = 3 independent array preparations) (c) Homo-FRET compaction assay of arrays containing H2B-Ub (black) and H2B-Ub2 (orange). Error bars, s.d. (n = 3 independent array preparations). (d) Oligomerization assay of unmodified nucleosome arrays (blue), and arrays containing H2B-Ub (black), H2B-Ub1 (green) and H2B-Ub2 (purple). The arrays were incubated with increasing amounts of Mg2+, the oligomers were removed by centrifugation, and the percentage of arrays remaining in solution was determined using SybrGold fluorescence. Error bars, s.e.m. (n = 3 – 6 independent array preparations). Ub1 designates ubiquitin with E16A and E18A mutations, and Ub2 contains I44A and F45A substitutions.
To probe the role of the ubiquitin surfaces in higher-order chromatin structure, as manifest by nucleosome array oligomerization, we carried out precipitation experiments15,28,29. We incubated nucleosome arrays with increasing concentrations of MgCl2, removed the precipitated material by centrifugation and quantified the presence of unassociated fibers in the supernatant (Fig. 3d). Similar to previous findings15,16, the introduction of wild-type ubiquitin into arrays hindered oligomerization. Arrays containing Ub2 displayed intermediate behavior, while H2B-Ub1 arrays oligomerized to the same extent as non-ubiquitylated fibers. Therefore, the ability of ubiquitin to hinder nucleosome fiber compaction and fiber association rests on the presence of Glu16 and/or Glu18. The canonical hydrophobic patch centered at Ile44 and Phe45 has no detectable effect on intra-fiber compaction, but does contribute, albeit modestly, to fiber oligomerization. The decompaction interactions mediated by the two glutamate residues are relevant under higher monovalent salt conditions (Supplementary Fig. 12a), and are specific for H2B ubiquitylation as alanine substitution at these positions in the context of H2A-Ub had no effect on fiber oligomerization (Supplementary Fig. 12b).
Ubiquitin decompacts through electrostatic interactions
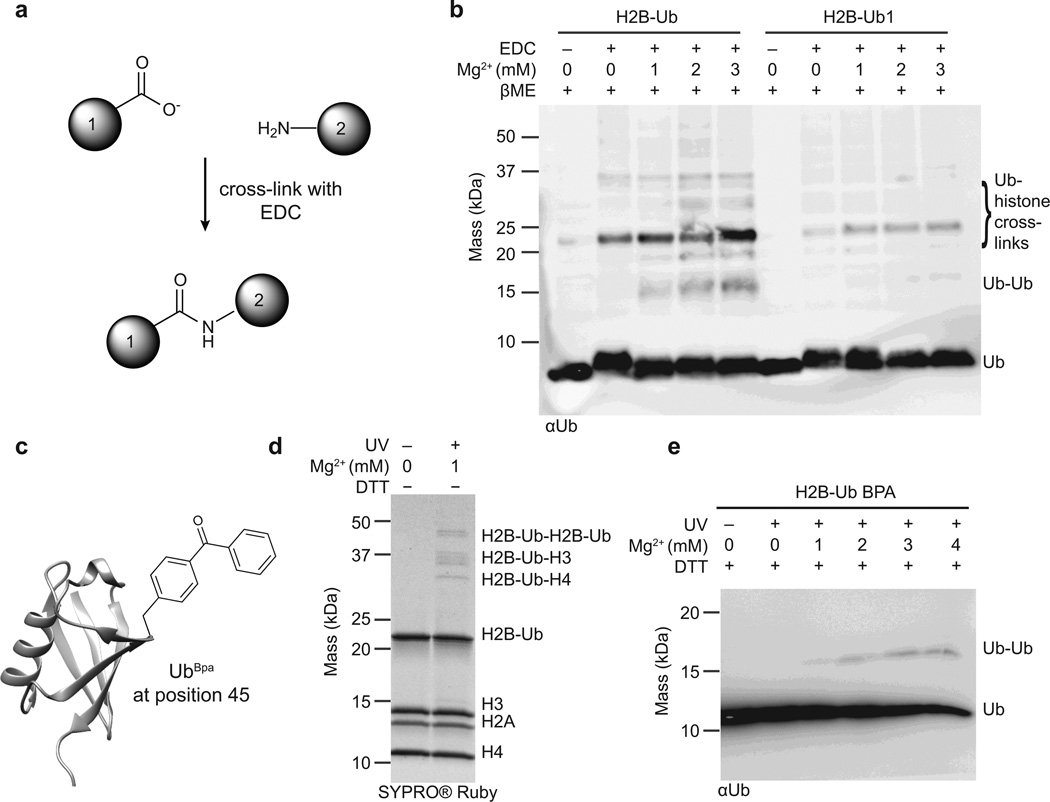
Having established the identity of the ubiquitin residues that impact chromatin fiber compaction, we next sought to determine their interaction partners in the nucleosome array environment. To this end, we relied on two types of cross-linking experiment, each designed to probe complementary aspects of ubiquitin-chromatin interactions. In the first approach, we used the zero-length cross-linker 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), a reagent that couples carboxyl groups to juxtaposed primary amines to yield amide bonds (Fig. 4a). While non-specific in nature, we hypothesized that this cross-linking strategy would be well-suited to inform on potential electrostatic interactions involving the carboxyl side-chain groups of Glu16 and Glu18, and nearby lysine residues in the chromatin environment. Therefore, we performed cross-linking experiments of H2B-Ub or H2B-Ub1 nucleosome arrays in the presence or absence of MgCl2, reduced the disulfide bond between ubiquitin and H2B, and employed Western blot analysis against ubiquitin to detect the corresponding interactions (Fig. 4b). Comparison of the H2B-Ub and H2B-Ub1 samples revealed a much stronger cross-linking pattern for the former, indicating that the presence of Glu16 and Glu18 promotes more promiscuous sampling of the chromatin environment, presumably driven by electrostatic interactions. Based on the size of the crosslinked bands, it is evident that ubiquitin interacts with both histones and other copies of itself. Notably, the ubiquitin-ubiquitin band was detected only in the H2B-Ub sample and was dependent on the presence of Mg2+, suggesting that ubiquitin-ubiquitin interactions are an important part of the chromatin decompaction mechanism.
Figure 4.
Ubiquitin-chromatin interactions probed with cross-linking experiments. (a) The zero-length cross-linker EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide) couples carboxyl groups to primary amines to form a stable amide bond. (b) EDC-based cross-linking of nucleosome arrays containing H2B-Ub and H2B-Ub1. Cross-linking was performed in the presence of increasing concentrations of Mg2+, and all samples were subsequently reduced with β-mercaptoethanol to detach ubiquitin and ubiquitin cross-linked species from H2B. Analysis was performed by SDS-PAGE followed by western blotting with an antibody against ubiquitin. (c) The site-specific cross-linker Bpa (p-benzoyl-phenylananine) was installed at position 45 on ubiquitin using amber codon suppression. (d) UV-induced cross-linking of H2B-UbBpa arrays in the presence of 1 mM Mg2+. The cross-linked bands were resolved by SDS-PAGE under non-reducing conditions. (e) UV-induced cross-linking of H2B-UbBpa arrays in the presence of increasing concentrations of Mg2+. Samples were reduced with 100 mM dithiothreitol, separated by SDS-PAGE, and analyzed by western blotting against ubiquitin. Representative data from three independent experiments are shown and full images are presented in Supplementary Fig. 15.
