Abstract
Purpose
To successfully metastasize, tumor cells must respond appropriately to biological stressors encountered during metastatic progression. We sought to test the hypothesis that enhanced efficiency of mRNA translation during periods of metastatic stress is required for metastatic competence of osteosarcoma and that this metastasis-specific adaptation is amenable to therapeutic intervention.
Experimental Design
We employ novel reporter and proteomic systems that enable tracking of mRNA translation efficiency and output in metastatic osteosarcoma cells as they colonize the lungs. We test the potential to target mRNA translation as an anti-metastatic therapeutic strategy through pharmacokinetic studies and pre-clinical assessment of the prototypic mTOR inhibitor, rapamycin, across multiple models of metastasis.
Results
Metastatic osteosarcoma cells translate mRNA more efficiently than non-metastatic cells during critical stressful periods of metastatic colonization of the lung. Rapamycin inhibits translational output during periods of metastatic stress, mitigates lung colonization, and prolongs survival. mTOR-inhibiting exposures of rapamycin are achievable in mice using treatment schedules that correspond to human doses well below the maximum tolerated doses defined in human patients, and as such are very likely to be tolerated over long exposures alone and in combination with other agents.
Conclusions
Metastatic competence of osteosarcoma cells is dependent on efficient mRNA translation during stressful periods of metastatic progression and the mTOR inhibitor, rapamycin, can mitigate this translation and inhibit metastasis in vivo. Our data suggest that mTOR pathway inhibitors should be reconsidered in the clinic using rationally designed dosing schedules and clinical metrics related to metastatic progression.
Keywords: metastasis, osteosarcoma, mTOR, rapamycin, translation
Introduction
Decades of study have demonstrated that the biological drivers of metastatic progression are distinct from those of primary tumor formation(1). As cancer cells egress from primary tumors, travel to and arrest within distant secondary organs, and colonize those organs to form overt metastases, they encounter a number of cellular stresses not present in their primary tumor microenvironments. We and others have previously shown that the greatest stress and impediment to metastatic progression occurs after cells have extravasated into the microenvironments of distant secondary sites(2, 3). Therefore, in order to complete the steps of metastasis, cancer cells must possess the capacity to overcome these stresses in a dynamic fashion. One mechanism by which metastatic tumor cells may surmount metastatic stresses is through efficient translation of mRNA into protein in the face of nutrient deprivation and exogenous microenvironmental insults(4-6). Indeed, enhanced mRNA translation has been proposed to be a key mechanism endowing tumor cells with metastatic capacity(7). Previous studies by our group have shown the metastatic phenotype of osteosarcoma cells to be associated with an enhanced capacity to translate mRNA containing complex structure in 5’ untranslated regions (UTRs) in vitro, but not with global increases in translational output(8, 9). mRNA transcripts with complex 5’ UTR structure require the translation initiation machinery to actively deconvolute this structure before cap-dependent translation can occur(10, 11). As such, proteins whose transcripts contain highly structured 5’ UTRs have been termed “weakly translated” proteins based on their reduced rate of protein synthesis at baseline(8). Neither the timing of weakly translated protein synthesis during metastatic progression nor the molecular pathways regulating this biology have been described in osteosarcoma or other cancers.
The mammalian target of rapamycin (mTOR) kinase represents a key signaling node for cellular response to nutrient availability. mTOR acts downstream of the phosphatidylinositol-3-OH kinase (PI(3)K)-AKT signaling pathway through the formation of two distinct complexes: mTORC1 and mTORC2(12). The mTORC1 complex regulates protein synthesis through direct phosphorylation of 4EBP1 and p70S6K. Phosphorylation of these substrates leads to enhanced translational output and promotes cell growth, proliferation, and survival. mTOR pathway activation has been implicated in the development and progression of myriad human cancers including osteosarcoma(13). As such, mTOR inhibitors have been the subject of intensive pre-clinical and clinical investigations across tumor types.
Dysregulation of the PI3K/mTOR pathway has been implicated in osteosarcoma(14) and clinical trials of mTOR pathway inhibitors have been completed in osteosarcoma patients(15-17). However, these studies have largely focused on primary tumor biology and have not rigorously assessed effects of mTOR pathway inhibition on metastatic progression. Despite intensified combination chemotherapy regimens, clinical outcomes for osteosarcoma patients have not improved for over 25 years(18, 19). There is thus a pressing clinical need for the development of novel anti-metastatic therapeutic approaches. In the presented studies we test whether mTOR pathway inhibition may offer such an approach.
We hypothesize that enhanced mRNA translation during critical periods of metastatic stress is a key component of the osteosarcoma metastatic phenotype. Based on the knowledge that mTOR activation results in enhanced mRNA translation, we further hypothesize that dynamic translation during metastasis occurs in an mTOR-dependent fashion and that mTOR pathway inhibitors may have specific anti-metastatic properties.
Materials and Methods
Cell Culture
HOS, MG63, and MNNG were purchased from ATCC. MG63.3 cells were derived from MG63.2 (obtained from Dr. Hue Luu, University of Chicago, Chicago, IL)(20). AS1.46 and K7M2 were developed in our lab(20, 21). All cells were cultured as previously described(22). Cell lines were authenticated by STR profiling by the International Cell Line Authentication Committee in 2015(22).
Destabilized Stem-loop Reporter Production and Use
Cloning and lentiviral production completed on contract by the Advanced Technology Program, SAIC-Frederick, Inc., NCI, NIH.
Plasmid Construction and Lentiviral Production
Gateway Entry clone was produced containing a stem-loop sequence (5’-GATATCCCCTTGGACCTCCGTACCCTGCAGGGTACGGAGGTCCAAGGGGATATC-3’) with a predicted ΔG = −52.43kcal/mol preceding mCherry with a protein destabilizing PEST sequence. Expression clone was produced by incorporating the CMV promoter sequence upstream of the reporter construct. Lentivirus produced by standard methods.
Lentiviral Transduction and Selection
Transduction was performed in the presence of 8ug/ml polybrene. Cells were selected with 2ug/ml puromycin.
Mouse Studies
Animals were housed and handled in accordance with protocols approved by the NCI IACUC. 8-10 week old female BALB/c mice (Charles River) were used for allograft experiments. 8-10 week old female SCID-beige (Charles River) mice were used for xenograft experiments. Pharmacokinetic studies were completed in 8-10 week old female Athymic Nude mice (Charles River).
Ex Vivo Lung Metastasis Assay
The ex vivo lung metastasis assay is described elsewhere(23).
In Vivo/Ex Vivo Lung Metastasis Imaging
The in vivo/ex vivo lung metastasis assay is described elsewhere(2).
Characterization of Endogenous mRNA Translation by Highly Metastatic Cells in the Lung
Labeling of Newly Synthesized Proteins
Newly synthesized proteins were identified using the bioorthogonal non-canonical amino acid tagging, BONCAT, approach as previously described(24). Two highly metastatic human osteosarcoma cell lines (MG63.3 and MNNG) were compared to one non-metastatic cell line (MG63). For all cell lines, in vitro and ex vivo lung culture arms of the experiment were completed in triplicate. For the in vitro arm, 1×106 cells were plated in T75 flasks and grown in standard culture media with the methionine surrogate Click-iT L-azidohomoalanine, L-AHA, (Invitrogen) substituted for methionine. For the ex vivo lung culture arm, 5×105 cells were tail vein injected and lung sections were generated as described elsewhere(23). Custom lung culture media was made with L-AHA supplemented for methionine. Cells in each experimental arm were labeled over a 24hr growth period. Cells were then isolated and labeled protein was harvested for downstream tandem mass spectrometry analysis. Full description provided in supplement.
Isolation of Newly Synthesized Proteins
To isolate newly synthesized proteins labeled with L-AHA, a click reaction was performed followed by strepavadin bead pull down. A custom Biotin-Flag-Alkyne tag (NH2-Biocytin-GGADYKDDDDK-propargylglycine-CONH2) was synthesized on contract (GenScript Corporation) and made into 25mM stock solution in 1 × PBS. This peptide was used in combination with the Click-iT Protein Reaction Buffer Kit (Invitrogen) to covalently bind the peptide to AHA-labeled proteins. Click reaction, protein precipitation, and washing were performed according to the manufacturer’s protocol.
