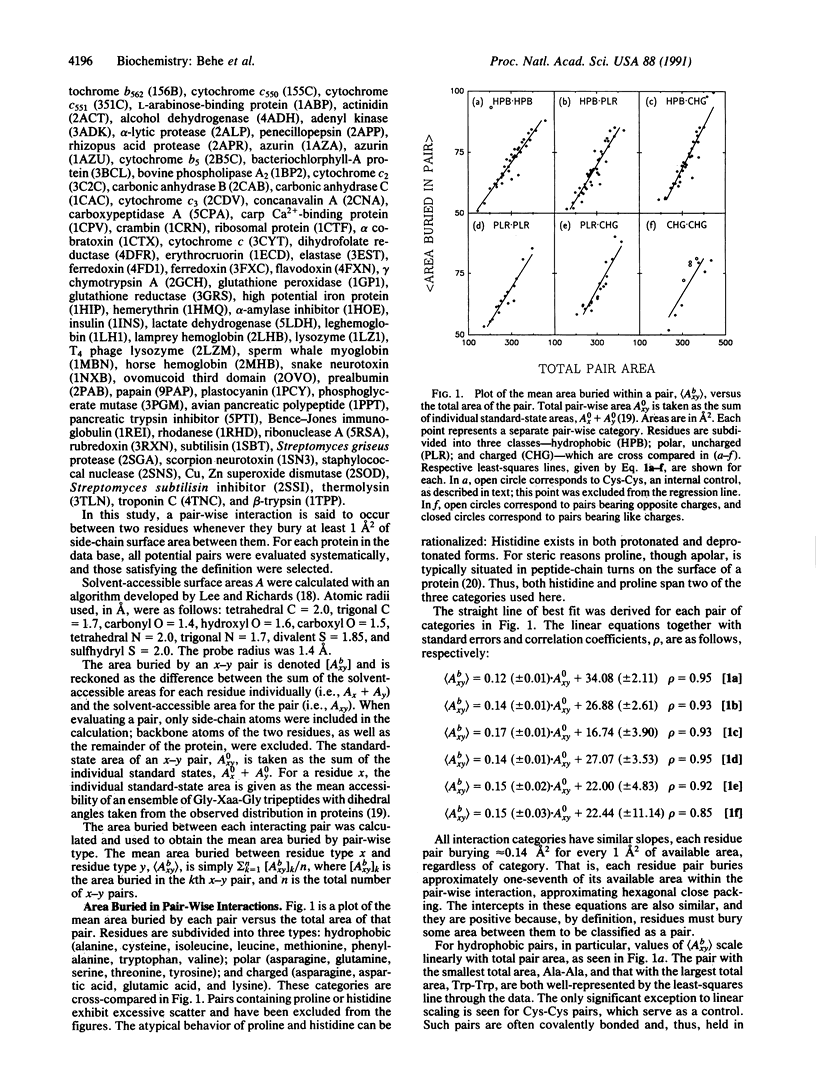
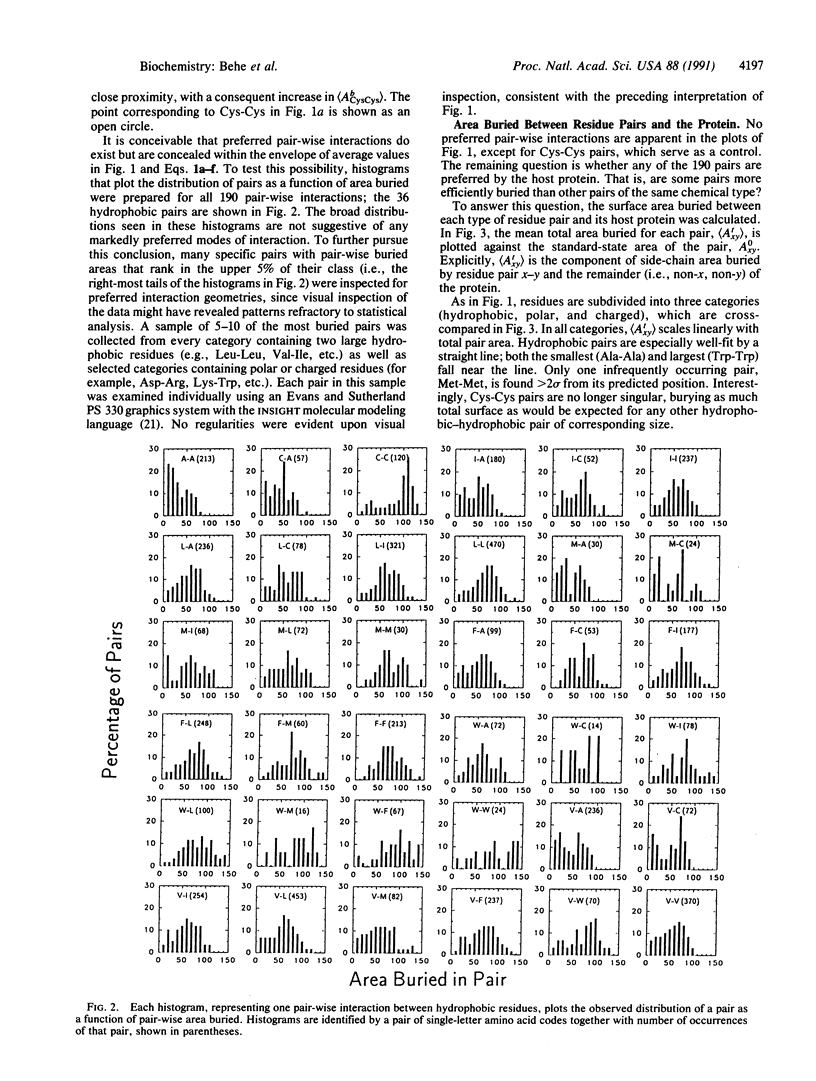
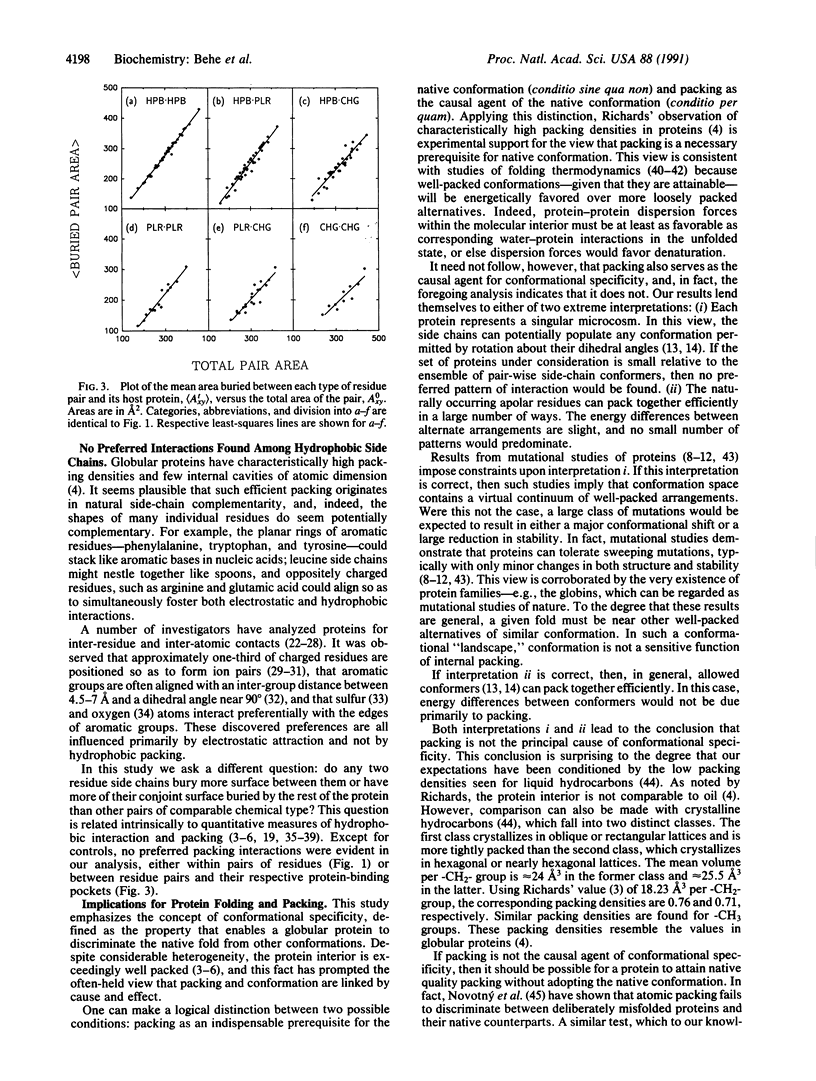
Abstract
A globular protein adopts its native three-dimensional structure spontaneously under physiological conditions. This structure is specified by a stereochemical code embedded within the amino acid sequence of that protein. Elucidation of this code is a major, unsolved challenge, known as the protein-folding problem. A critical aspect of the code is thought to involve molecular packing. Globular proteins have high packing densities, a consequence of the fact that residue side chains within the molecular interior fit together with an exquisite complementarity, like pieces of a three-dimensional jigsaw puzzle [Richards, F. M. (1977) Annu. Rev. Biophys. Bioeng. 6, 151]. Such packing interactions are widely viewed as the principal determinant of the native structure. To test this view, we analyzed proteins of known structure for the presence of preferred interactions, reasoning that if side-chain complementarity is an important source of structural specificity, then sets of residues that interact favorably should be apparent. Our analysis leads to the surprising conclusion that high packing densities--so characteristic of globular proteins--are readily attainable among clusters of the naturally occurring hydrophobic amino acid residues. It is anticipated that this realization will simplify approaches to the protein-folding problem.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alber T. Mutational effects on protein stability. Annu Rev Biochem. 1989;58:765–798. doi: 10.1146/annurev.bi.58.070189.004001. [DOI] [PubMed] [Google Scholar]
- Anfinsen C. B. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Barlow D. J., Thornton J. M. Ion-pairs in proteins. J Mol Biol. 1983 Aug 25;168(4):867–885. doi: 10.1016/s0022-2836(83)80079-5. [DOI] [PubMed] [Google Scholar]
