Abstract
Over the past decade, there has been both great interest and confusion about whether recent demographic events—notably the Out-of-Africa-bottleneck and recent population growth—have led to differences in mutation load among human populations. The confusion can be traced to the use of different summary statistics to measure load, which lead to apparently conflicting results. We argue, however, that when statistics more directly related to load are used, the results of different studies and data sets consistently reveal little or no difference in the load of non-synonymous mutations among human populations. Theory helps to understand why no such differences are seen, as well as to predict in what settings they are to be expected. In particular, as predicted by modeling, there is evidence for changes in the load of recessive loss of function mutations in founder and inbred human populations. Also as predicted, eastern subspecies of gorilla, Neanderthals and Denisovans, who are thought to have undergone reductions in population sizes that exceed the human Out-of-Africa bottleneck in duration and severity, show evidence for increased load of non-synonymous mutations (relative to western subspecies of gorillas and modern humans, respectively). A coherent picture is thus starting to emerge about the effects of demographic history on the mutation load in populations of humans and close evolutionary relatives.
Introduction
The recent demographic history of human populations is reflected in their distributions of genetic variation. For instance, Europeans and Asians harbor a greater fraction of high frequency variants compared to Africans, likely due to an ancient “Out-of-Africa” bottleneck [1–4], and all of these populations harbor numerous rare variants resulting from more recent explosive growth [4–9]. Genetic variation in human populations has also been affected by founder events [10–12], by inbreeding [13,14], and by extensive admixture among populations [15,16] and with archaic humans [17,18].
It is therefore natural to ask whether recent demographic events also affected the burden of deleterious mutations, leading it to differ among extant human populations. In addition to the observation that overall patterns of genetic diversity vary among populations with different demographic histories, theory suggests that, at equilibrium, the efficiency with which purifying selection removes deleterious variation is profoundly affected by population size and degree of inbreeding [19–23]. With data now available to address the question, the possibility that there exist differences in the burden of deleterious mutation among human populations has garnered considerable attention.
Answers to this question have been confusing, because many studies appear to reach conflicting conclusions (cf. [24–26]). In trying to sort out the source of the conflicts, we start by reviewing what is meant by the burden of deleterious mutations. Traditionally, this burden has been quantified in terms of the mutation load (sometime abbreviated by load below), defined as the proportional reduction in average fitness due to deleterious mutations [19,23,27–29]. Under a simple model that assumes one locus with fitnesses 1, 1-hs and 1-s, the load is
where p and q are the “normal” and “deleterious” allele frequencies. This reduces to
| (1) |
in the additive (semi-dominant) case and to
| (2) |
in the recessive case. More generally, load takes the form
where Wmax is the fitness of a mutation-free individual and W̄ is the average fitness. In practice, we cannot measure fitness and we know little about the distribution of selection and dominance coefficients, let alone about how the effects of deleterious mutations combine across loci. This is why recent studies have relied on population genetic summaries meant as proxies for load.
The choice of proxy turns out to be key in what the studies found. Notably, Lohmueller and colleagues (2008) introduced two summaries in order to compare individuals of European and African descent [30]. Using the first, they found the ratio of non-synonymous to synonymous segregating sites to be greater in the European sample than in the African one, which they interpreted as evidence for a reduced “efficacy of selection” in Europeans. Using the second summary, they found that European individuals carry (on average) more sites that are homozygous for non-synonymous derived alleles than do Africans, which they took as suggesting that Europeans likely suffer from a greater burden of recessive deleterious non-synonymous mutations. More recently, Simons et al. [31] and Do et al. [32] introduced a third summary (defined slightly differently in the two studies), the average number of derived non-synonymous variants per individual. They found no significant differences between European and African populations, and interpreted the pattern as indicating little or no difference in load. These studies and others [4,10,11, 33–42] applied the same methodologies to subsets of non-synonymous variants classified according to their predicted severity (e.g., using computational tools that rely on phylogenetic conservation and protein structure [43]), as well as to other human populations (see also [44,45]). With few exceptions (see below), analyses relying on the Lohmueller et al. summaries found substantial differences among populations whereas those that relied on the Simons et al. and Do et al. summaries did not.
Comparing proxies for load
Given that the answer seems to depend on the summary, the question becomes which summary is most appropriate. The ideal summary would relate to load as directly as possible but also be insensitive to other differences among populations.