In the second cross-linking approach, we employed amber suppression methods to site-specifically incorporate the unnatural amino acid, p-benzoyl-L-phenylananine (Bpa), into the ubiquitin sequence30 (Fig. 4c). Irradiation of Bpa generates an excited triplet state that preferentially reacts with nearby C-H bonds31. Thus, we imagined that use of this cross-linker would provide independent and complementary information on ubiquitin-protein interactions in the chromatin setting. We substituted residue Phe45 in ubiquitin for Bpa (UbBpa), as this position was implicated in ubiquitin-chromatin interactions by H/D exchange/NMR and shown to play a role in fiber oligomerization (Fig. 1c and Fig. 3d). Moreover, this substitution is structurally conservative and was expected to have a minimal impact on ubiquitin function. We generated H2B-UbBpa in excellent yield using the disulfide strategy (Supplementary Fig. 13) and subsequently incorporated it into nucleosome arrays. Following UV-mediated cross-linking, we resolved the resulting interactions by SDS-PAGE, revealing a well-resolved cross-linked band pattern (Fig. 4d). The assignment of these bands (by size and Western blot analysis) confirmed the observation that ubiquitin interacts both with histone proteins, and with other ubiquitin moieties in a Mg2+-dependent fashion (Fig. 4e, Supplementary Fig. 14 and 15).
Ubiquitin-ubiquitin interactions block fiber association
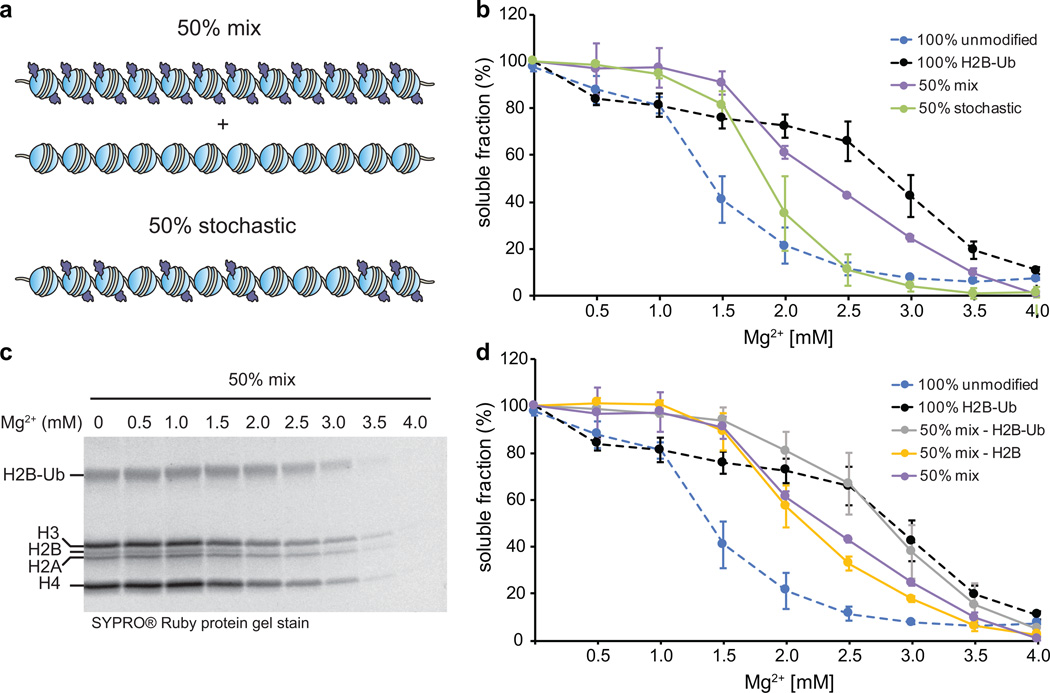
Next, we asked whether the ubiquitin-ubiquitin interactions observed in the cross-linking experiments were required for impairment of fiber oligomerization, and if so, whether intra- or inter-array contacts were more important. To explore these questions, we prepared and compared two array samples (Fig. 5a) – one containing a 50/50 mixture of unmodified and fully ubiquitylated arrays (50% mixed sample), and another containing a 50% stochastic distribution of unmodified and modified octamers on the same array (50% stochastic sample). Importantly, the total number of ubiquitin moieties was identical in the two samples while their spatial distribution differed (Supplementary Fig. 16). We incubated these samples with increasing amounts of Mg2+ and measured the percentage of arrays remaining in solution, similar to the experiment presented in Fig. 3d. A comparison of the two datasets revealed different oligomerization behavior, with the 50% mixed array sample requiring higher Mg2+ concentrations for fiber association (Fig. 5b). Thus, clustering of many ubiquitin moieties on the same fiber appears to have a more dominant effect on association than the stochastic distribution of the same number of ubiquitin moieties in trans.
Figure 5.
Intra-array ubiquitin-ubiquitin interactions impede oligomerization. (a) A schematic depicting the composition of 50% mix and 50% stochastic arrays. (b) Comparison of the oligomerization of 50% mix and 50% stochastic arrays. (c) SDS-PAGE analysis of the supernatant fractions of 50% mix array samples as a function of Mg2+ concentration. Full gel image is given in Supplementary Fig. 16. (d) Quantitative analysis of the H2B-Ub and H2B band intensities shown in (c) compared to the oligomerization of 100% unmodified, 100% ubiquitylated and 50% mix arrays. (b) and (d) Error bars, s.e.m. (n = 3). The data for the unmodified and 100% ubiquitylated arrays are the same as in Fig. 3. All array samples contain the equivalent of 250 nM total 601 sites.
The individual components within the 50% mixed sample exhibited different structures at low Mg2+ concentrations (Fig. 3b) – unmodified arrays were more compact, and ubiquitylated arrays had a more open structure. We wondered whether these structural differences would be translated into a differential association behavior of the components, i.e. would compact unmodified polymers prefer to associate with other compact unmodified polymers, and similarly, would less folded ubiquitylated polymers interact preferentially with other ubiquitylated fibers, or, alternatively, would co-assemblies between modified and unmodified components be favored? To answer this question, we used non-reducing SDS-PAGE to analyze the soluble fractions of the 50% mixed sample as a function of Mg2+ concentration (Fig. 5c). Since H2B-Ub and H2B have a significantly different molecular weigth (22 kDa vs. 14 kDa), and they were present exclusively only in one array type, this provided a convenient readout of the association behavior of the two components within the mix. Quantitative analysis of the H2B-Ub and H2B band intensities (Fig. 5c) revealed that unmodified arrays precipitated at much higher Mg2+ concentrations when in the presence of ubiquitylated arrays than in the absence (Fig. 5d), and that this effect is not due to the reduced effective concentration of each array type in the mix (Supplementary Fig. 17). Thus, ubiquitylated chromatin polymers can solubilize unmodified fibers by obstructing their association.