- Barlow D. J., Thornton J. M. The distribution of charged groups in proteins. Biopolymers. 1986 Sep;25(9):1717–1733. doi: 10.1002/bip.360250913. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Bowie J. U., Clarke N. D., Pabo C. O., Sauer R. T. Identification of protein folds: matching hydrophobicity patterns of sequence sets with solvent accessibility patterns of known structures. Proteins. 1990;7(3):257–264. doi: 10.1002/prot.340070307. [DOI] [PubMed] [Google Scholar]
- Bryant S. H., Amzel L. M. Correctly folded proteins make twice as many hydrophobic contacts. Int J Pept Protein Res. 1987 Jan;29(1):46–52. doi: 10.1111/j.1399-3011.1987.tb02228.x. [DOI] [PubMed] [Google Scholar]
- Bryant S. H. PKB: a program system and data base for analysis of protein structure. Proteins. 1989;5(3):233–247. doi: 10.1002/prot.340050307. [DOI] [PubMed] [Google Scholar]
- Burley S. K., Petsko G. A. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985 Jul 5;229(4708):23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Chothia C. Structural invariants in protein folding. Nature. 1975 Mar 27;254(5498):304–308. doi: 10.1038/254304a0. [DOI] [PubMed] [Google Scholar]
- Connelly P. R., Varadarajan R., Sturtevant J. M., Richards F. M. Thermodynamics of protein-peptide interactions in the ribonuclease S system studied by titration calorimetry. Biochemistry. 1990 Jun 26;29(25):6108–6114. doi: 10.1021/bi00477a031. [DOI] [PubMed] [Google Scholar]
- Correa P. E. The building of protein structures from alpha-carbon coordinates. Proteins. 1990;7(4):366–377. doi: 10.1002/prot.340070408. [DOI] [PubMed] [Google Scholar]
- Dill K. A. Dominant forces in protein folding. Biochemistry. 1990 Aug 7;29(31):7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., McLachlan A. D. Solvation energy in protein folding and binding. Nature. 1986 Jan 16;319(6050):199–203. doi: 10.1038/319199a0. [DOI] [PubMed] [Google Scholar]
- Estell D. A., Graycar T. P., Miller J. V., Powers D. B., Wells J. A., Burnier J. P., Ng P. G. Probing steric and hydrophobic effects on enzyme-substrate interactions by protein engineering. Science. 1986 Aug 8;233(4764):659–663. doi: 10.1126/science.233.4764.659. [DOI] [PubMed] [Google Scholar]