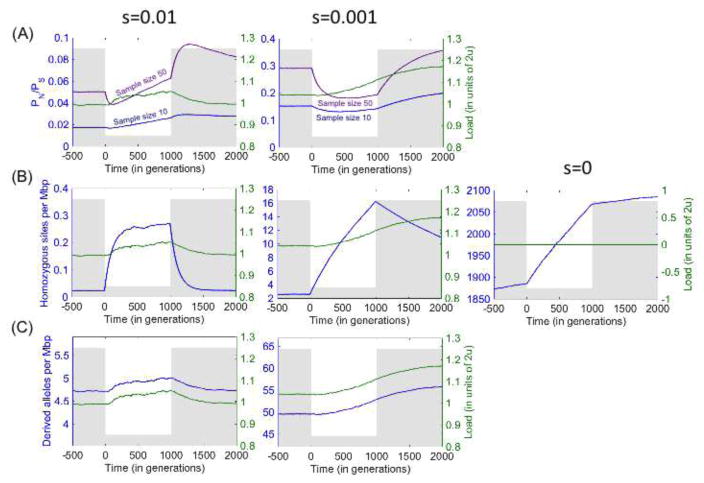
With these criteria in mind, we first consider the ratio of the number of non-synonymous to synonymous sites segregating in a population sample, PN/PS (or subsets of non-synonymous sites). The idea is that PS measures neutral diversity levels, and therefore PN/PS measures an effective proportion of neutral non-synonymous mutations [46]; increased PN/PS then reflects relaxed selection on non-synonymous mutations [25,30]. This interpretation applies at demographic equilibrium, e.g., when the population size is constant, or when non-synonymous mutations are either neutral or strongly selected, but it breaks down when neither assumption holds, which is precisely the case of interest. Under a population bottleneck, for example, PN/PS first exhibit drastic changes because increased drift affects PN and PS differently, due to the different initial nonsynonymous and synonymous frequency spectra [30,32]; then PN and PS approach equilibrium at different rates because selected alleles have faster turnover than neutral ones [26,31,47,48]. Neither of these effects is related to relaxation of selection or increased load. To complicate the interpretation of PN/PS even further, this statistic is extremely sensitive to the sample size (again because of the different synonymous and nonsynonymous frequency spectra). As a result, changes to PN/PS do not correspond to changes to load in any obvious way, even under straightforward demographic scenarios and assumptions about selection (Fig. 1A and [24]).
Figure 1.
Load and summaries used to measure load under simulations with a bottleneck and additive selection. The population size (grey) drops from 10,000 to 1,000 at time 0 and recovers a 1,000 generations later. The number of sites simulated in each case was chosen to achieve standard errors below 1%. For further simulation details see [31].
We next consider the behavior of the average number of homozygous derived non-synonymous sites. For recessive deleterious mutations, individual load is related directly to this number (cf. Eq. 2). However, not all mutations that contribute to this summary are recessive or deleterious. Notably, many derived non-synonymous alleles may be neutral, and because they reach higher frequencies than deleterious ones, they would contribute disproportionally to the number of homozygous, derived sites, swamping any underlying signal. Moreover, demographic events often have marked effects on the number of neutral derived homozygous sites (Fig. 1B). For instance, bottlenecks increase the variance of neutral allele frequencies, V(p2)=E(p2)-E2(p), without affecting their mean, E(p), thus increasing the frequency of homozygotes, E(p2). Comparing European populations that experienced the Out-of-Africa bottleneck to African ones that have not, we would therefore expect a large excess of homozygous, derived neutral sites in Europeans, even in the absence of a difference in load. Restricting the analysis to subsets of variants that are predicted to be more damaging will not solve this problem: while such subsets will include fewer neutral alleles, those neutral alleles that remain will contribute proportionally more homozygous sites, because more damaging variants have lower average frequencies [31]. Moreover, even if considering more damaging classes of variants helps to weed out neutral variants, non-recessive, deleterious variants continue to contribute, again complicating the relationship between the number of homozygous sites and load (Fig. 1B). Thus, the utility of this summary is severely compromised by its sensitivity to factors that have little to do with load.
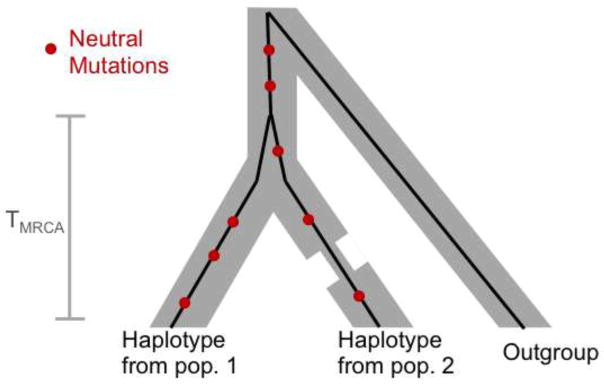
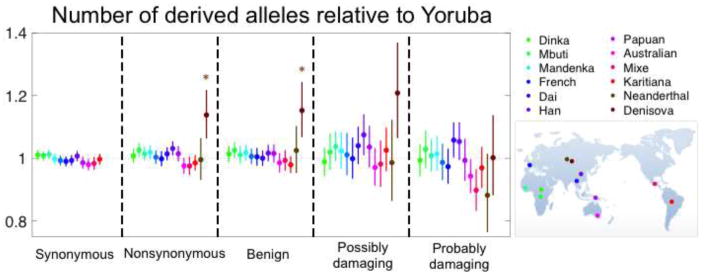
Lastly, we consider the average number of non-synonymous derived alleles. This number is directly related to individual load when derived alleles are deleterious and additive (i.e., semi-dominant) (Fig. 1C). Moreover, for this summary, comparisons between populations are not confounded by the presence of neutral alleles [31,32]. This advantage becomes clear by considering a single sample from each population at a non-recombining locus (Fig. 2): if the mutation rate on the lineages leading from their common ancestor to both samples is the same then the expected number of neutral mutations on each lineage should also be the same. Using this statistic, no significant differences between human populations are seen for (putatively neutral) synonymous derived alleles (Fig. 3; [31,32] but see [49]). As we discuss below, non-synonymous variation is likely to be dominated by additive (or at least approximately additive) and neutral mutations, for which this summary is particularly well-suited.
Figure 2.