The samples were resolubilized in 50mM Tris, pH 7.5, 0.01% SDS. Labeled proteins bound to the biotin tag were then isolated using streptavidin coupled magnetic beads (Dynabeads® M-280 Streptavidin, Invitrogen). Each sample was added to 50uL of beads and incubated with rocking at room temperature for 5hrs. Beads were washed 5× 20min with 0.5% SDS in PBS. Streptavidin-biotin interaction was reversed by incubating beads in 0.5M Tris-HCL (pH 6.8), 4.4% SDS, 2% 2-mercaptoethanol.
Identification of Newly Synthesized Proteins
Peptide generation and mass spectrometric analysis was performed on contract by the Laboratory of Proteomics and Analytical Technologies, NCI. Full description of methods provided in supplement.
Analysis of Newly Translated Proteins
To increase proteome coverage, triplicates were pooled into a single data set for each condition. Pooled data sets were used to identify proteins commonly and specifically synthesized by highly metastatic cells within the lung microenvironment. Proteins identified in both highly metastatic cell lines (MG63.3 and MNNG) growing in ex vivo lung culture, but not in the in vitro controls for these cell lines nor the non-metastatic cell line (MG63) data sets (in vitro or ex vivo lung culture) were considered metastasis-specific. The structure and ΔG of the 5’ UTR of these genes was predicted using the ViennaRNA Package 2.0 software(25).
Immunoblotting
Western blot analysis was performed as described previously(26).
p70S6K Analysis
Cells were treated with rapamycin (0.1μM or 1μM) or vehicle for 4 hours. Blots were probed with mouse phospho-p70S6K (Thr389) (1:1000; Cell Signaling #9206) and rabbit total p70S6K (1:1000; Cell Signaling #9202) antibodies.
PTCHD2 Analysis
Blots were probed with rabbit PTCHD2 (ab113414, Abcam; 1μg/mL) antibody.
Rapamycin Pharmacokinetic Studies
Chronic Rapamycin Dosing Studies
Mice were treated three times per week with 5mg/kg rapamycin administered IP for two weeks. On Day 14 whole blood was drawn from mice just before rapamycin administration and at 2 and 6 hours post-treatment.
Single Rapamycin Dose Studies
Mice were given a single IP dose of 5mg/kg rapamycin (n=3 mice per condition).
Whole blood was extracted from each mouse at 0, 1, 2, 6, 24, and 48 hour time points.
Quantification of Rapamycin in Whole Blood
Rapamycin concentrations in whole blood were determined via a validated LC/MS/MS assay previously published(27).
Pharmacokinetic Analysis
Pharmacokinetic parameters were calculated by noncompartmental methods using Phoenix WinNonlin 6.3.0.395 (Certara USA). The Cmax, and trough levels were determined directly from the data and AUC0-48h after the first dose of rapamycin was calculated with the log-linear trapezoidal method.
In Vitro Assessment of Rapamycin Effect on mRNA Translation
K7M2 and MG63.3 cells were treated with either 0.1μM rapamycin or vehicle control in triplicate for 5 days, changing the media every other day. At the conclusion of this 5 day period, cells were imaged by inverted fluorescent microscopy (Leica DM IRB) at a magnification of 10×. 15 images × 3 replicates per condition were taken. Image analysis was performed using ImageJ software to quantify average ds-SL-mCherry intensity within GFP+ cell areas.
In Vivo Assessment of Rapamycin Effect on mRNA Translation
5×105 tumor cells were injected into the tail vein of each mouse (n=3 mice per condition). Tumors were allowed to develop for 21 days in the mice. Starting on day 22, mice were treated with daily IP doses of 5mg/kg rapamycin or vehicle control for 5 days (days 22-26). On day 27, lungs were extracted and imaged as described above. Five images were captured per mouse and ds-SL-mCherry expression was assessed within GFP+ tumor area. Following imaging, lungs were formalin fixed and embedded in paraffin for PTCHD2 staining as described below.
PTCHD2 Lung Metastasis Staining
In vivo expression of PTCHD2 was assessed in multiple experiments. Full description of sample preparation is described in the supplement. Sections were stained for PTCHD2 (Rabbit polyclonal IgG PTCHD2 antibody; ab113414, Abcam; 5μg/mL).
Phospho-S6 Staining
Lungs of mice receiving tail vein injection of 5×105 MG63 or MG63.3 cells were harvested 3 days after tail injection. Lung sections were prepared as described in supplement. Cells growing in vitro were also fixed and stained. Samples were stained to assess phosphorylation of Ser235/236 of S6 ribosomal protein (Rabbit polyclonal phospho-S6 antibody; Cell Signaling #2211), diluting primary antibody 1:80.
Assessment of Anti-Metastatic Properties of Rapamycin
Ex Vivo Lung Culture
5×105 tumor cells were injected into the tail vein of each mouse and lung sections were prepared as described elsewhere(23). Media of rapamycin-treated cultures was supplemented to a final concentration of 0.1μM or 1μM rapamycin. Vehicle treated culture media was supplemented with DMSO volumes matching rapamycin treatment. Media was changed and fresh rapamycin or DMSO was added every 2 days. A total of 8 lung sections were imaged for each condition. Lung sections were imaged and metastatic burden quantified as described above.
Experimental Metastasis Model
1-5×105 tumor cells were injected into the tail vein of each mouse (n=15 per condition). Mice were treated with daily IP doses of 5mg/kg rapamycin or vehicle control 5 days per week. Experiments were continued until mice died or became moribund and required euthanasia. All mice underwent necropsy examination to confirm metastasis.
Spontaneous Metastasis Model
1×105 to 1×106 tumor cells were injected orthotopically to the para-osseous proximal tibia as previously described(20). Mice were treated with daily IP doses of 5mg/kg rapamycin or vehicle control 5 days per week. Tumors were measured every 3-4 days. The volume of orthotopic tumor was calculated as previously reported (28). Tumor bearing limbs were amputated at a tumor size of 1.5-1.7 cm3. Experiments were continued until mice died or became moribund and required euthanasia. All mice underwent necropsy examination to confirmation metastasis. To control for differences in rate of primary tumor growth, survival times were calculated from the time of primary limb amputation.
Results
Highly metastatic osteosarcoma cells translate mRNA more efficiently than non-metastatic cells during critical periods of metastatic progression
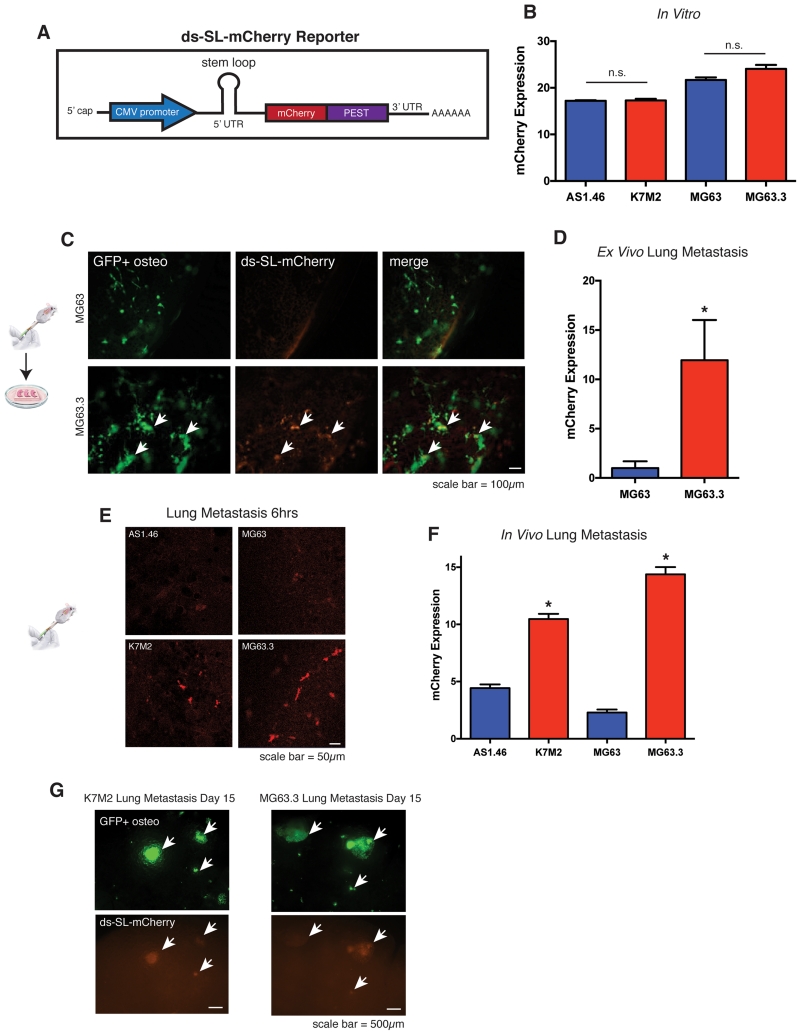
We first sought to determine whether highly metastatic osteosarcoma cells translate mRNA more efficiently than non-metastatic cells as they arrive to and colonize the lung microenvironment. We developed a novel fluorescent reporter to track the translational output of osteosarcoma cells in real-time in vivo. It has been shown that complex secondary structure within the 5’ UTR of mRNA transcripts reduces the rate of cap-dependent mRNA translation and this reduction is directly correlated with the predicted free energy (ΔG) of stem-loop structures in the 5’ UTR(10, 11). Based on this finding, we created a fluorescent mCherry reporter containing a stem-loop in the 5’ UTR with a predicted ΔG = −52.43 kcal/mol (Fig 1A). This degree of secondary structure was predicted to significantly inhibit translation of the reporter at baseline and was based on a previously published stem-loop sequence demonstrated to dramatically inhibit protein synthesis of a downstream cistron(8, 9). Thus, we used the reporter as a surrogate for increased translational output. We also incorporated a PEST sequence into the construct to reduce the half-life of the protein. We term this destabilized stem-loop reporter of mRNA translation ds-SL-mCherry.