- Finney J. L. Volume occupation, environment and accessibility in proteins. The problem of the protein surface. J Mol Biol. 1975 Aug 25;96(4):721–732. doi: 10.1016/0022-2836(75)90148-5. [DOI] [PubMed] [Google Scholar]
- Gribskov M., McLachlan A. D., Eisenberg D. Profile analysis: detection of distantly related proteins. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAUZMANN W. Some factors in the interpretation of protein denaturation. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- Kuntz I. D., Crippen G. M. Protein densities. Int J Pept Protein Res. 1979 Feb;13(2):223–228. doi: 10.1111/j.1399-3011.1979.tb01872.x. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lee B., Richards F. M. The interpretation of protein structures: estimation of static accessibility. J Mol Biol. 1971 Feb 14;55(3):379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- Lesser G. J., Rose G. D. Hydrophobicity of amino acid subgroups in proteins. Proteins. 1990;8(1):6–13. doi: 10.1002/prot.340080104. [DOI] [PubMed] [Google Scholar]
- Lim W. A., Sauer R. T. Alternative packing arrangements in the hydrophobic core of lambda repressor. Nature. 1989 May 4;339(6219):31–36. doi: 10.1038/339031a0. [DOI] [PubMed] [Google Scholar]
- Malcolm B. A., Wilson K. P., Matthews B. W., Kirsch J. F., Wilson A. C. Ancestral lysozymes reconstructed, neutrality tested, and thermostability linked to hydrocarbon packing. Nature. 1990 May 3;345(6270):86–89. doi: 10.1038/345086a0. [DOI] [PubMed] [Google Scholar]
- Manavalan P., Ponnuswamy P. K. A study of the preferred environment of amino acid residues in globular proteins. Arch Biochem Biophys. 1977 Dec;184(2):476–487. doi: 10.1016/0003-9861(77)90457-x. [DOI] [PubMed] [Google Scholar]
- Matouschek A., Kellis J. T., Jr, Serrano L., Fersht A. R. Mapping the transition state and pathway of protein folding by protein engineering. Nature. 1989 Jul 13;340(6229):122–126. doi: 10.1038/340122a0. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Genetic and structural analysis of the protein stability problem. Biochemistry. 1987 Nov 3;26(22):6885–6888. doi: 10.1021/bi00396a001. [DOI] [PubMed] [Google Scholar]