The genealogy at a locus of one sample from each of two populations, illustrating that the expected number of derived neutral alleles on each sample is the same, and depends only on the time to the most recent common ancestor (TMRCA) of the two lineages and not on the demographic history of the populations.
Figure 3.
No significant difference between human populations in the mean number of derived alleles per individual. Each population sample was compared with the Yoruba sample, using data from [32]. Nonsynonymous SNPs were classified into benign, possibly and probably damaging using PolyPhen 2.0 as described in [32]. The numbers of derived alleles for each comparison were counted at sites that were segregating in the joint sample from the two populations, and significance and the shown 95% confidence intervals were evaluated as described in Box 1, dividing the genome into 1,000 blocks. While there are multiple tests performed, it is not obvious how to correct for them, because population samples are also not independent. However, if we assume that at minimum 6 tests were performed then none of the comparisons among human populations is significant at the 5% level. The comparisons with Neanderthal and Denisova incorporate the modifications described in [32], where Denisova are significantly different regardless of the correction for multiple testing.
These considerations suggest that among summaries used to date, the number of derived non-synonymous alleles is a much more appropriate measure of load. Importantly then, studies that use this summary (or equivalent ones) found no significant differences among human populations (see Box 1), even though they analyzed large datasets from many populations and considered a variety of classifications of non-synonymous variants (Fig. 3). In the two exceptions of which we are aware, one study performed no statistical test of significance [40] and the other used a test that we believe does not fully account for uncertainty [36] (see Box 1). Of course, other factors could potentially affect this summary (recessivity of deleterious mutations is considered below). However, in order for them to result in there being no difference among populations, they would have to almost perfectly cancel out with any true signal. We therefore conclude that there is currently no reliable evidence for differences in load among human populations driven by differences in demographic history, at least at non-synonymous sites.
Box 1. Statistical tests for demographically induced differences in load.
Having chosen the number of derived alleles (of a given class) per individual as our proxy for load, the next question becomes how to test for significant differences between populations. One option is to test for a significant difference between population samples in the average counts per individual (e.g., using a Mann-Whitney test). The problem with this approach is that even if we were to run the exact evolutionary experiment twice, with the same demographic history, we would expect our summary to take different values, due to stochasticity in the mutational and genealogical processes. Thus, it is unclear whether a significant result reflects a difference in the demographic history of the two populations. An alternative approach is to divide the genomes into blocks, and bootstrap over these blocks to test for differences between the populations. The idea here is that different blocks represent independent evolutionary experiments with the same demographic history. The blocks need to be sufficiently large for different blocks to represent independent genealogies. They also need to be resampled at random from the union of the two population samples to control for the shared part of the genealogical history of the two populations and to ensure that other factors that vary along the genome (e.g., gene density) are the same in the population samples compared. It turns out that the two approaches to assessing statistical significance yield different answers: when the former approach is applied to sufficiently large samples, it indicates significant difference between populations [36], whereas the latter approach, which we argue is more appropriate, shows none.
Should we expect substantial differences in load among human populations?
This question has been tackled using a combination of simulations and analytical tools. Most studies focused on changes to population size and specifically on bottlenecks and recent growth (but see [40]), with parameters based on rapidly improving demographic inferences [4,50–53]. Because much less is known about the distribution of selection and dominance coefficients, let alone about how the effects of deleterious mutations combine across loci, most studies considered single locus models under a range of selection and dominance coefficients (but see [25,36,53]). Even in this case, a general understanding of demographic effects on load requires reviewing many cases, notably different selection regimes (cf. [31]). Here we focus on some general insights that have emerged.
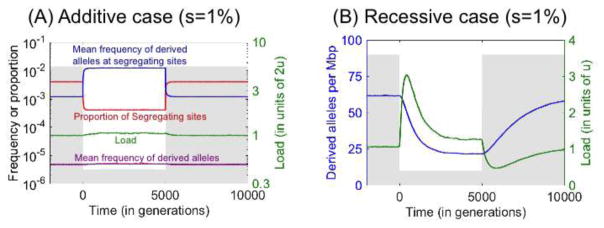
One important conclusion is that while bottlenecks and growth can dramatically affect both the number and frequencies of deleterious (and neutral) mutations, these shifts roughly cancel each other out, resulting in much subtler effects on load (Fig. 4A). In particular, for mutations that are effectively neutral (2Ns≪1) or partially dominant and strongly selected (2Nhs≫1) throughout the period being considered, changes to population size have been shown to have no effect on load [31]. Thus, it is quite possible to see little or no differences in load among populations, despite clear differences in overall patterns of genetic variation.
Figure 4.
Bottleneck effects on load and the number of derived alleles in additive (A) and recessive (B) cases. The population size (grey) drops from 10,000 to 1,000 at time 0 and recovers 5,000 generations later. The number of sites simulated in each case was chosen to achieve standard errors below 1%. For further simulation details see [31].