DISCUSSION
Using well-defined semi-synthetic nucleosome arrays that emulate the chromatin environment and a suite of biophysical and biochemical experiments, we have explored the molecular mechanism by which H2B ubiquitylation induces chromatin decompaction.
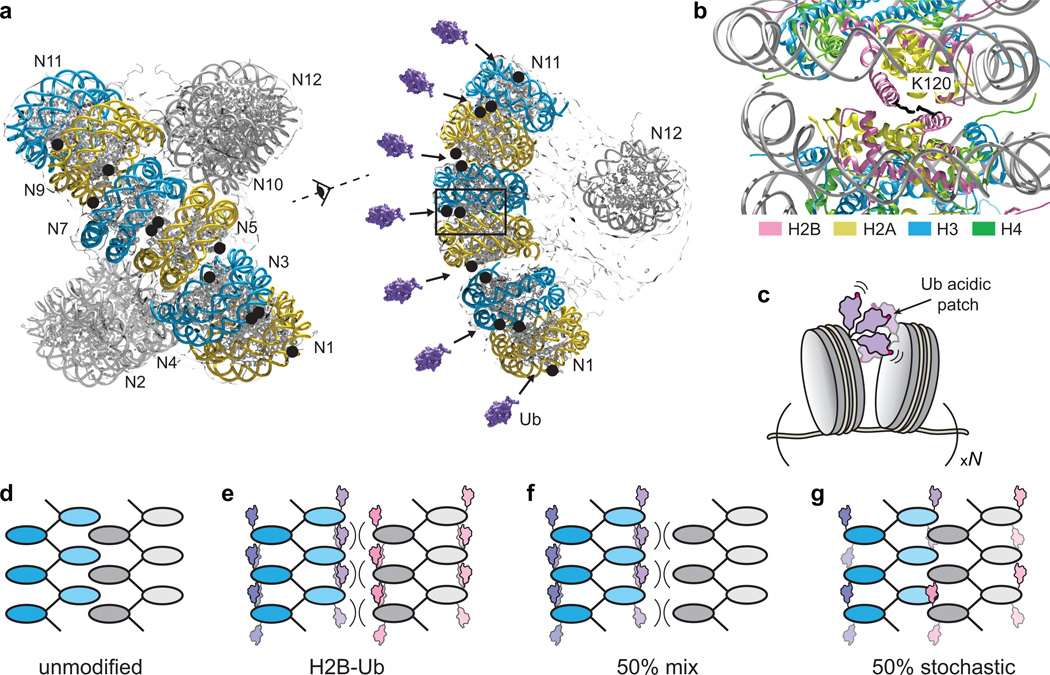
Previous studies have suggested that features on the ubiquitin surface and not necessarily the addition of steric bulk are responsible for decompaction15, and our work confirms this hypothesis and implicates a small acidic patch on ubiquitin in this process. This negatively charged patch, comprised of Glu16 and Glu18, impacts both local and higher order chromatin interactions, with other ubiquitin surface features (e.g. hydrophobic residues such as Ile44 and Phe45) imparting a supporting role in oligomerization. The prominent role of the glutamate patch implies that the decompaction mechanism is mediated by electrostatic interactions. Furthermore, our chemical cross-linking experiments indicate that ubiquitin promiscuously samples its environment and interacts with histone proteins and, surprisingly, other ubiquitin molecules. This finding is consistent with the published cryo-electron microscopy structural model of a compacted 12-mer nucleosome array32, which positions the H2B ubiquitin attachment sites in close spatial proximity (Fig. 6a), and in some of the modeled interfaces even directly next to each other (Fig. 6b). Thus, ubiquitin moieties from neighboring nucleosomes may act as a wedge to prevent the establishment of a close interface between nucleosomes (Fig. 6c). In this process, ubiquitin potentially utilizes its acidic patch as a “hook” to form transient interactions with different basic residues such as lysine and arginine side-chains from histone proteins or proximal ubiquitin moieties. The dynamic sampling of the nucleosome environment by the ubiquitin modification is also suggested by a recent crystal structure of a ubiquitylated mononucleosome in which electron density for ubiquitin could not be detected, presumably due to conformational flexibility16. As indicated by our cross-linking experiments ubiquitin-nucleosome interactions appear to be Mg2+-dependent: in the absence of Mg2+ neighboring nucleosomes could be too far apart for the establishment of electrostatic interactions involving the ubiquitin moiety; the addition of divalent salt, however, would bring the nucleosome surfaces closer to each other thus enabling the engagement of the ubiquitin acidic patch. In addition, we note that the dynamic ubiquitin wedge mechanism proposed here is in contrast to the action of H4 acetylation, a well-known decompaction PTM that is thought to function by weakening the electrostatic interactions between nucleosomes.29
Figure 6.
Mechanism of chromatin decompaction induced by H2B ubiquitylation. (a) Cryo-electron microscopy structural model of unmodified 12-mer nucleosome arrays (EMD_2600).32 The nucleosome surface43 (PDB ID: 1AOI) was fitted to the electron density map using the program Chimera44. H2B Lys120, the attachment site for ubiquitin, is denoted as black circles. Ubiquitin (PDB ID: 1UBQ, purple, Ref.42) is represented to scale. (b) Zoom in of the modeled nucleosome-nucleosome interface depicting the ubiquitin attachment sites and the surrounding histone environment. (c) A cartoon illustrating the dynamic ubiquitin “wedge” that prevents the establishment of close intra-molecular nucleosome-nucleosome interfaces. The ubiquitin acidic patch is depicted in pink. (d–g) Model of ubiquitin-mediated chromatin polymer disassociation. Intra-array ubiquitin-ubiquitin interactions prevent the interdigitation of nucleosome surfaces from different polymers, thus “solubilizing” chromatin.
The structure of highly compacted chromatin, as manifest in vitro by nucleosome array oligomerization, is still debated1,33–37. A leading hypothesis posits that oligomerization is achieved through the interdigitation of 10 nm chromatin fibers in a polymer melt-like structure36,37 with nucleosome-nucleosome interactions formed between different fibers. (Fig. 6d). In this framework, a dynamic ubiquitin wedge may sterically block the interdigitation of nucleosome surfaces (Fig. 6e). Furthermore, as evident from our oligomerization experiments performed with mixed and stochastically modified array samples, it appears that the impairment is much stronger when ubiquitin is present on neighboring nucleosomes, i.e. in cis (Fig. 6f), rather than when it is randomly distributed throughout the array (Fig. 6g). This situation could be especially important in chromatin regions expected to have a high density of H2B-Ub such as in gene bodies10,12 and near the sites of double-stranded DNA breaks38,39. Interestingly, ubiquitylated chromatin polymers, expected to have a more open and accessible structure, appear to co-precipitate with unmodified polymers, driving their association behavior towards higher Mg2+-concentrations. Thus, H2B ubiquitylation in the cell may impact chromatin structural interactions in trans by solubilizing regions of the chromatin fiber that are in close spatial proximity.