Figure 1.
Highly metastatic osteosarcoma cells translate mRNA more efficiently than non-metastatic cells during critical periods of metastatic progression.
A) Schematic of ds-SL-mCherry reporter of mRNA translation illustrating CMV promoter, 5’ UTR stem-loop, mCherry reporter, and PEST protein destabilization sequence.
B) In vitro expression of ds-SL-mCherry, measured as mCherry pixel intensity within GFP+ cell area, in non-metastatic and highly metastatic murine (AS1.46 and K7M2) and human (MG63 and MG63.3) cell lines. Non-metastatic cell lines indicated by blue bars, highly metastatic cell lines indicated by red bars. Values represent averages of at least 50 individual cells +/− SEM. P-value does not reach significance using unpaired t-test with Welch’s correction.
C) Representative images of GFP+ ds-SL-mCherry MG63 (non-metastatic) and MG63.3 (metastatic) ex vivo lung sections at day 4. Images were captured using a 10× lens on a Leica-DM IRB inverted fluorescent microscope.
D) Quantification of ds-SL-mCherry expression, measured as mCherry pixel intensity within GFP+ tumor area, for MG63 and MG63.3 lung sections at day 4. Values represent averages of 10 lung sections (5 sections per mouse × 2 mice) +/− SD. * P-value calculated using unpaired t-test with Welch’s correction < 0.05.
E) In Vivo images of ds-SL-mCherry expression in AS1.46 and MG63 (non-metastatic) and K7M2 and MG63.3 (metastatic) cells in the lung 6hrs following tail-vein injection of 1×106 cells. Images were captured using a 20× lens on a Zeiss LSM-710 laser scanning confocal microscope.
F) Quantification of ds-SL-mCherry expression, measured as mCherry pixel intensity within GFP+ tumor area, for AS1.46 and MG63 (non-metastatic) and K7M2 and MG63.3 (metastatic) cells 6 hours after tail-vein injection. Values represent averages of 5 images per mouse × 5 mice per condition +/− SD. * P-value calculated using unpaired t-test with Welch’s correction < 0.05.
G) In Vivo images of GFP+ ds-SL-mCherry K7M2 (murine) and MG63.3 (human) highly metastatic cells in the lung 15 days following tail-vein injection of 1×106 cells. Images were captured using a 2.5× lens on a Leica-DM IRB inverted fluorescent microscope.
We transduced the ds-SL-mCherry reporter into highly metastatic murine and human osteosarcoma cells (K7M2 and MG63.3, respectively) as well as clonally related non-metastatic cell lines (AS1.46 and MG63, respectively). We show that the highly metastatic osteosarcoma cell lines, K7M2 and MG63.3, expressed this reporter at similar levels to their non-metastatic counterparts, AS1.46 and MG63, in vitro (Fig 1B). In order to investigate translational efficiency of highly metastatic cells during metastasis, we first imaged cells transduced with the ds-SL-mCherry reporter in ex vivo lung culture as previously described(23). We found that MG63.3 cells expressed the ds-SL-mCherry construct at higher levels than MG63 cells as they grow within the lung microenvironment in our ex vivo model (Fig. 1C and 1D). To test whether highly metastatic cells expressed the ds-SL-mCherry reporter at higher levels during critical periods of metastatic stress in fully in vivo models, we performed in vivo experimental metastasis assays. For these experiments, osteosarcoma cells were injected into the tail vein of mice followed by lung extraction and imaging 6 hours after injection. We found that both K7M2 and MG63.3 cells expressed higher levels of the reporter than AS1.46 and MG63 cells, as they arrive to and invade the lung microenvironment (Fig 1E and 1F). Further, we found that K7M2 and MG63.3 cells continue to express high levels of the reporter at later time points of metastatic lung colonization as they begin to form overt metastases (Fig 1G). These results indicate that increased translational output during stressful periods of metastatic invasion and colonization of the lung is associated with the osteosarcoma metastatic phenotype.
Proteins uniquely synthesized by highly metastatic osteosarcoma cells during colonization of the lung contain complex 5’ UTR structure
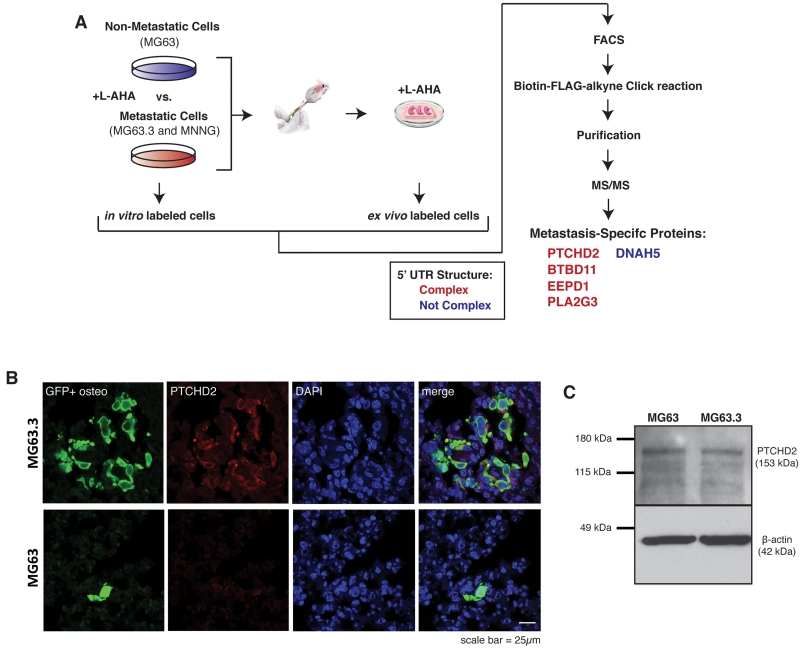
We next sought to identify endogenous proteins that were translated more efficiently by highly metastatic cells than by non-metastatic cells during metastatic colonization. To this end, we performed a proteomic experiment using an approach known as bioorthogonal non-canonical amino acid tagging (BONCAT)(24) to identify proteins uniquely synthesized by highly metastatic osteosarcoma cells as they arrive to and grow within the metastatic microenvironment of the lung (Fig 2A). Using the ex vivo model of pulmonary metastasis described, we grew lung sections containing GFP+ tail vein injected human osteosarcoma cells in methionine-free conditions supplemented with the methionine surrogate, L-azidohomoalanine (L-AHA). Lung sections were grown in these conditions for 24 hours so that any newly synthesized proteins would incorporate L-AHA in place of methionine. In parallel, the same cells were grown in standard in vitro culture with L-AHA substituted for methionine. At the conclusion of this labeling period, GFP+ tumor cells were sorted from in vitro culture and lung tissue, protein was isolated and newly synthesized proteins were enriched using a Click reaction of the azide group on L-AHA with a custom biotin-Flag-alkyne tag. Biotin-Flag-bound proteins were purified by streptavidin pull down and analyzed by tandem mass spectrometry. This approach was performed in triplicate for the highly metastatic human osteosarcoma cell lines MG63.3 and MNNG as well as the non-metastatic human osteosarcoma cell line MG63. Triplicate samples for each cell line were pooled to increase coverage. Pooling was necessary as the stringent selection of newly synthesized proteins substantially reduced the amount of protein isolated from sorted cells. 5 proteins were identified by unique peptides in both highly metastatic cell lines but no peptides in the non-metastatic cell line (Fig 2A). The structure and ΔG of the 5’ UTR of these genes was predicted using the ViennaRNA Package 2.0 software (Supplemental Fig 1)(25). Peptide counts and 5’ UTR ΔG values for identified proteins are reported in Supplemental Table 1. Based on previous studies(10), genes with predicted 5’ UTR ΔG’s of −30 kcal/mol or less were considered complex/weakly translated. Surprisingly, we found that mRNA of 4 out of 5 of these proteins possess complex 5’ UTR structure suggesting that their translation requires highly active translational machinery.