Observing little or no differences in load among populations might seem at odds with theoretical predictions. Specifically, theory predicts that at demographic equilibrium, a considerable portion of deleterious alleles for which 2Nes ≤ 1 will be fixed, leading to a much greater load in smaller populations [19,22,54]. Consistent with the reduced efficacy of selection in smaller populations, lineages that tended to have smaller effective population sizes over long evolutionary timescales (e.g., since the split between rodents and primates) show evidence for relaxed constraint at coding and regulatory regions [55,56]. One might therefore expect a substantial increase in load, due to the additive mutations that the Out-of-Africa bottleneck turned from strongly to weakly selected. In fact, the duration of the bottleneck was too short to have led to many deleterious fixations, and therefore the increase is predicted to be minor (Fig. 1) [31]. A similar argument applies to the effects of explosive growth, which is much too recent to impact load [24,31,57]. More generally, the presumed duration of the demographic events that differ among human populations are much shorter than the timescales required for weakly selected variation to equilibrate (roughly on the order of one over the mutation rate; cf. [31]), which explains why the differences expected at equilibrium are not seen in data.
In contrast to the additive case, in the fully recessive case, the mutation load and average number of deleterious alleles can change rapidly and potentially dramatically [31,32,58–61]. As an illustration, consider variation that is strongly selected throughout a bottleneck and subsequent expansion (Fig. 4B). When the population size drops, the increase in genetic drift leads to a loss of variation and a consequent drop in the average number of deleterious alleles per individual. However, some deleterious alleles drift to higher frequencies, contributing disproportionally to the number of homozygous sites and causing a surge in load. The response to changing population size is faster when selection is stronger, with a new equilibrium approached on the timescale of allelic turnover ( generations [31,60]). The load at equilibrium is insensitive to the population size, but the average number of deleterious alleles per individual scales with . The expansion after the bottleneck is also accompanied by rapid changes to load and to the number of deleterious alleles (Fig. 4B).
Observing changes in recessive load
Given that bottlenecks and expansions occurred in the recent history of many human populations (e.g., [2,10–12]), why do we not see significant differences among populations in the number of derived non-synonymous alleles due to recessive variants? One possibility is that the proportion of recessive variants among derived alleles is too small for them to generate significant differences among populations. If this were the case, then we might hope to see such a difference when applying the test to subsets that are enriched for recessive deleterious variants.
Population genetics theory and experiments suggest that alleles with large effects are more likely to be recessive, whereas alleles with smaller effect are likely to be additive [62–67]. While non-synonymous variation is likely dominated by mutations of smaller effects, Loss of Function (LoF) mutations are plausibly enriched for mutations of large effects, and indeed there are numerous examples of recessive LoF Mendelian diseases [68,cf. 69]. In turn, as discussed above, theory predicts that strongly selected recessive variation responds to changes in population size fairly quickly. In accordance with theory, founder populations, such as Ashkenazi Jews and Finns, are known to have their own disease heritage, i.e., a particularly high incidence of specific recessive diseases, but fewer segregating recessive alleles underlying these and other recessive diseases than in neighboring outgroup populations ([12,70–75], cf. [69]). We may therefore expect to see differences in the average number of LoF alleles per individual between founder and outgroup populations.
Providing strong evidence to this effect in humans, Narasimhan et al. [14] found significantly fewer LoF alleles per individual in Finns than in other (non-founder) populations. They also found significantly fewer LoF mutations in the Birmingham/Bradford Pakistani heritage population than in other populations (except Finns), likely due to a recent surge in inbreeding (which, similar to a bottleneck followed by an expansion, increases homozygosity [76–78], leading to the more efficient purging of recessive deleterious mutations [79,80]). In gorillas, Xue et al. [81] inferred that eastern lowland and mountain subspecies experienced a more drastic reduction in population size than their western lowland and cross river counterparts, with recent population sizes in eastern subspecies so small that autozygosity levels exceed those found in the most inbred human populations. As expected from this demographic history, they also find evidence for markedly and significantly fewer LoF mutations per individual in the two eastern subspecies than in their western counterparts.
Observing a reduction in the number of LoF mutations per individual does not necessarily imply that the load is currently lower. For example, as illustrated in Fig. 4B, the relationship between the average number of alleles per individual and recessive load is also affected by the demographic history since the bottleneck. Nonetheless, such an observation likely indicates a past change in load and, in conjunction with other information, e.g., about changes to population size or levels of inbreeding, inferences about the current load may be feasible.
Observing changes in additive load
While theory predicts that the increase to the load of additive mutations following a bottleneck would be extremely slow, it may still be detectable if the drop in population size was sufficiently long and severe. One example is provided by the increased reduction in population size in eastern compared to western gorillas, which was estimated to have been more severe and longer lasting than the Out-of-Africa bottleneck in humans [52,81]. Eastern gorilla subspecies also carry significantly more non-synonymous alleles per individual than their western counterparts, suggesting that the greater reduction in population size also led to an increase in load due to additive mutations [81].
Similarly, both Neanderthals and Denisovans are inferred to have had an extremely low population size since their main separation from humans, which was longer ago than the separation of gorilla subspecies [18,82]. These estimates suggest that they should have also incurred a substantial increase in load. In fact, their effective population sizes are among the lowest measured in any taxon [83], leading to speculation that a corresponding accumulation of deleterious mutations may have contributed to their demise [84]. To test the hypothesis that their load was higher, Do et al. [32] compared the number of derived non-synonymous alleles per individual between these archaic hominins and modern humans, using a modified summary aimed to control for the branch shortening and DNA degradation in ancient samples [32]. They found a markedly and significantly greater number in Denisovans, indicative of increased load, but not in Neanderthals (Fig. 3).