In conclusion, the work described herein brings to light a novel, chromatin-related aspect of the “ubiquitin code” that is distinct from the hydrophobic-type interactions that typically mediate ubiquitin molecular recognition processes2,13,14. Our findings highlight the value of performing detailed biophysical and biochemical studies on reconstituted designer chromatin. For example, yeast genetics studies failed to identify a role of the ubiquitin acidic patch in cell viability40, likely due to the inability of such assays to tease apart more subtle structural effects involving ubiquitin (we note that H2B ubiquitylation itself is not essential for yeast survival9,41). We propose that the attachment of ubiquitin to a specific site on the nucleosome target simultaneously restricts and orients the protein, exposing distinctive surface features to the chromatin environment and exploiting unique modes of ubiquitin-protein interactions to control gene function in the cell. We imagine that the NMR based H/D exchange strategy developed herein, which we show can be applied to multiple ubiquitin attachment sites on chromatin, will be broadly useful for studying ubiquitin-mediated recognition processes.
Online methods
General laboratory methods
All commonly used chemical reagents and solvents were purchased from Sigma-Aldrich or Fisher Scientific, while isotopically enriched reagents were purchased from Cambridge Isotope Laboratories. Restriction enzymes were obtained from New England BioLabs, while primer synthesis and gene sequencing were performed by Integrated DNA Technologies and Genewiz, respectively. Gene mutagenesis was achieved using the QuickChange II Site-Directed Mutagenesis Kit (Agilent), and PCR purification kits were obtained from Qiagen. Slide-A-Lyzer dialysis cassettes and mini-dialysis units were purchased from Thermo Fisher Scientific. Size exclusion chromatography was performed on an AKTA FPLC system from GE Healthcare equipped with a P-920 pump, and an UPC-900 monitor. Analytical reversed-phase HPLC (RP-HPLC) was performed on an Agilent 1200 series instrument with a Vydac C18 column (5 micron, 4 × 150 mm), employing 0.1% TFA in water (HPLC solvent A), and 90% acetonitrile, 0.1% TFA in water (HPLC solvent B), as the mobile phases. Preparative scale purifications were conducted on a Waters prep LC system comprised of a Waters 2545 Binary Gradient Module and a Waters 2489 UV detector. A Vydac C18 preparative column (15–20 micron, 20 × 250 mm) or a semi-preparative column (12 micron, 10 mm × 250 mm) was employed at a flow rate of 18 mL/min or 4 mL/min, respectively. ESI-MS analysis was conducted on a MicrOTOF-Q II ESI-Qq-TOF mass spectrometer (Bruker Daltonics). UV spectrometry was performed on an Agilent 8453 UV-Vis spectrophotometer. Gel and Western blot images were acquired with an ImageQuant LAS4000 instrument (GE Healthcare). Molecular graphics and analyses were performed with the UCSF Chimera package. (Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311))44.
Mutagenesis
To prepare ubiquitin-aminoethanethiol (Ub-SH), we used a previously described pTXB1-Ub-AvaDnaE-AAFN-His6 plasmid.24 Ubiquitin double mutants (Ub1 – E16A, E18A; Ub2 – I44A, F45A) were generated using a Quick Change Mutagenesis kit with the Pfu Ultra II polymerase (Agilent) and the following primers: forward Ub1 5-TTCTCAATGGTGTCACTCGGAGCCACAGCGAGAGTGATGGTCTTACCAG-3; reverse Ub1 5-CTGGTAAGACCATCACTCTCGCTGTGGCTCCGAGTGACACCATTGAGAA-3; forward Ub2 5-CATCTTCCAGCTGTTTCCCAGCAGCAGCCAACCTCTGCTGGTCAGGAGGG-3; reverse Ub2 5-CCCTCCTGACCAGCAGAGGTTGGCTGCTGCTGGGAAACAGCTGGAAGATG-3; forward UbBpa 5-CCAGCTGTTTCCCAGCCTAGATCAACCTCTGCTGG-3; reverse UbBpa 5-CCAGCAGAGGTTGATCTAGGCTGGGAAACAGCTGG-3; forward K119C 5-GTCCGTGCTGCTGCCCAAGTGTACCGAGAGTTCCAAGTCGG-3; reverse K119C 5-CCGACTTGGAACTCTCGGTACACTTGGGCAGCAGCACGGAC-3.
The correct sequence was confirmed by gene sequencing. The plasmids for wild-type Xenopus histones, H2B K120C, H2A N110C, and all DNA constructs have been described before.15,25,45,46 pEVOL-pBpF (used for UbBpa expression) was a gift from Peter Schultz (Addgene plasmid # 31190).30
Expression of recombinant histones
Recombinant Xenopus laevis histones were expressed and purified as previously described with some modifications15. Briefly, BL21(DE3) pLysS E. coli cells were transfected with the appropriate plasmids, grown in 6 L of LB at 37 °C until OD600 = 0.6, and induced with 0.5 mM IPTG for 2 hr. Cells were harvested by centrifugation at 5000 × g, and resuspended in lysis buffer (50 mM Tris, 100 mM NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, pH 7.5). The cells were lysed by sonication, centrifuged at 30,000 × g, and the insoluble pellet was resuspended and washed twice in lysis buffer also containing 1% triton X-100. After a final wash without detergent, the inclusion body pellet was resuspended and incubated for 1 hr at 4 °C in 6 M guanidine HCl, 20 mM Tris, 1 mM EDTA, 100 mM NaCl, 1 mM DTT, pH 7.5). The solution was dialyzed against 7 M urea, 10 mM Tris, 1 mM EDTA, 100 mM NaCl, 1 mM DTT, pH 7.5. After dialysis, cation exchange was performed using an SP HiTrap HP 5 mL column (GE Healthcare), and the fractions containing the appropriate histone protein were combined and further purified using a preparative RP-HPLC column using a 30–70% solvent B gradient. The fractions were assessed for purity by analytical RP-HPLC and ESI-MS. (H4: calculated 11,236 Da, observed 11,236.1 Da; H3 C110A: calculated 15,239 Da, observed 15,238.7 Da; H2A: calculated 13,950 Da, observed 13,949.8 Da; H2A N110C: calculated 13,940 Da, observed 13939.2 Da; H2A K119C: calculated 13,925 Da, observed 13,925.2 Da; H2B: calculated 13,817 Da, observed 13,816.8 Da; H2B K120C: calculated 13,792, observed 13,791.8 Da).