Figure 2.
Proteins uniquely synthesized by highly metastatic osteosarcoma cells during colonization of the lung contain complex 5’ UTR structure.
A) Schematic of experimental design for proteomic identification of newly synthesized proteins in ex vivo lung culture by bioorthogonal non-canonical amino acid tagging (BONCAT). Highly metastatic (MG63.3 and MNNG) and non-metastatic (MG63) human osteosarcoma cells were grown in vitro and in ex vivo lung culture in methionine-deplete media supplemented with the methionine surrogate L-azidohomoalanine (L-AHA) to label newly synthesized proteins over 24 hour growth period. GFP+ cells were isolated by FACS, newly synthesized proteins were purified and detected by tandem mass spectrometry. Proteins uniquely identified in highly metastatic cells growing in ex vivo lung culture are listed. The structure and ΔG of the 5’ UTR of mRNA for these genes were predicted using the ViennaRNA Package 2.0 software(25). Genes with predicted 5’ UTR ΔG’s of −30 kcal/mol or less were considered complex/weakly translated.
B) In Vivo images of GFP+ non-metastatic (MG63) and highly metastatic (MG63.3) cells in the lung 7 days following tail-vein injection of 1×106 cells. Sections stained for GFP, PTCHD2, and DAPI. Images were captured using a 63× lens on a Zeiss LSM-710 laser scanning confocal microscope.
C) Immunoblot analysis of PTCHD2 expression in non-metastatic (MG63) and highly metastatic (MG63.3) cells growing in in vitro culture. Images cropped to show relevant bands.
We confirmed increased expression of one of these genes, PTCHD2, by immunohistochemistry of in vivo metastatic lesions (Fig 2B). We found that PTCHD2 was expressed at high levels in MG63.3 cells growing within the lung, but was barely detectable in MG63 cells. We next tested whether PTCHD2 expression is increased in highly metastatic cells at baseline by immunoblotting protein isolated from MG63.3 and MG63 cells growing in vitro (Fig 2C). We found that outside of the metastatic microenvironment these cells express PTCHD2 at similar levels. PTCHD2 is a soluble activator of hedgehog signaling(29). While the result suggests that hedgehog signaling may play a role in osteosarcoma metastasis, for the purposes of this study, we simply used this protein as a surrogate for endogenous translation of an mRNA with complex 5’ UTR structure. Collectively, these results confirm that highly metastatic osteosarcoma cells translate a set of endogenous proteins more efficiently than non-metastatic cells as they arrive to and grow within the lung microenvironment.
mTOR-inhibiting rapamycin concentrations are achievable in vivo
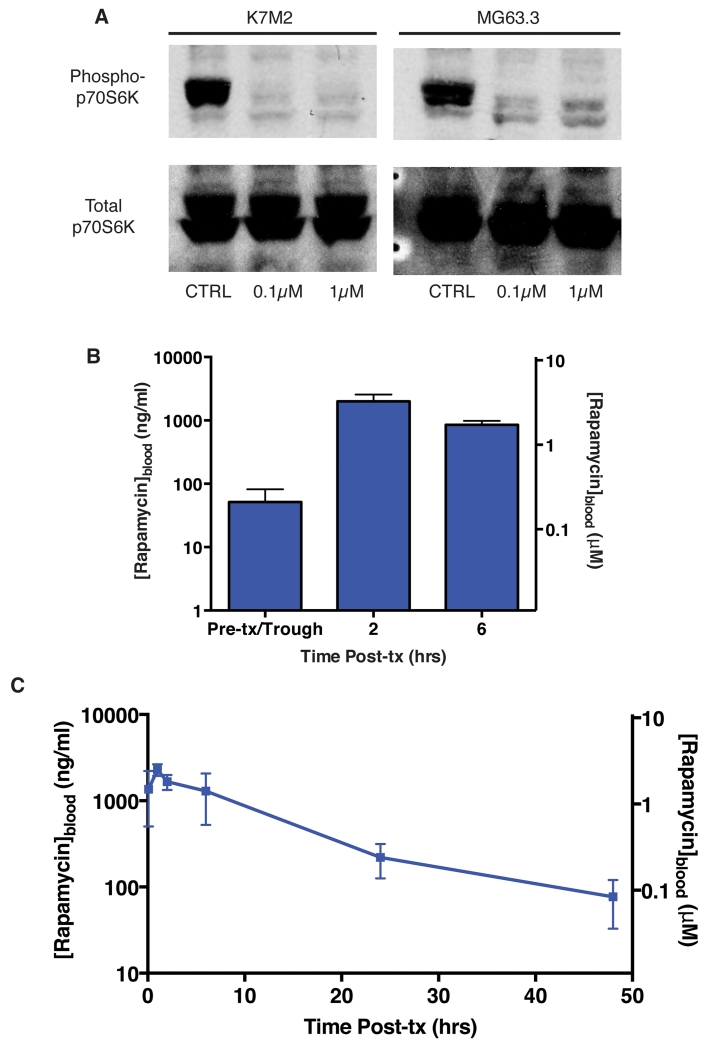
We next sought to test whether mTOR inhibition might interfere with mRNA translation during stressful periods of metastasis and therefore mitigate metastatic progression. Before initiating such studies, we performed in vitro studies of mTOR pathway inhibition in our cell lines and in vivo pharmacokinetic studies to ensure feasibility. We first established rapamycin exposures necessary to inhibit mTOR pathway activation. Phosphorylation of threonine 389 of p70S6K by the mTORC1 complex serves as a hallmark indicator of mTOR pathway activity(12). In order to test the inhibitory effect of rapamycin on mTOR pathway activity, we treated highly metastatic K7M2 and MG63.3 osteosarcoma cells with low (0.1μM) and high (1μM) concentrations of rapamycin. We demonstrate that rapamycin effectively inhibited phosphorylation of p70S6K at concentrations as low as 0.1μM (Fig 3A). Next, we completed pharmacokinetic studies following intraperitoneal (IP) rapamycin dosing in mice. We demonstrate that IP dosing of rapamycin at 5mg/kg three times per week for two weeks resulted in a trough exposure near 0.1μM (Fig 3B). We then treated naïve mice with a single IP dose of 5mg/kg rapamycin IP (Fig 3C). We found that this dosing schedule was sufficient to maintain whole blood concentrations above 0.1μM for 24hrs. Collectively, these results demonstrated that 5mg/kg IP dosing of rapamycin was sufficient to maintain whole blood concentrations of rapamycin above 0.1μM, an exposure shown to effectively inhibit p70S6K phosphorylation, for at least 24 hours between doses.
Figure 3.
Active exposures of rapamycin are achievable in vivo at low doses.
A) Immunoblot for phospho- and total p70S6K in K7M2 (murine) and MG63.3 (human) highly metastatic osteosarcoma cells treated with vehicle, 0.1μM rapamycin, or 1μM rapamycin for 4 hours. Images cropped to show relevant bands.