Strong evidence for increased load in Neanderthals comes from another source: the distribution of Neanderthal ancestry along the genomes of modern humans. Current estimates suggest that 2–4% of the genome of non-African individuals is the result of introgression from Neanderthals and that Neanderthal haplotypes collectively span ~30% of the genome [17,85,86]. This Neanderthal ancestry is, on average, markedly lower in regions of high gene density and low recombination, indicating that, with some notable exceptions (cf. [87]), selection has been acting to remove Neanderthal DNA [85,86,88,89]. A couple of recent studies have shown that these ancestry patterns are plausibly explained by selection against deleterious mutations from Neanderthals [53,84] (not found by Do et al. [32], possibly due to technical complications in the use of ancient samples). Using the ancestry patterns to infer the strength of selection against non-synonymous Neanderthal mutations, Juric et al. [84] found that it accords with the range that would have been effectively neutral in Neanderthals but effectively selected in humans. These studies further suggest, albeit speculatively, that some residual “admixture load” may remain in non-Africans today.
Conclusion
The contradictory conclusions about the impact of recent demographic events on the mutation load in humans can be traced back to the use of different summary statistics and to how they are affected by factors other than load. We contend that of the summaries used thus far, the number of derived alleles per individual is both more sensitive and specific to load. Applying this summary and appropriate statistical tests to data yields findings that accord with theoretical expectations for modern human populations, archaic humans and subspecies of gorilla. Specifically, the findings follow from the plausible assumption that non-synonymous variation is dominated by mutations of small additive effects, whose contribution to load should changes slowly in response to changes in population size, whereas LoF mutations are likely enriched for large recessive effects, whose contribution to load should respond rapidly to changes in population size or surges of inbreeding. Of course, future advances, such as improvements in our classification of variants, may reveal differences in load that evade us at present. However, current findings suggest that differences among extant human populations are likely to be small.
Acknowledgments
We thank Molly Przeworski, Guy Amster, Jeff Ross-Ibarra, Aylwyn Scally and Jonathan Pritchard for comments on the manuscript. We owe special thanks to Ron Do, for his considerable help in producing Figure 2, and to Vagheesh Narasimhan, for answering many questions about his analyses. This work was funded by NIH grant GM115889 to GS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Wall JD, Przeworski M. When did the human population size start increasing? Genetics. 2000;155:1865–1874. doi: 10.1093/genetics/155.4.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci U S A. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S, McGee S, Do R, Liu X, Jun G, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coventry A, Bull-Otterson LM, Liu X, Clark AG, Maxwell TJ, Crosby J, Hixson JE, Rea TJ, Muzny DM, Lewis LR, et al. Deep resequencing reveals excess rare recent variants consistent with explosive population growth. Nat Commun. 2010;1:131. doi: 10.1038/ncomms1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marth GT, Yu F, Indap AR, Garimella K, Gravel S, Leong WF, Tyler-Smith C, Bainbridge M, Blackwell T, Zheng-Bradley X, et al. The functional spectrum of low-frequency coding variation. Genome Biol. 2011;12:R84. doi: 10.1186/gb-2011-12-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keinan A, Clark AG. Recent explosive human population growth has resulted in an excess of rare genetic variants. Science. 2012;336:740–743. doi: 10.1126/science.1217283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C, Shen J, Tang Z, Bacanu SA, Fraser D, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature. 2013;493:216–220. doi: 10.1038/nature11690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Casals F, Hodgkinson A, Hussin J, Idaghdour Y, Bruat V, de Maillard T, Grenier J-C, Gbeha E, Hamdan FF, Girard S, et al. Whole-exome sequencing reveals a rapid change in the frequency of rare functional variants in a founding population of humans. PLoS Genet. 2013;9:e1003815. doi: 10.1371/journal.pgen.1003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carmi S, Hui KY, Kochav E, Liu X, Xue J, Grady F, Guha S, Upadhyay K, Ben-Avraham D, Mukherjee S, et al. Sequencing an Ashkenazi reference panel supports population-targeted personal genomics and illuminates Jewish and European origins. Nat Commun. 2014:5. doi: 10.1038/ncomms5835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim ET, Würtz P, Havulinna AS, Palta P, Tukiainen T, Rehnström K, Esko T, Mägi R, Inouye M, Lappalainen T, et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saleheen D, Natarajan P, Zhao W, Rasheed A, Khetarpal S, Won H-H, Karczewski KJ, ODonnell-Luria AH, Samocha KE, Gupta N, et al. Human knockouts in a cohort with a high rate of consanguinity. bioRxiv. 2015 doi: 10.1038/nature22034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14**.Narasimhan VM, Hunt KA, Mason D, Baker CL, Karczewski KJ, Barnes MR, Barnett AH, Bates C, Bellary S, Bockett NA, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352:474–477. doi: 10.1126/science.aac8624. This study takes advantage of high levels of inbreeding in a population of Pakistani heritage to study the effects of LoF mutations in the human genome. The authors find that the average number of LoF mutations in this and in the Finnish founder-population is significantly lower than in other human populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellenthal G, Busby GBJ, Band G, Wilson JF, Capelli C, Falush D, Myers S. A genetic atlas of human admixture history. Science. 2014;343:747–751. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pickrell JK, Reich D. Toward a new history and geography of human genes informed by ancient DNA. Trends Genet. 2014;30:377–389. doi: 10.1016/j.tig.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19**.Kimura M, Maruyama T, Crow JF. The mutation load in small populations. Genetics. 1963;48:1303–1312. doi: 10.1093/genetics/48.10.1303. A foundational paper on the effect of drift on mutation load in finite populations, showing that mildly deleterious alleles are expected to contribute substantially more to load than strongly deleterious ones. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura M. The neutral theory of molecular evolution. Cambridge University Press; 1984. [Google Scholar]