Preparation of Ub-SH
The preparation of Ub-SH was based on the protocol published in Ref.23 with the following modifications. Isotopically labeled Ub-Ava-His6 was expressed in BL21(DE3) cells grown in minimal M9 media supplemented with 15N-labeled ammonium chloride (15N-labeled samples), or 13C-labeled glucose and 15N-labeled ammonium chloride (15N,13C-labeled sample used for assignments). Natural abundance Ub, Ub1 and Ub2-SH were prepared in LB media. Cells were grown to OD600 = 0.6 at 37 °C, and protein expression was induced using 0.5 mM IPTG for 14–16 hr at 18 °C. The cells were harvested by centrifugation, and the cell pellet was resuspended in lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 5 mM imidazole, no thiols, pH 8.0) supplemented with cOmplete protease inhibitor cocktail (Roche). The cells were lysed by sonication, and the soluble fraction was recovered by centrifugation (30,000 rcf, 30 min). The Ub-Ava-His6 protein was purified using a Ni-NTA column at 4 °C, and the fractions containing the protein were supplemented with 150 mM 2-aminoethanethiol, 10 mM TCEP and 2.5 mM EDTA immediately after elution. The pH was re-adjusted to 8.0 and the sample was left to incubate overnight at 22.5 °C. The final ubiquitin-aminoethanethiol product (Ub-SH) was purified over a prep-scale RP-HPLC using a 30–70% gradient of HPLC solvent B, and the purity of each protein was confirmed by analytical HPLC and ESI-MS (15N-Ub-SH: calculated 8,728 Da average, observed 8727.9 Da; 15N,13C-Ub-SH: calculated 9,103 Da average, observed 9,095 Da; Ub-SH: calculated 8,624 Da, observed 8,623.5 Da; Ub1-SH: calculated 8,508 Da, observed 8,507.6 Da; Ub2-SH: calculated 8506 Da, observed 8,505.5 Da).
Preparation of UbBpa
Bpa (p-benzoyl-L-phenylananine) was purchased from Bachem, and the expression protocol was based on protocols described in Ref.47,48 BL21 (DE3) E. coli cells were doubly transfected with the pEVOL-pBpF plasmid and the pTXB1-Ub-AvaDnaE-AAFN-His6 plasmid (containing the appropriate site-specific mutation for amber suppression), and grown at 37 °C in 1 L of LB media supplemented with the appropriate antibiotics (100 µg/mL ampicillin, and 35 µg/mL chloramphenicol). At OD600 = 0.4, 1 mL of 1M Bpa dissolved in 1M NaOH was added to the media for final Bpa concentration of 1 mM. The cells were moved to 18 °C, grown to OD600 = 0.7, and protein expression was induced using 0.5 mM IPTG and 0.2% arabinose for 18 hr. UbBpa-SH was purified as described above while minimizing the exposure to ambient light. Yields were ~ 10 mg/L of culture, and the purity was confirmed by analytical HPLC and ESI-MS (UbBpa-SH: calculated 8,728.0 Da average, observed 8,727.7 Da).
Activation of H2B K120C and H2A K119C
Activation of H2B K120C was performed according to the protocol described in ref.23. Briefly, 14 mg of 2,2’-dithiobis(5-nitropyridine) were dissolved with sonication in 4.5 mL of a 3:1 (v/v) acetic acid/water solution. The mixture was added to 21 mg H2B K120C and the reaction was allowed to proceed for 12 hr at 22.5 °C. The activated protein was purified on a semi-prep RP-HPLC column at 50 °C using a 42–48% gradient of HPLC solvent B, and the collected fractions were analyzed by analytical HPLC and ESI-MS (calculated 13,946 Da, observed 13,945.8 Da). H2A K119C activation, purification and analysis were performed identically (calculated MW 14,079 Da, observed 14,078.9 Da).
Synthesis of disulfide-linked H2B-Ub
Synthesis of H2B-Ub was performed according to the protocol described in ref.23. Briefly, 1 equivalent of HPLC-purified activated H2B (3 mg) and 2 equivalents of HPLC-purified Ub-SH or mutants thereof (~ 4.0 mg) were dissolved in 1.5 mL reaction buffer (6 M guanidine hydrochloride, 1 M HEPES, pH 6.93). The reaction was allowed to proceed for 1 hr at 22.5 °C, and the sample was mixed with 0.5 mL 25% HPLC solvent B. Disulfide-linked H2B-Ub was purified on a semi-prep RP-HPLC column using a 30–70% gradient of HPLC solvent B. Synthesis of H2A-15N-Ub and H2A-Ub1 was performed using the same protocol. The purity of the proteins was assessed using analytical HPLC and ESI-MS (Supplementary Fig. 2, 6 and 13).
Labeling of H2A
To perform steady-state anisotropy measurements, H2A was labeled with fluorescein-5-maleimide at position N110C according to a previously described protocol.15 Approximately 10 mg HPLC-purified H2A N110C were dissolved in 4 mL labeling buffer (20 mM Tris, pH 7.8, 6 M guanidine hydrochloride, 0.5 mM TCEP), the solution was incubated at 22.5 °C for 1 hr, and 1.5 mg fluorescein-5-maleimide (dissolved in 50 µL N,N-dimethylformamide) was added slowly to the protein solution. The mixture was stirred for 1 hr in the dark, and the reaction was quenched with 1 mM 2-mercaptoethanol. Fluorescently labeled H2A was purified using a semi preparative C18 RP-HPLC column using a 30–70% HPLC solvent B gradient. The protein was characterized using analytical HPLC and ESI-MS (calculated 14368 Da, observed 14366.5 Da).
DNA preparation
The plasmid containing the 12×601 sequence (177 bp repeat) was purified from 6 L of DH5α cells following the protocol described in Ref25. Total plasmid yields were 30 – 50 mg. The 12×601 sequence was excised from the plasmid using the EcoRV restriction enzyme, and purified using 6% PEG6000 precipitation (at 1 mg/mL of DNA). After purification, the proteins and PEG were removed using phenol-chloroform extraction and ethanol precipitation, and the 12×601-containing pellet was dissolved in 10 mM Tris pH 8.0, 0.1 mM EDTA buffer, aliquoted and stored at −20°C. DNA fragment yields were 10 – 15 mg. Each 601 repeat has the following sequence: CTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGTAGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCGCGTTTTAACCGCCAAGGGGATTACTCCCTAGTCTCCAGGCACGTGTCAGATATATACATCCTGTGCATGTAAGATCCAGTACTACGCGGCCGCC
A similar protocol was used to prepare large amounts of the weak nucleosome binding MMTV sequence46 (“buffer DNA”, see below) with the following modification: 7% PEG6000 was used for purification where the 145 bp MMTV fragment remained in the supernatant, while the plasmid backbone was separated into the pellet fraction. The MMTV sequence is as follows: ACTTGCAACAGTCCTAACATTCACCTCTTGTGTGTTTGTGTCTGTTCGCCATCCCGTCTCCGCTCGTCACTTATCCTTCACTTTCCAGAGGGTCCCCCCGCAGACCCCGGCGACCCTGGTCGGCCGACTGCGGCACAGTTTTTTG
Octamer assembly
Histone octamers were prepared using established protocols.15,45 Briefly, lyophilized histones were dissolved at 2 mg/mL in 6 M guanidine hydrochloride, 20 mM Tris pH 7.5, and the concentration of the solutions was determined by UV absorbance. The four histone proteins (ubiquitylated or wild-type) were combined in equimolar amounts and the final concentration was adjusted to 1 mg/mL. The solution was dialyzed against 2 M NaCl, 10 mM Tris, 1 mM EDTA, and the octamers were purified from the tetramers and dimer species using size-exclusion chromatography on a Superdex 200 10/300 column (GE Healthcare Life Sciences). The purity of the fractions was analyzed by SDS-PAGE and pure fractions were combined, concentrated and stored in 50% glycerol at −20°C.