B) Whole blood rapamycin levels on day 14 in mice receiving 5mg/kg IP rapamycin three times per week. Blood was drawn on day 14 prior to treatment (trough) as well as 2 and 6 hours following IP administration of 5mg/kg rapamycin. N=3 mice.
C) Whole blood rapamycin levels in naïve mice treated with a single dose of 5mg/kg rapamycin IP. Whole blood was collected immediately following rapamycin administration (0 hours) and at 2, 6, 24, and 48 hour time points. N=3 mice.
Rapamycin inhibits mRNA translation by highly metastatic osteosarcoma cells within the lung microenvironment
We next sought to determine whether mTOR activity correlated with metastatic progression of osteosarcoma cells. To test this, we assessed phosphorylation of ribosomal protein S6, a robust marker of mTORC1 activity(30). Consistent with our in vitro reporter studies (Fig 1B), we found that phospho-S6 levels were similar between highly and non-metastatic cells in vitro (Supplemental Fig 2A and 2B), but increased during metastatic progression in highly metastatic cells (Supplemental Fig 2C and 2D). These findings support our hypothesis that mTOR pathway activity is likely necessary for efficient mRNA translation during metastatic progression and provide a biological rationale for further investigation of mTOR pathway inhibitors as therapeutic agents in the treatment of metastatic osteosarcoma.
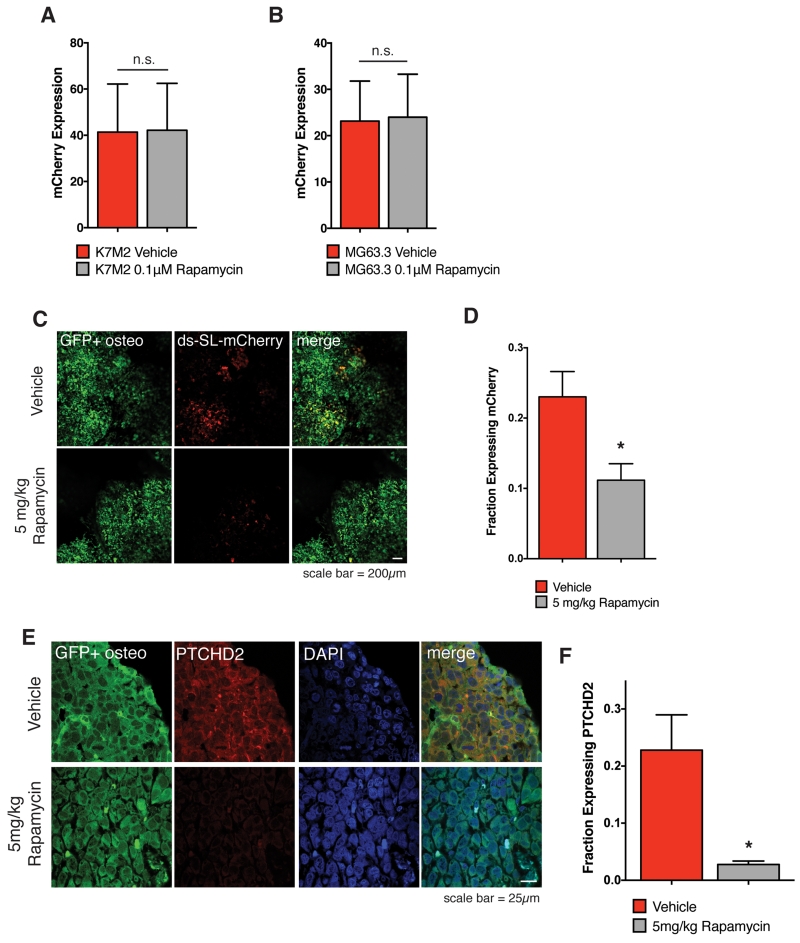
We next tested the effect of rapamycin treatment on mRNA translation within the metastatic microenvironment of the lung. We first used the ds-SL-mCherry reporter as a surrogate for translational output. We found that 1μM rapamycin had no effect on reporter expression in vitro (Fig 4A and 4B). To test the in vivo effect of rapamycin on ds-SL-mCherry expression during metastatic colonization, we completed an experimental metastasis assay with GFP+ MG63.3 cells transduced with the ds-SL-mCherry reporter. We initially tested the effect of mTOR inhibition on translation at early points of metastatic colonization. However, due to high rates of cell attrition that resulted from rapamycin treatment, we were not able to adequately assess effects on mRNA translation at these early time points. Therefore, to test the biological activity of mTOR inhibition, we treated cells at later time points of metastatic colonization with rapamycin. We allowed metastases to develop for 21 days and then treated mice with daily IP doses of 5mg/kg rapamycin or vehicle control for 5 consecutive days (days 22-26). On day 27, we extracted the lungs and imaged them to assess ds-SL-mCherry expression. We found that IP rapamycin treatment of 5mg/kg significantly reduced expression of the construct relative to vehicle control (Fig 4C and 4D). Finally, to test the effects of rapamycin treatment on endogenous mRNA translation, we assessed lung metastases from vehicle and 5mg/kg rapamycin treated mice for PTCHD2 expression. Similar to the ds-SL-mCherry reporter, we found that rapamycin treatment significantly reduced expression of PTCHD2 (Fig 4E and 4F). The continued expression of the GFP reporter lacking a stem-loop structure in the 5’ UTR by rapamycin treated cells suggests that rapamycin may be most potent at inhibiting translation of protein with highly structured 5’ UTRs. These results demonstrate that mRNA translation by highly metastatic cells within the lung microenvironment is attenuated by rapamycin treatment. We reasoned that if this translation is critical to metastatic success then rapamycin may indeed have unique activity in the setting of metastasis that would support its reconsideration as a useful therapeutic agent for the treatment of osteosarcoma.
Figure 4.
Rapamycin inhibits translation of complex mRNA transcripts by highly metastatic osteosarcoma cells within the lung microenvironment.
A) Expression of ds-SL-mCherry reporter in highly metastatic murine K7M2 cells exposed to 0.1μM rapamycin or vehicle for 5 days in culture. Values represent averages from 15 images per plate × 3 plates per condition +/− SD. P-value does not reach significance using unpaired t-test with Welch’s correction.
B) Expression of ds-SL-mCherry reporter in highly metastatic human MG63.3 cells exposed to 0.1μM rapamycin or vehicle for 5 days in culture. Values represent averages of 15 images per plate × 3 plates per condition +/− SD. P-value does not reach significance using unpaired t-test with Welch’s correction.
C) In Vivo images of GFP+ ds-SL-mCherry MG63.3 lung metastases on day 27 of an experimental metastasis assay following 5 daily IP doses of vehicle or 5mg/kg rapamycin (days 22-26). Images were captured using a 5× lens on a Leica-DM IRB inverted fluorescent microscope.
D) Quantification of fraction of GFP+ tumor area expressing ds-SL-mCherry reporter in lung metastases following 5 daily IP doses of vehicle or 5mg/kg rapamycin. Values represent averages of 5 images per mouse × 3 mice per condition +/− SD. * P-value calculated using unpaired t-test with Welch’s correction < 0.05.
E) Images of PTCHD2 in GFP+ MG63.3 lung metastases on day 27 of an experimental metastasis assay following 5 daily IP doses of vehicle or 5mg/kg rapamycin (days 22-26). Images were captured using a 63× lens on a Zeiss LSM-710 laser scanning confocal microscope.
F) Quantification of fraction of GFP+ tumor area expressing PTCHD2 in lung metastases following 5 daily IP doses of vehicle or 5mg/kg rapamycin. Values represent averages of 2 images per mouse × 3 mice per condition +/− SD. * P-value calculated using unpaired t-test with Welch’s correction < 0.05.