- 21.Kondrashov AS, Crow JF. King’s formula for the mutation load with epistasis. Genetics. 1988;120:853–856. doi: 10.1093/genetics/120.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohta T, Gillespie JH. Development of neutral and nearly neutral theories. Theor Popul Biol. 1996;49:128–142. doi: 10.1006/tpbi.1996.0007. [DOI] [PubMed] [Google Scholar]
- 23**.Agrawal AF, Whitlock MC. Mutation load: The fitness of individuals in populations where deleterious alleles are abundant. Annu Rev Ecol Evol Syst. 2012;43:115–135. An up to date and broad review about the determinants of mutation load and its implications. [Google Scholar]
- 24.Lohmueller KE. The distribution of deleterious genetic variation in human populations. Curr Opin Genet Dev. 2014;29:139–146. doi: 10.1016/j.gde.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Henn BM, Botigue LR, Bustamante CD, Clark AG, Gravel S. Estimating the mutation load in human genomes. Nat Rev Genet. 2015;16:333–343. doi: 10.1038/nrg3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Brandvain Y, Wright SI. The limits of natural selection in a nonequilibrium world. Trends Genet. 2016;32:201–210. doi: 10.1016/j.tig.2016.01.004. A recent, phylogenetically broad perspective about the interaction of selection and non-equilibrium demography. [DOI] [PubMed] [Google Scholar]
- 27*.Haldane JBS. The effect of variation on fitness. Am Nat. 1937;71:337–349. In this seminal paper, Haldane calculates the fitness cost of mutation, which we now call load, at mutation-selection balance. [Google Scholar]
- 28*.Muller HJ. Our load of mutations. Am J Hum Genet. 1950;2:111–176. In this seminal paper, Muller coins the term load and argues that deleterious mutations contribute substantially to human mortality and disease. [PMC free article] [PubMed] [Google Scholar]
- 29.Crow JF. Mutation, mean fitness, and genetic load. Oxf Surv Evol Biol. 1993;9:3–42. [Google Scholar]
- 30*.Lohmueller KE, Indap AR, Schmidt S, Boyko AR, Hernandez RD, Hubisz MJ, Sninsky JJ, White TJ, Sunyaev SR, Nielsen R, et al. Proportionally more deleterious genetic variation in European than in African populations. Nature. 2008;451:994–997. doi: 10.1038/nature06611. This paper gave rise to the current interest in the effects of recent human demography on load, and in particular suggested that Europeans suffer from greater load than Africans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nat Genet. 2014;46:220–224. doi: 10.1038/ng.2896. This study uses population genetic models to show that recent human demography likely had little impact on load, and introduces a measure for load based on the number of derived non-synonymous alleles (in parallel to [32]) to show that individuals of west African and European ancestry carry very similar burdens of damaging mutations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32**.Do R, Balick D, Li H, Adzhubei I, Sunyaev S, Reich D. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat Genet. 2015;47:126–131. doi: 10.1038/ng.3186. This study introduces measures of load based on the number of derived non-synonymous alleles per individual (in parallel to [31]) to find no evidence for differences in load among diverse human populations, but higher load in Denisovans compared to modern humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kidd JM, Gravel S, Byrnes J, Moreno-Estrada A, Musharoff S, Bryc K, Degenhardt JD, Brisbin A, Sheth V, Chen R, et al. Population genetic inference from personal genome data: impact of ancestry and admixture on human genomic variation. Am J Hum Genet. 2012;91:660–671. doi: 10.1016/j.ajhg.2012.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodgkinson A, Casals F, Idaghdour Y, Grenier JC, Hernandez RD, Awadalla P. Selective constraint, background selection, and mutation accumulation variability within and between human populations. BMC Genomics. 2013;14:495. doi: 10.1186/1471-2164-14-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pierron D, Cortes NG, Letellier T, Grossman LI. Current relaxation of selection on the human genome: tolerance of deleterious mutations on olfactory receptors. Mol Phylogenet Evol. 2013;66:558–564. doi: 10.1016/j.ympev.2012.07.032. [DOI] [PubMed] [Google Scholar]
- 36.Fu W, Gittelman RM, Bamshad MJ, Akey JM. Characteristics of neutral and deleterious protein-coding variation among individuals and populations. Am J Hum Genet. 2014;95:421–436. doi: 10.1016/j.ajhg.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cocca M, Pybus M, Palamara PF, Garrison E, Traglia M, Sala CF, Ulivi S, Memari Y, Kolb-Kokocinski A, Durbin R, et al. Purging of deleterious variants in Italian founder populations with extended autozygosity. bioRxiv. 2015 [Google Scholar]