Nucleosome array reconstitution
The reconstitution of nucleosome arrays followed the protocols described in Ref.15,49 with some modifications. Generally, the reconstitution protocol started with the assembly of octamers, 12-mer DNA (12 repeats of the 601 DNA sequence separated by 30 bp linkers), and buffer DNA (155 bp of the weak nucleosome binding MMTV sequence; necessary to ensure good reproducibility of the array reconstitution on a variety of scales46). This was done in 2 M TEK buffer (2 M KCl, 10 mM Tris pH 7.5, 0.1 mM EDTA). Small-scale reconstitutions (e.g. for homo-FRET or Mg2+ precipitation experiments) were done in 50–100 µL volume, containing 0.5–2.0 µM 601 sites, 0.3 eq. of the MMTV DNA, and an optimized ratio of octamers (1.2–1.5 eq. depending on the preparation). Large-scale assembly (for H/D exchange and NMR) was performed in 3 mL total volume, at 2.5 µM 601 sites, 0.3 eq. of MMTV DNA and an appropriate ratio of octamers. The samples were placed in 3.5 kDa cutoff Slide-A-Lyzer MINI diaysis units (small-scale assembly) or Slide-A-Lyzer dialysis cassettes (large-scale), both available from Thermo Fisher Scientific. The samples were then dialyzed at 4°C against 200 mL (small-scale) or 1 L (large-scale) 1.4 M TEK buffer (1.4 M KCl, 10 mM Tris pH 7.5, 0.1 mM EDTA). After 1 hr, gradual salt dilution dialysis was performed, during which 350 mL (small-scale) or 1.75 L (large-scale) 10 mM TEK buffer (10 mM KCl, 10 mM Tris pH 7.5, 0.1 mM EDTA) were added at 1 or 5 mL/min respectively to bring the overall KCl concentration to ~ 0.5 M. Final dialysis against 10 mM TEK was performed overnight. After assembly, MMTV nucleosomes and free MMTV DNA were purified from the 12mer nucleosome arrays using selective MgCl2 precipitation (2–3 mM for WT and Ub-H2A arrays, 4–5 mM for Ub-H2B arrays). After the addition of MgCl2, the samples were incubated on ice for 10 min, centrifuged at 17,000×g for 10 min, and the pellet and supernatant were separated. An appropriate volume of 10 mM TEK was added to the pellet, and the resuspended arrays were dialyzed overnight against fresh 10 mM TEK to remove residual MgCl2. Arrays were usually used for experiments within 1–2 days after assembly. The reconstituted arrays were analyzed on native 1% agarose/2% polyacrylamide (APAGE) gels stained with Sybr Gold nucleic acid gel stain (Life Technologies). Since the 601 repeats in the 12mer DNA sequence are separated by ScaI restriction enzyme sites, the full occupancy of the 601 sites and the nucleosome positioning could be assessed by ScaI digestion. Briefly, the equivalent of 1 pmole nucleosomes were digested in 3.75 µL total volume containing 10 U of ScaI-HF (New England BioLabs), 100 mM KCl, 10 mM Tris pH 7.5, 0.5 mM MgCl2, 1 mM DTT. Digestion was performed for 4 hr at 37°C or overnight at 25°C. Full 601 site occupancy was detected by the presence of a well-defined nucleosome band and minimal amounts of free 601 DNA or dinucleosomes.
H/D exchange
After reconstitution and Mg2+ precipitation, the pelleted arrays were resuspended in 1.2 mL cold 10 mM TEK to final concentration of ~ 5 µM (601 sites). The sample was divided into three Eppendorf tubes, and Mg2+ precipitation was performed to reduce the volume of the samples (4 mM Mg2+ for H2B-15N-Ub arrays, 2.5 mM for H2A-15N-Ub arrays). The pellets were carefully resuspended in 100 µL TKM buffer (10 mM Tris pH 7.5 at room temperature, 10 mM KCl, 1 mM MgCl2, 100% protonated). Care was taken that each pellet was completely dissolved until the solution was transparent and the measured A260 was consistent with > 95% resolubilization of the arrays. The samples were equilibrated at room temperature for 15 min, and 900 µL of 100% deuterated TKM buffer were added quickly to each tube to give 1:9 protonated/deuterated buffer and final concentration of 2 µM 601 sites (167 nM arrays). H/D exchange was performed at room temperature for 30 sec, 15 min or 60 min, and quenched by flash freezing in liquid N2 and lyophilization to remove the solvent. As control, 15N-labeled Ub-SH was refolded in 10 mM TEK buffer, followed by dialysis in protonated TKM buffer to match the buffer used for H/D exchange of the arrays. Three tubes containing 100 µL each of 15N-Ub-SH in protonated TKM buffer were prepared where the Ub-SH concentration was equivalent to the concentration of Ub in the array samples (40 µM, taking into account that there are 2 Ub molecules per nucleosome). H/D exchange was performed as described above, usually in parallel with each array exchange experiment.
NMR spectroscopy of H/D exchanged samples
NMR experiments of the lyophilized H/D exchanged samples (arrays or the Ub-SH control) were performed in the following solvent: 95% DMSO-d6, 4.5% D2O, 0.5% dichloroacetic acid-d2, 10 mM triphenylphosphine, pD adjusted to 5.0 with 4% NaOD. A volume of 75 µL was added to each sample tube (3 tubes per experiment). Under these conditions, all proteins were expected to dissolve, while the DNA remained insoluble. To remove the insoluble fraction, the samples were centrifuged for 2 min at 17,000×g and the supernatant was transferred quickly to a 5 mm DMSO-susceptibility matched NMR tube (~ 210 µL total volume, Shigemi Inc). The sample processing and NMR spectrometer optimization time were kept to a minimum to avoid back exchange of the amide protons (~ 15 min for all sample preps). NMR experiments were performed on a Bruker Avance III 800 MHz NMR spectrometer equipped with a 5 mm QCI 1H/13C/15N probe. 2D HSQC experiments were performed at 298 K, with 16 scans per point, 0.8 sec scan delay, 4096 points and 11.16 kHz sweep width in the 1H dimension, and 128 points and 1.6 kHz sweep width in the 15N dimension. The center frequency was set at 3.7 ppm for 1H, and 113 ppm for 15N. Each HSQC experiment took ~ 30 min. Spectra were processed with the program NMRPipe50 and visualized with the Sparky software51.