Rapamycin inhibits growth of highly metastatic murine and human osteosarcoma cells in the lung microenvironment
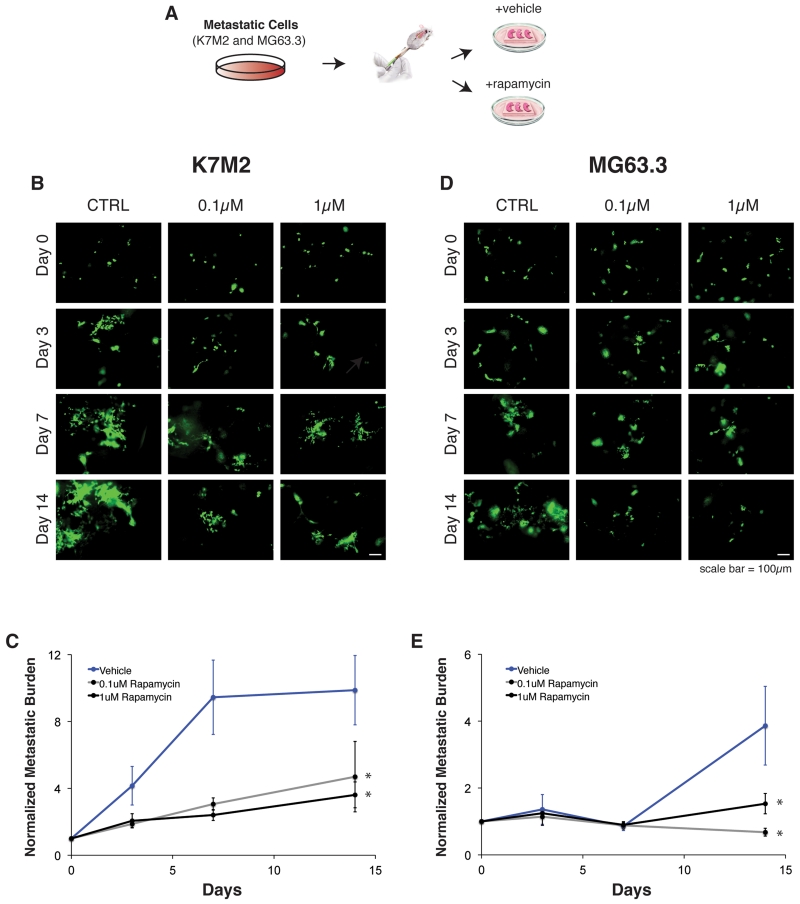
Based on the premise that mTOR-mediated mRNA translation is uniquely involved in the metastatic progression of osteosarcoma, we sought to test the anti-metastatic effects of mTOR inhibition. We first tested this in our ex vivo model of pulmonary metastasis (Fig 5A). We found that 0.1μM and 1μM concentrations of rapamycin robustly inhibited metastatic outgrowth of highly metastatic murine, K7M2, (Fig 5B and 5C) and human, MG63.3, (Fig 5D and 5E) osteosarcoma cell lines. We observed that rapamycin had nearly identical anti-metastatic effects at low (0.1μM) and high (1μM) concentrations. This was consistent with our in vitro results showing that p70S6K phosphorylation was inhibited to an equivalent degree across these concentrations (Fig 3A). These results indicate that inhibition of mTOR signaling is sufficient to mitigate outgrowth of highly metastatic osteosarcoma cells within the lung microenvironment and support additional pre-clinical testing of mTOR inhibition as an anti-metastatic therapeutic strategy.
Figure 5.
Rapamycin inhibits growth of highly metastatic cells in the lung microenvironment.
A) Schematic of experimental design.
B) Representative images of vehicle (DMSO), 0.1μM rapamycin, and 1μM rapamycin treated lung sections containing K7M2 cells. Images were captured using a 10× lens on a Leica-DM IRB inverted fluorescent microscope.
C) Kinetics of metastatic outgrowth of metastatic K7M2 cells in ex vivo lung culture with vehicle (DMSO), 0.1μM rapamycin, or 1μM rapamycin treatment. Metastatic burden measured as total GFP+ area per lung section normalized to GFP+ area on day 0. Values represent averages of 8 lung sections +/−SEM. * P-value calculated using unpaired t-test with Welch’s correction < 0.05.
D) Representative images of vehicle (DMSO), 0.1μM rapamycin, and 1μM rapamycin treated lung sections containing MG63.3 cells. Images were captured using a 10× lens on a Leica-DM IRB inverted fluorescent microscope.
E) Kinetics of metastatic outgrowth of metastatic MG63.3 cells in ex vivo lung culture with vehicle (DMSO), 0.1μM rapamycin, or 1μM rapamycin treatment. Metastatic burden measured as total GFP+ area per lung section normalized to GFP+ area on day 0. Values represent averages of 8 lung sections (4 sections per mouse × 2 mice) +/−SEM. * P-value calculated using unpaired t-test with Welch’s correction < 0.05.
Rapamycin shows anti-metastatic activity in murine allograft and human xenograft models of osteosarcoma
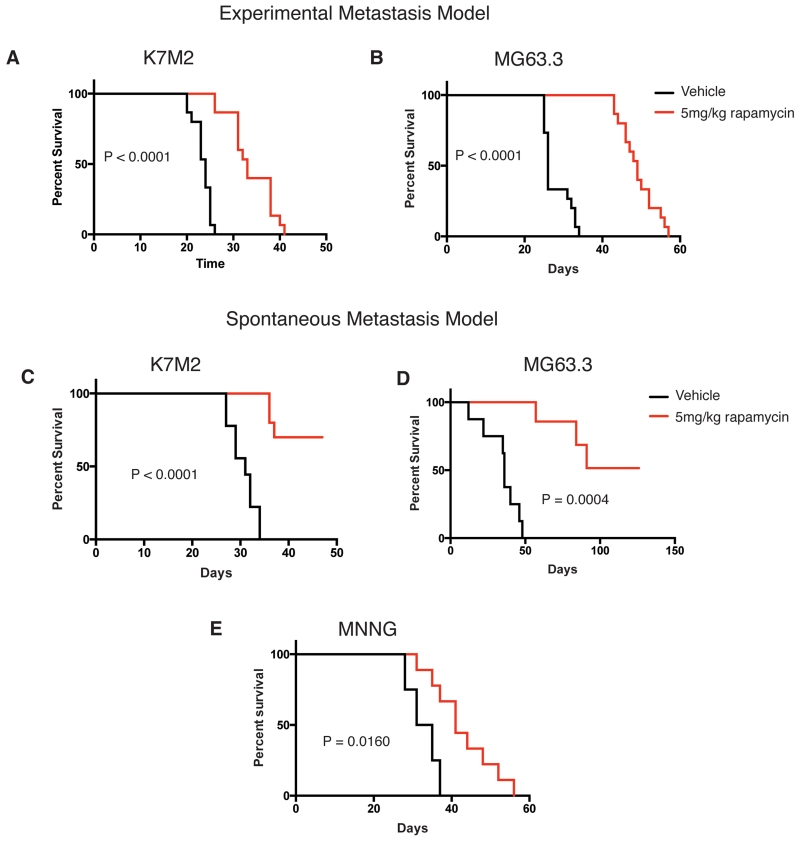
To test the anti-metastatic effect of rapamycin in vivo, we used multiple allograft and xenograft models of highly metastatic murine and human osteosarcoma. Based on the pharmacokinetic data, we chose to administer daily IP doses of 5mg/kg rapamycin or vehicle control for these experiments. This dosing schedule was chosen to ensure that pharmacologically active exposures of rapamycin were maintained between doses. First, we tested the effect of rapamycin treatment in an experimental metastasis assay with highly metastatic murine (K7M2) and human (MG63.3) cells (Fig 6A and 6B). We show that rapamycin treatment significantly prolonged survival of mice relative to vehicle controls.
Figure 6.
Rapamycin inhibits osteosarcoma lung metastasis and prolongs survival in vivo.
A) Kaplan-Meier plot of vehicle and 5mg/kg treated mice tail-vein injected with 5×105 K7M2 cells (N=15 mice per condition). P-value calculated by Gehan-Breslow-Wilcoxon Test.
B) Kaplan-Meier plot of vehicle and 5mg/kg treated mice tail-vein injected with 1×105 MG63.3 cells (N=15 mice per condition). P-value calculated by Gehan-Breslow-Wilcoxon Test.
C) Kaplan-Meier plot of vehicle and 5mg/kg treated mice orthotopically injected with 1×106 K7M2 cells (N = at least 9 mice per condition). Tumor bearing limbs were amputated at a tumor size of 1.5-1.7cm3. Survival times were calculated from the time of primary limb amputation. P-value calculated by Gehan-Breslow-Wilcoxon Test.