- 38.Kaiser VB, Svinti V, Prendergast JG, Chau YY, Campbell A, Patarcic I, Barroso I, Joshi PK, Hastie ND, Miljkovic A, et al. Homozygous loss-of-function variants in European cosmopolitan and isolate populations. Hum Mol Genet. 2015;24:5464–5474. doi: 10.1093/hmg/ddv272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kehdy FS, Gouveia MH, Machado M, Magalhaes WC, Horimoto AR, Horta BL, Moreira RG, Leal TP, Scliar MO, Soares-Souza GB, et al. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc Natl Acad Sci U S A. 2015;112:8696–8701. doi: 10.1073/pnas.1504447112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henn BM, Botigué LR, Peischl S, Dupanloup I, Lipatov M, Maples BK, Martin AR, Musharoff S, Cann H, Snyder MP, et al. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci U S A. 2016;113:E440–E449. doi: 10.1073/pnas.1510805112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeroncic A, Memari Y, Ritchie GR, Hendricks AE, Kolb-Kokocinski A, Matchan A, Vitart V, Hayward C, Kolcic I, Glodzik D, et al. Whole-exome sequencing in an isolated population from the Dalmatian island of Vis. Eur J Hum Genet. 2016 doi: 10.1038/ejhg.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subramanian S. Europeans have a higher proportion of highfrequency deleterious variants than Africans. Hum Genet. 2016;135:1–7. doi: 10.1007/s00439-015-1604-z. [DOI] [PubMed] [Google Scholar]
- 43.Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628–640. doi: 10.1038/nrg3046. [DOI] [PubMed] [Google Scholar]
- 44.Sudmant PH, Mallick S, Nelson BJ, Hormozdiari F, Krumm N, Huddleston J, Coe BP, Baker C, Nordenfelt S, Bamshad M, et al. Global diversity, population stratification, and selection of human copy-number variation. Science. 2015;349:aab3761. doi: 10.1126/science.aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Hsi-Yang Fritz M, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 47.Gordo I, Dionisio F. Nonequilibrium model for estimating parameters of deleterious mutations. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;71:031907. doi: 10.1103/PhysRevE.71.031907. [DOI] [PubMed] [Google Scholar]
- 48.Pennings PS, Kryazhimskiy S, Wakeley J. Loss and Recovery of Genetic Diversity in Adapting Populations of HIV. PLoS Genet. 2014;10:e1004000. doi: 10.1371/journal.pgen.1004000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris K. Evidence for recent, population-specific evolution of the human mutation rate. Proc Natl Acad Sci U S A. 2015;112:3439–3444. doi: 10.1073/pnas.1418652112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gutenkunst RN, Hernandez RD, Williamson SH, Bustamante CD. Inferring the joint demographic history of multiple populations from multidimensional SNP frequency data. PLoS Genet. 2009;5:e1000695. doi: 10.1371/journal.pgen.1000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schiffels S, Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53**.Harris K, Nielsen R. The genetic cost of Neanderthal introgression. Genetics. 2016 doi: 10.1534/genetics.116.186890. This study (and [84]) argues that the distribution of Neanderthal ancestry along the genomes of modern humans is best explained by selection against deleterious mutations from Neanderthals, strongly suggesting that Neanderthals suffered from a higher load than do modern humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kondrashov AS. Contamination of the genome by very slightly deleterious mutations: why have we not died 100 times over? J Theor Biol. 1995;175:583–594. doi: 10.1006/jtbi.1995.0167. [DOI] [PubMed] [Google Scholar]
- 55.Ohta T. An examination of the generation-time effect on molecular evolution. Proc Natl Acad Sci U S A. 1993;90:10676–10680. doi: 10.1073/pnas.90.22.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keightley PD, Lercher MJ, Eyre-Walker A. Evidence for widespread degradation of gene control regions in hominid genomes. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gazave E, Chang D, Clark AG, Keinan A. Population growth inflates the per-individual number of deleterious mutations and reduces their mean effect. Genetics. 2013;195:969–978. doi: 10.1534/genetics.113.153973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Caballero A, Keightley PD, Hill WG. Bottleneck effect on genetic variance. A theoretical investigation of the role of dominance. Genetics. 1998;150:435–447. doi: 10.1093/genetics/150.1.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kirkpatrick M, Jarne P. The effects of a bottleneck on inbreeding depression and the genetic load. Am Nat. 2000;155:154–167. doi: 10.1086/303312. [DOI] [PubMed] [Google Scholar]
- 60.Balick DJ, Do R, Cassa CA, Reich D, Sunyaev SR. Dominance of deleterious alleles controls the response to a population bottleneck. PLoS Genet. 2015;11:e1005436. doi: 10.1371/journal.pgen.1005436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gravel S. When Is selection effective? Genetics. 2016;203:451–462. doi: 10.1534/genetics.115.184630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62**.Wright S. Fisher’s theory of dominance. Am Nat. 1929;63:274–279. Wright argues against an evolutionary theory of dominance, and proposes a physiological explanation for why mutations of large effect tend to be recessive. [Google Scholar]
- 63.Greenberg R, Crow JF. A comparison of the effect of lethal and detrimental chromosomes from drosophila populations. Genetics. 1960;45:1153–1168. doi: 10.1093/genetics/45.8.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64**.Simmons MJ, Crow JF. Mutations affecting fitness in Drosophila populations. Annu Rev Genet. 1977;11:49–78. doi: 10.1146/annurev.ge.11.120177.000405. A seminal review of empirical work about the fitness effects of mutations in Drosophila, arguing for an inverse relationship between dominance and selection effects. [DOI] [PubMed] [Google Scholar]
- 65.Charlesworth B. Evidence against Fisher’s theory of dominance. Nature. 1979;278:848–849. [Google Scholar]
- 66.Phadnis N, Fry JD. Widespread correlations between dominance and homozygous effects of mutations: implications for theories of dominance. Genetics. 2005;171:385–392. doi: 10.1534/genetics.104.039016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67*.Agrawal AF, Whitlock MC. Inferences about the distribution of dominance drawn from yeast gene knockout data. Genetics. 2011;187:553–566. doi: 10.1534/genetics.110.124560. A recent study of the distribution of dominance effects and their relationship to selection effects for gene knockouts in yeast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD): Online Mendelian Inheritance in Man, OMIM®.