Resonance assignments
Resonance assignments were performed with 13C,15N-labeled Ub-SH refolded and lyophilized in the same manner as the Ub-SH control. After lyophilization, the sample was dissolved in the DMSO-based solvent described above, with the exception that 4.5% H2O was substituted for D2O. The final sample concentration was 1 mg/mL. Assignments were based on 3D HNCA and HNCACB experiments using the following parameters: 298 K, 16 scans, 0.7 sec scan delay; 80 ms acquisition time, 2048 points and 4.73 ppm center frequency for the 1H dimension; 14.8 ms acquisition time, 48 points and 114 ppm center frequency for the 15N dimension; 9 ms acquisition time, 240 points and 45 ppm center frequency for the 13C dimension (HNCACB experiment); 10.6 ms acquisition time, 128 points and 54 ppm center frequency for the 13C dimension (HNCA experiment); JNH = 90 Hz, JNCa = 20 Hz, J’NH = 108 Hz, and JCACB = 69 Hz. Spectra were processed with the program NMRPipe50 and assigned with the Sparky software51.
Steady-state anisotropy measurements
Steady-state anisotropy measurements were performed following the protocol described in Ref.15. Briefly, array samples were diluted to 50 nM eq. of 601 sites in 10 mM Tris, 10 mM KCl, pH 7.8 with the appropriate concentration of Mg2+ (0 mM, 0.2 mM, 0.4 mM, 0.6 mM, 0.8 mM and 1 mM). The 0 mM Mg2+ sample contained 0.1 mM EDTA. Samples were equilibrated at 22.5 °C for 15 min, and measurements were performed on a Fluorolog3-11 instrument equipped with an L-format dual polarizer (HORIBA Jobin Yvon) with excitation and emission wavelengths of 480 nm (5 nm bandwidth) and 520 nm (5 nm bandwidth), respectively. Six technical replicates, each collected with 5 sec integration time, were averaged for each point, and the steady-state anisotropy was calculated according to the formula SSA = (IVV − G×IVH)/(IVV + 2G×IVH), where G = IHV/IHH is an instrument dependent parameter, I is the light intensity, V and H are the vertical and horizontal positions of the excitation/emission polarizers. The final data represent the average of three measurements using independently prepared array samples.
Mg2+ precipitation experiments
The Mg2+ precipitation assay was performed at 0.25 µM eq. of 601 sites in 10 mM Tris, 10 mM KCl, pH 7.8. Briefly, 6 µL of 2× array sample (0.5 µM eq. of 601) were incubated with 6 µL of 2× Mg2+ solution for 10 min at 22.5 °C. Samples were centrifuged at 17,000×g for 10 min at 15 °C, and the supernatants were transferred to a fresh tube. 5 µL of the supernatant was added to 200 µL of SYBR® Gold nucleic acid stain (Life Technologies, 1:10,000 dilution in H2O), and the dye fluorescence was recorded in a plate reader format (Spectramax M3, Molecular Devices) for each Mg2+ concentration. Two technical replicates were performed for each precipitation experiment, and the data for 3–6 precipitation experiments were averaged. For precipitation experiments analyzed by SDS-PAGE (Fig. 5c,d), 20 µL of solution containing 0.25 µM of total 601 sites (0.125 µM for ubiquitylated arrays and 0.125 µM for unmodified arrays) and the appropriate concentration of Mg2+ were prepared. Samples were centrifuged and the supernatant was loaded on a 15% Tris-HCl gel (non-reducing conditions). The gel was stained using SYPRO® Ruby protein stain (Thermo Fisher Scientific) and the gel bands were quantified using ImageStudioLite (LI-COR Biosciences).
Circular dichroism
CD measurements were performed with Ub-SH, Ub1-SH and Ub2-SH samples refolded in 10 mM phosphate, 100 mM KF, 1 mM DTT, pH 7.3 at a concentration of 20 µM. Experiments were performed using a Chirascan CD spectrometer (Applied Photophysics).
Cross-linking with EDC
1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDC) was purchased from Bachem and the cross-linking protocol was based on Ref.52. The array samples were dialyzed against 10 mM HEPES, 10 mM KCl, pH 7.0 buffer, and the sample volume and concentration were adjusted to 20 µL and 1.5 µM 601 sites respectively. An appropriate amount of MgCl2 was added to adjust the concentration to 0 mM, 1 mM, 2 mM or 3 mM, and the samples were incubated at 22.5 °C for 15 min. The pH of the samples was adjusted to 6.0 using 1 µL of 0.025N HCl, and EDC was added to a final concentration of 2.2 mM. Cross-linking was performed for 75 min at 22.5 °C, and quenched with SDS gel loading buffer containing 2-mercaptoethanol. The samples were boiled for 5 min, and loaded onto a 12% BisTris gel (BioRad). Western blotting was performed using the mouse monoclonal P4D1/P4G7 anti-ubiquitin antibody (BioLegend/Covance, #838701, 1:1000 dilution), followed by incubation with a secondary goat anti-mouse antibody conjugated to horseradish peroxidase (Biorad Laboratories).
Cross-linking with BPA
Cross-linking was performed in 15 µL volume at an H2B-UbBpa array concentration corresponding to 1 µM 601 sites, in the presence of 1 mM MgCl2, 10 mM Tris, 10 mM KCl, pH 7.5. Experiments were performed with and without UV irradiation (5 min) at 350 nm. Analysis of the cross-linking experiments was performed by SDS-PAGE (15% Tris-HCl gel) followed by staining with SYPRO® Ruby protein stain gel, or Western blot analysis. The antibody details are given in Supplementary Fig. 14, and all experiments used goat anti-rabbit antibody conjugated to horseradish peroxidase for detection (Biorad Laboratories).
Supplementary Material
Acknowledgments
We thank M. Müller, I. Pelczer, B. Fierz, L. Guerra, N. Shah, S. Kilic, M. Holt, U. Nguyen, J. Bos and K. Jani for stimulating discussions and help regarding the preparation of materials. This work was funded by US National Institutes of Health grant R01-GM107047 to T.W.M.