D) Kaplan-Meier plot of vehicle and 5mg/kg treated mice orthotopically injected with 1×105 MG63.3 cells (N= at least 7 mice per condition). Tumor bearing limbs were amputated at a tumor size of 1.5-1.7cm3. Survival times were calculated from the time of primary limb amputation. P-value calculated by Gehan-Breslow-Wilcoxon Test.
E) Kaplan-Meier plot of vehicle and 5mg/kg treated mice orthotopically injected with 1×106 MNNG cells (N= at least 4 mice per condition). Tumor bearing limbs were amputated at a tumor size of 1.5-1.7cm3. Survival times were calculated from the time of primary limb amputation. P-value calculated by Gehan-Breslow-Wilcoxon Test.
We next tested the effect of rapamycin on metastasis in fully orthotopic spontaneous metastasis models of murine and human osteosarcoma. To control for heterogeneous primary tumor growth, we amputated tumor-bearing limbs of mice when tumors reached a size of 1.5-1.7 cm3 and calculated survival times from the time of limb amputation. We demonstrate that 5mg/kg daily IP doses of rapamycin significantly prolonged survival of mice orthotopically injected with highly metastatic murine (K7M2) and human (MG63.3) cells and verified this result in an additional highly metastatic human osteosarcoma cell line, MNNG (Fig 6C-6E). These results confirmed that mTOR inhibition via rapamycin treatment is sufficient to delay metastatic progression in spontaneous, fully orthotopic models of osteosarcoma metastasis. Further, the results demonstrated that anti-metastatic effects of mTOR pathway inhibition are independent of effects on primary tumor growth. Collectively, these findings support the notion that mTOR pathway inhibition may indeed have unique anti-metastatic activity in human osteosarcoma and argue in favor of reconsideration of rapamycin and other mTOR inhibitors in the clinical arena.
Given the observed anti-metastatic activities of rapamycin in pre-clinical models of osteosarcoma, we sought to identify and develop a therapeutic rapamycin exposure that would be expected to exert anti-metastatic activities in animal models of osteosarcoma, both inducible and naturally occurring. To this end, we utilized the pharmacokinetic profiles of IP rapamycin generated in mice treated with a dosage of rapamycin found to exert anti-metastatic activities, 5mg/kg (Fig. 3), and calculated total rapamycin exposure as determined by noncompartmental methods related to dosage. Derived from the tandem pharmacokinetic and anti-metastatic data, it was predicted that the achievement of an AUC/dose equivalent value of 3,555 ([ng*hr/ml]/[mg/kg]) would be sufficient to inhibit p70S6K phosphorylation and exert anti-metastatic activities in animal models of osteosarcoma. This data is centrally important for the rational derivation of future rapamycin dosing strategies intended to delay metastatic progression in pediatric and adult osteosarcoma patients.
Discussion
In the described studies we show that the osteosarcoma metastatic phenotype is associated with mTOR pathway activation and enhanced mRNA translation during periods of cellular stress encountered as cells arrive to and grow within the metastatic microenvironment of the lung. We demonstrate that 0.1μM exposures of rapamycin are sufficient to inhibit mTOR activity and that this inhibition is associated with mitigated translational output of highly metastatic cells during metastatic progression. Finally, we find that dosing schedules of rapamycin achieving mTOR-inhibiting exposures are sufficient to abrogate osteosarcoma metastasis and prolong survival across multiple pre-clinical experimental model systems.
While our studies have identified new aspects of the osteosarcoma metastatic phenotype that are amenable to therapeutic intervention, they also raise additional questions about the mechanism and consequence of enhanced translation during metastatic progression. First, the specific stressors encountered by tumor cells within the metastatic microenvironment that require metastatic cells to alter their translational output remain to be identified. While we have investigated a number of possible stressors including anchorage independent growth, serum starvation, and altered pH among others, we have yet to identify the stimulus of this biology. Second, it is possible that specific translation factors are overexpressed in highly metastatic cells that result in the observed enhanced rates of protein synthesis of mRNA transcripts with highly structured 5’ UTRs. Others have demonstrated that a number of factors including eIF4A, eIF4B, eIF4E, eIF4H, and eIF4G(9, 11, 31) may play a role in this biology. Previous studies by our group investigating the role of some of these factors in osteosarcoma metastasis have failed to provide a definitive answer to this question(32). It is possible that genetic or epigenetic alterations to the genomes of highly metastatic osteosarcoma cells may lead to altered expression and/or function of such translation factors. However, neither highly recurrent genetic nor epigenetic changes have been associated with the osteosarcoma metastatic phenotype to date(19). Finally, it would be informative to more fully understand the biological consequence of enhanced mRNA translation during metastatic progression. Specifically, understanding the functions of proteins synthesized more efficiently by highly metastatic cells than non-metastatic cells during metastatic colonization would not only shed new light on the molecular drivers of metastasis but may provide further opportunities for therapeutic intervention. Future studies may reasonably seek to provide answers to these questions and are supported by the data presented in this manuscript.
Collectively, our results indicate that mTOR inhibition is an effective means of delaying metastatic progression of osteosarcoma and provide an argument in favor of reconsidering the adjuvant use of rapamycin and other mTOR inhibitors in the clinical setting. As the majority of osteosarcoma deaths are caused by metastatic progression to the lung, the most pressing clinical need for these patients is the development of treatment strategies to effectively slow, prevent, and/or reverse metastatic progression of this aggressive cancer. We recently published a set of drug development guidelines with other experts in the clinical and research communities outlining pre-clinical and clinical approaches for the evaluation of potential therapeutic agents that uniquely target molecular and cellular processes leading to osteosarcoma metastasis(33). This paradigm should reasonably be applied not only to the investigation of novel therapeutic agents, but also to compounds that may not have undergone rigorous assessment for anti-metastatic properties in prior pre-clinical or clinical studies, such as mTOR inhibitors.
Based upon our pre-clinical pharmacokinetic data, we derived what we believe to be a justifiable AUC/dose equivalent target for slowing micrometastatic disease progression through rapamycin’s inhibition of mTOR. Validation of our proposed AUC/dose equivalent target for delaying metastatic progression will require further evaluation in additional tumor models that faithfully recapitulate the inherent cellular stressors encountered by metastatically successful osteosarcoma cells; and such an opportunity currently exists through the inclusion of dogs with osteosarcoma for evaluating rapamycin’s anti-metastatic activities. The data presented in this study have supported the launch of a prospective adjuvant study of rapamycin treatment in dogs with osteosarcoma. Based on the outcome of this study it would be our intent to propose a similar study in pediatric osteosarcoma patients. These pre-human trials in dogs will be informative of the potential benefit to osteosarcoma patients and of proper dosing strategies to maximize anti-metastatic effects while minimizing toxicity. Should such dog trials be successful, human trials carefully designed to test the anti-metastatic effects of mTOR inhibition in osteosarcoma may be warranted.
Supplementary Material
Translational Relevance.
In this study, we identify enhanced mRNA translation during stressful periods of metastatic progression as a therapeutic vulnerability in osteosarcoma. We show that the mTOR inhibitor, rapamycin, effectively targets this biology and provides substantial therapeutic benefit in multiple pre-clinical metastatic progression models. Our data provide a novel biological rationale for the therapeutic inhibition of mTOR that has not previously been considered in the clinical development and use of these agents. Our findings suggest that mTOR inhibitors may be best developed as anti-metastatic agents, and that a failure to do so will preclude their true value from being demonstrated in the clinic. Furthermore, we present the first pharmacokinetic data for rapamycin dosing in mice with correlation to metastatic outcome that poise our findings for immediate translation to rationally designed clinical trials.
Acknowledgements
The authors thank Joseph W. Briggs, Rosandra N. Kaplan, and Lee J. Helman for technical discussions of this project and Patricia A. Hebda for critical review of the manuscript.
Financial support:
This work was supported by funding from the NIH CA186633 (J.J.M.); GM007250 (J.J.M.); NIH Intramural Visiting Fellow Program #15335 (M.M.L.), R01CA160356 (P.C.S.); 1R01CA193677 (P.C.S); P30CA046934 (R.J.H and D.L.G.); and the NIH Intramural Research Program (C.K.).
Footnotes
Author’s Contributions
Conception and design: J. Morrow and C. Khanna
Development of methodology: J. Morrow
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): J. Morrow, A. Mendoza, A. Koyen, M. Lizardo, L. Ren, T. Waybright, R. Hansen, D. Gustafson, M. Zhou.