- 69.Jobling M, Hurles M, Tyler-Smith C. Human evolutionary genetics: origins, peoples & disease. Garland Science; 2013. [Google Scholar]
- 70.Peltonen L, Jalanko A, Varilo T. Molecular genetics the Finnish disease heritage. Hum Mol Genet. 1999;8:1913–1923. doi: 10.1093/hmg/8.10.1913. [DOI] [PubMed] [Google Scholar]
- 71.Ostrer H. A genetic profile of contemporary Jewish populations. Nat Rev Genet. 2001;2:891–898. doi: 10.1038/35098506. [DOI] [PubMed] [Google Scholar]
- 72.Norio R. Finnish disease heritage I. Hum Genet. 2003;112:441–456. doi: 10.1007/s00439-002-0875-3. [DOI] [PubMed] [Google Scholar]
- 73.Norio R. The Finnish disease heritage III: the individual diseases. Hum Genet. 2003;112:470–526. doi: 10.1007/s00439-002-0877-1. [DOI] [PubMed] [Google Scholar]
- 74.Risch N, Tang H, Katzenstein H, Ekstein J. Geographic distribution of disease mutations in the Ashkenazi Jewish population supports genetic drift over selection. Am J Hum Genet. 2003;72:812–822. doi: 10.1086/373882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slatkin M. A population-genetic test of founder effects and implications for Ashkenazi Jewish diseases. Am J Hum Genet. 2004;75:282–293. doi: 10.1086/423146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bundey S, Alam H. A five-year prospective study of the health of children in different ethnic groups, with particular reference to the effect of inbreeding. Eur J Hum Genet. 1993;1:206–219. doi: 10.1159/000472414. [DOI] [PubMed] [Google Scholar]
- 77.Tadmouri GO, Nair P, Obeid T, Al Ali MT, Al Khaja N, Hamamy HA. Consanguinity and reproductive health among Arabs. Reprod Health. 2009;6:1–9. doi: 10.1186/1742-4755-6-17. Tadmouri2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bittles AH, Hamamy HA. Endogamy and consanguineous marriage in Arab populations. In: Teebi SA, editor. Genetic Disorders Among Arab Populations. Springer; Berlin Heidelberg: 2010. pp. 85–108. Bittles 2010. [Google Scholar]
- 79.Wang J, Hill WG, Charlesworth D, Charlesworth B. Dynamics of inbreeding depression due to deleterious mutations in small populations: mutation parameters and inbreeding rate. Genet Res. 1999;74:165–178. doi: 10.1017/s0016672399003900. [DOI] [PubMed] [Google Scholar]
- 80.Garcia-Dorado A. A simple method to account for natural selection when predicting inbreeding depression. Genetics. 2008;180:1559–1566. doi: 10.1534/genetics.108.090597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81**.Xue Y, Prado-Martinez J, Sudmant PH, Narasimhan V, Ayub Q, Szpak M, Frandsen P, Chen Y, Yngvadottir B, Cooper DN, et al. Mountain gorilla genomes reveal the impact of long-term population decline and inbreeding. Science. 2015;348:242–245. doi: 10.1126/science.aaa3952. This study finds evidence for changes in the load of non-synonymous and loss of function mutations in eastern compared to western sub-species of gorilla, as expected from their inferred demographic histories. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leffler EM, Bullaughey K, Matute DR, Meyer WK, SÈgurel L, Venkat A, Andolfatto P, Przeworski M. Revisiting an old riddle: What determines genetic diversity levels within species? PLoS Biol. 2012;10:e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84**.Juric I, Aeschbacher S, Coop G. The strength of selection against Neanderthal introgression. bioRxiv. 2015 doi: 10.1371/journal.pgen.1006340. This study (and [53]) argues that the distribution of Neanderthal ancestry along the genomes of modern humans is best explained by selection against deleterious mutations from Neanderthals, strongly suggesting that Neanderthals suffered from a higher load than do modern humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sankararaman S, Mallick S, Dannemann M, Prufer K, Kelso J, Paabo S, Patterson N, Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, Dannemann M, Grote S, McCoy RC, Norton H, et al. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science. 2016;352:235–239. doi: 10.1126/science.aad9416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Racimo F, Sankararaman S, Nielsen R, Huerta-Sanchez E. Evidence for archaic adaptive introgression in humans. Nat Rev Genet. 2015;16:359–371. doi: 10.1038/nrg3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vernot B, Akey JM. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 2014;343:1017–1021. doi: 10.1126/science.1245938. [DOI] [PubMed] [Google Scholar]
- 89.Fu Q, Posth C, Hajdinjak M, Petr M, Mallick S, Fernandes D, Furtwängler A, Haak W, Meyer M, Mittnik A, et al. The genetic history of Ice Age Europe. Nature. 2016;534:200–205. doi: 10.1038/nature17993. [DOI] [PMC free article] [PubMed] [Google Scholar]