Footnotes
Author Contributions
G.T.D. designed experiments, prepared materials, performed H/D exchange and NMR experiments, compaction assays and cross-linking experiments, analyzed data and wrote the manuscript; K.G. prepared materials, performed compaction assays and analyzed data; T.W.M. designed experiments, analyzed data and wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Komander D, Rape M. The ubiquitin code. Annu. Rev. Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 3.Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L. Distinct functional surface regions on ubiquitin. J. Biol. Chem. 2001;276:30483–30489. doi: 10.1074/jbc.M103248200. [DOI] [PubMed] [Google Scholar]
- 4.Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nat. Rev. Mol. Cell. Biol. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun S, Madhani HD. Shaping the landscape: mechanistic consequences of ubiquitin modification of chromatin. EMBO Rep. 2012;13:619–630. doi: 10.1038/embor.2012.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 7.Zhou W, et al. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol. Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 9.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 10.Minsky N, et al. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell. Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs G, Hollander D, Voichek Y, Ast G, Oren M. Cotranscriptional histone H2B monoubiquitylation is tightly coupled with RNA polymerase II elongation rate. Genome Res. 2014;24:1572–1583. doi: 10.1101/gr.176487.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt MT, et al. Identification of a functional hotspot on ubiquitin required for stimulation of methyltransferase activity on chromatin. Proc. Natl. Acad. Sci. USA. 2015;112:10365–10370. doi: 10.1073/pnas.1504483112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou L, et al. Evidence that ubiquitylated H2B corrals hDot1L on the nucleosomal surface to induce H3K79 methylation. Nat. Commun. 2016;7:10589. doi: 10.1038/ncomms10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fierz B, et al. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Machida S, Sekine S, Nishiyama Y, Horikoshi N, Kurumizaka H. Structural and biochemical analyses of monoubiquitinated human histones H2B and H4. Open Biol. 2016;6 doi: 10.1098/rsob.160090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paterson Y, Englander SW, Roder H. An antibody binding site on cytochrome c defined by hydrogen exchange and two-dimensional NMR. Science. 1990;249:755–759. doi: 10.1126/science.1697101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raschke TM, Marqusee S. Hydrogen exchange studies of protein structure. Curr. Opin. Biotechnol. 1998;9:80–86. doi: 10.1016/s0958-1669(98)80088-8. [DOI] [PubMed] [Google Scholar]
- 19.Englander SW. Protein folding intermediates and pathways studied by hydrogen exchange. Annu. Rev. Biophys. Biomol. Struct. 2000;29:213–238. doi: 10.1146/annurev.biophys.29.1.213. [DOI] [PubMed] [Google Scholar]
- 20.Zhang YZ, Paterson Y, Roder H. Rapid amide proton exchange rates in peptides and proteins measured by solvent quenching and two-dimensional NMR. Protein. Sci. 1995;4:804–814. doi: 10.1002/pro.5560040420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoshino M, et al. Mapping the core of the beta(2)-microglobulin amyloid fibril by H/D exchange. Nat. Struct. Biol. 2002;9:332–336. doi: 10.1038/nsb792. [DOI] [PubMed] [Google Scholar]
- 22.Kato H, Gruschus J, Ghirlando R, Tjandra N, Bai Y. Characterization of the N-terminal tail domain of histone H3 in condensed nucleosome arrays by hydrogen exchange and NMR. J. Am. Chem. Soc. 2009;131:15104–15105. doi: 10.1021/ja9070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterjee C, McGinty RK, Fierz B, Muir TW. Disulfide-directed histone ubiquitylation reveals plasticity in hDot1L activation. Nat. Chem. Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 24.Shah NH, Dann GP, Vila-Perello M, Liu Z, Muir TW. Ultrafast protein splicing is common among cyanobacterial split inteins: implications for protein engineering. J. Am. Chem. Soc. 2012;134:11338–11341. doi: 10.1021/ja303226x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 26.Pan Y, Briggs MS. Hydrogen exchange in native and alcohol forms of ubiquitin. Biochemistry. 1992;31:11405–11412. doi: 10.1021/bi00161a019. [DOI] [PubMed] [Google Scholar]
- 27.Going CC, Xia Z, Williams ER. Real-time HD exchange kinetics of proteins from buffered aqueous solution with electrothermal supercharging and top-down tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2016;27:1019–1027. doi: 10.1007/s13361-016-1350-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarz PM, Felthauser A, Fletcher TM, Hansen JC. Reversible oligonucleosome self-association: dependence on divalent cations and core histone tail domains. Biochemistry. 1996;35:4009–4015. doi: 10.1021/bi9525684. [DOI] [PubMed] [Google Scholar]
- 29.Shogren-Knaak M, et al. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 30.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galardy RE, Craig LC, Printz MP. Benzophenone triplet: a new photochemical probe of biological ligand-receptor interactions. Nat. New Biol. 1973;242:127–128. doi: 10.1038/newbio242127a0. [DOI] [PubMed] [Google Scholar]
- 32.Song F, et al. Cryo-EM study of the chromatin fiber reveals a double helix twisted by tetranucleosomal units. Science. 2014;344:376–380. doi: 10.1126/science.1251413. [DOI] [PubMed] [Google Scholar]
- 33.Zheng C, Lu X, Hansen JC, Hayes JJ. Salt-dependent intra- and internucleosomal interactions of the H3 tail domain in a model oligonucleosomal array. J. Biol. Chem. 2005;280:33552–33557. doi: 10.1074/jbc.M507241200. [DOI] [PubMed] [Google Scholar]
- 34.Nishino Y, et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–1653. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joti Y, et al. Chromosomes without a 30-nm chromatin fiber. Nucleus. 2012;3:404–410. doi: 10.4161/nucl.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maeshima K, Imai R, Tamura S, Nozaki T. Chromatin as dynamic 10-nm fibers. Chromosoma. 2014;123:225–237. doi: 10.1007/s00412-014-0460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maeshima K, et al. Nucleosomal arrays self-assemble into supramolecular globular structures lacking 30-nm fibers. EMBO J. 2016;35:1115–1132. doi: 10.15252/embj.201592660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura K, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Moyal L, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roscoe BP, Thayer KM, Zeldovich KB, Fushman D, Bolon DN. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J. Mol. Biol. 2013;425:1363–1377. doi: 10.1016/j.jmb.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Batta K, Zhang Z, Yen K, Goffman DB, Pugh BF. Genome-wide function of H2B ubiquitylation in promoter and genic regions. Genes Dev. 2011;25:2254–2265. doi: 10.1101/gad.177238.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J. Mol. Biol. 1987;194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 43.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 44.Pettersen EF, et al. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 45.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J. Mol. Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 46.Flaus A, Richmond TJ. Positioning and stability of nucleosomes on MMTV 3'LTR sequences. J. Mol. Biol. 1998;275:427–441. doi: 10.1006/jmbi.1997.1464. [DOI] [PubMed] [Google Scholar]
- 47.Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat. Methods. 2005;2:377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 48.Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. J. Mol. Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 49.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 50.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 51.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; [Google Scholar]
- 52.Ueda T, Chou H, Kawase T, Shirakawa H, Yoshida M. Acidic C-tail of HMGB1 is required for its target binding to nucleosome linker DNA and transcription stimulation. Biochemistry. 2004;43:9901–9908. doi: 10.1021/bi035975l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.