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): J. Morrow, T. Waybright, R. Hansen, D. Gustafson, M. Zhou, T. Fan.
Writing, review, and/or revision of the manuscript: J. Morrow, A. Mendoza, A. Koyen, T. Waybright, R. Hansen, P. Scacheri, C. Khanna.
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): N/A
Study supervision: P. Scacheri, C. Khanna.
Conflicts of interest:
The authors have no conflicts of interest to disclose.
References
- 1.Vanharanta S, Massague J. Origins of metastatic traits. Cancer cell. 2013;24:410–21. doi: 10.1016/j.ccr.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong SH, Ren L, Mendoza A, Eleswarapu A, Khanna C. Apoptosis resistance and PKC signaling: distinguishing features of high and low metastatic cells. Neoplasia (New York, NY) 2012;14:249–58. doi: 10.1593/neo.111498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nature reviews Cancer. 2002;2:563–72. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 4.Leprivier G, Rotblat B, Khan D, Jan E, Sorensen PH. Stress-mediated translational control in cancer cells. Biochim Biophys Acta. 2015;1849:845–60. doi: 10.1016/j.bbagrm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Liu B, Qian SB. Translational reprogramming in cellular stress response. Wiley interdisciplinary reviews RNA. 2014;5:301–15. doi: 10.1002/wrna.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loreni F, Mancino M, Biffo S. Translation factors and ribosomal proteins control tumor onset and progression: how? Oncogene. 2014;33:2145–56. doi: 10.1038/onc.2013.153. [DOI] [PubMed] [Google Scholar]
- 7.Graff JR, Zimmer SG. Translational control and metastatic progression: enhanced activity of the mRNA cap-binding protein eIF-4E selectively enhances translation of metastasis-related mRNAs. Clinical & experimental metastasis. 2003;20:265–73. doi: 10.1023/a:1022943419011. [DOI] [PubMed] [Google Scholar]
- 8.Briggs JW, Ren L, Nguyen R, Chakrabarti K, Cassavaugh J, Rahim S, et al. The ezrin metastatic phenotype is associated with the initiation of protein translation. Neoplasia (New York, NY) 2012;14:297–310. doi: 10.1593/neo.11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang HS, Cho MH, Zakowicz H, Hegamyer G, Sonenberg N, Colburn NH. A novel function of the MA-3 domains in transformation and translation suppressor Pdcd4 is essential for its binding to eukaryotic translation initiation factor 4A. Molecular and cellular biology. 2004;24:3894–906. doi: 10.1128/MCB.24.9.3894-3906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–70. [PubMed] [Google Scholar]
- 11.Gray NK, Hentze MW. Regulation of protein synthesis by mRNA structure. Mol Biol Rep. 1994;19:195–200. doi: 10.1007/BF00986961. [DOI] [PubMed] [Google Scholar]
- 12.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–94. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Populo H, Lopes JM, Soares P. The mTOR signalling pathway in human cancer. International journal of molecular sciences. 2012;13:1886–918. doi: 10.3390/ijms13021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry JA, Kiezun A, Tonzi P, Van Allen EM, Carter SL, Baca SC, et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc Natl Acad Sci U S A. 2014;111:E5564–73. doi: 10.1073/pnas.1419260111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chawla SP, Staddon AP, Baker LH, Schuetze SM, Tolcher AW, D’Amato GZ, et al. Phase II study of the mammalian target of rapamycin inhibitor ridaforolimus in patients with advanced bone and soft tissue sarcomas. J Clin Oncol. 2012;30:78–84. doi: 10.1200/JCO.2011.35.6329. [DOI] [PubMed] [Google Scholar]
- 16.Schuetze SM, Zhao L, Chugh R, Thomas DG, Lucas DR, Metko G, et al. Results of a phase II study of sirolimus and cyclophosphamide in patients with advanced sarcoma. Eur J Cancer. 2012;48:1347–53. doi: 10.1016/j.ejca.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 17.Demetri GD, Chawla SP, Ray-Coquard I, Le Cesne A, Staddon AP, Milhem MM, et al. Results of an international randomized phase III trial of the mammalian target of rapamycin inhibitor ridaforolimus versus placebo to control metastatic sarcomas in patients after benefit from prior chemotherapy. J Clin Oncol. 2013;31:2485–92. doi: 10.1200/JCO.2012.45.5766. [DOI] [PubMed] [Google Scholar]
- 18.Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clinical advances in hematology & oncology : H&O. 2010;8:705–18. [PubMed] [Google Scholar]
- 19.Morrow JJ, Khanna C. Osteosarcoma Genetics and Epigenetics: Emerging Biology and Candidate Therapies. Crit Rev Oncog. 2015;20:173–97. doi: 10.1615/critrevoncog.2015013713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khanna C, Prehn J, Yeung C, Caylor J, Tsokos M, Helman L. An orthotopic model of murine osteosarcoma with clonally related variants differing in pulmonary metastatic potential. Clinical & experimental metastasis. 2000;18:261–71. doi: 10.1023/a:1006767007547. [DOI] [PubMed] [Google Scholar]
- 21.Khanna C, Wan X, Bose S, Cassaday R, Olomu O, Mendoza A, et al. The membrane-cytoskeleton linker ezrin is necessary for osteosarcoma metastasis. Nat Med. 2004;10:182–6. doi: 10.1038/nm982. [DOI] [PubMed] [Google Scholar]
- 22.Ren L, Mendoza A, Zhu J, Briggs J, Halsey C, Hong E, et al. Characterization of the metastatic phenotype of a panel of established osteosarcoma cells. Oncotarget. 2015 doi: 10.18632/oncotarget.5177. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendoza A, Hong SH, Osborne T, Khan MA, Campbell K, Briggs J, et al. Modeling metastasis biology and therapy in real time in the mouse lung. The Journal of clinical investigation. 2010;120:2979–88. doi: 10.1172/JCI40252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dieterich DC, Lee JJ, Link AJ, Graumann J, Tirrell DA, Schuman EM. Labeling, detection and identification of newly synthesized proteomes with bioorthogonal non-canonical amino-acid tagging. Nat Protoc. 2007;2:532–40. doi: 10.1038/nprot.2007.52. [DOI] [PubMed] [Google Scholar]
- 25.Lorenz R, Bernhart SH, Honer Zu Siederdissen C, Tafer H, Flamm C, Stadler PF, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren L, Hong SH, Cassavaugh J, Osborne T, Chou AJ, Kim SY, et al. The actin-cytoskeleton linker protein ezrin is regulated during osteosarcoma metastasis by PKC. Oncogene. 2009;28:792–802. doi: 10.1038/onc.2008.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larson JC, Allstadt SD, Fan TM, Khanna C, Lunghofer PJ, Hansen RJ, et al. Pharmacokinetics of orally administered low-dose rapamycin in healthy dogs. Am J Vet Res. 2016;77:65–71. doi: 10.2460/ajvr.77.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Werner S, Mendoza A, Hilger RA, Erlacher M, Reichardt W, Lissat A, et al. Preclinical studies of treosulfan demonstrate potent activity in Ewing’s sarcoma. Cancer Chemother Pharmacol. 2008;62:19–31. doi: 10.1007/s00280-007-0566-9. [DOI] [PubMed] [Google Scholar]
- 29.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14:416–29. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 30.Tabernero J, Rojo F, Calvo E, Burris H, Judson I, Hazell K, et al. Dose- and schedule-dependent inhibition of the mammalian target of rapamycin pathway with everolimus: a phase I tumor pharmacodynamic study in patients with advanced solid tumors. J Clin Oncol. 2008;26:1603–10. doi: 10.1200/JCO.2007.14.5482. [DOI] [PubMed] [Google Scholar]
- 31.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–80. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 32.Osborne TS, Ren L, Healey JH, Shapiro LQ, Chou AJ, Gorlick RG, et al. Evaluation of eIF4E expression in an osteosarcoma-specific tissue microarray. Journal of pediatric hematology/oncology. 2011;33:524–8. doi: 10.1097/MPH.0b013e318223d0c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khanna C, Fan TM, Gorlick R, Helman LJ, Kleinerman ES, Adamson PC, et al. Toward a drug development path that targets metastatic progression in osteosarcoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2014;20:4200–9. doi: 10.1158/1078-0432.CCR-13-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.