- Moult J., James M. N. An algorithm for determining the conformation of polypeptide segments in proteins by systematic search. Proteins. 1986 Oct;1(2):146–163. doi: 10.1002/prot.340010207. [DOI] [PubMed] [Google Scholar]
- Narayana S. V., Argos P. Residue contacts in protein structures and implications for protein folding. Int J Pept Protein Res. 1984 Jul;24(1):25–39. doi: 10.1111/j.1399-3011.1984.tb00924.x. [DOI] [PubMed] [Google Scholar]
- Novotný J., Rashin A. A., Bruccoleri R. E. Criteria that discriminate between native proteins and incorrectly folded models. Proteins. 1988;4(1):19–30. doi: 10.1002/prot.340040105. [DOI] [PubMed] [Google Scholar]
- Nozaki Y., Tanford C. The solubility of amino acids and two glycine peptides in aqueous ethanol and dioxane solutions. Establishment of a hydrophobicity scale. J Biol Chem. 1971 Apr 10;246(7):2211–2217. [PubMed] [Google Scholar]
- Ponder J. W., Richards F. M. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 1987 Feb 20;193(4):775–791. doi: 10.1016/0022-2836(87)90358-5. [DOI] [PubMed] [Google Scholar]
- Rashin A. A., Honig B. On the environment of ionizable groups in globular proteins. J Mol Biol. 1984 Mar 15;173(4):515–521. doi: 10.1016/0022-2836(84)90394-2. [DOI] [PubMed] [Google Scholar]
- Reid L. S., Thornton J. M. Rebuilding flavodoxin from C alpha coordinates: a test study. Proteins. 1989;5(2):170–182. doi: 10.1002/prot.340050212. [DOI] [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- Richards F. M. The interpretation of protein structures: total volume, group volume distributions and packing density. J Mol Biol. 1974 Jan 5;82(1):1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- Rose G. D., Geselowitz A. R., Lesser G. J., Lee R. H., Zehfus M. H. Hydrophobicity of amino acid residues in globular proteins. Science. 1985 Aug 30;229(4716):834–838. doi: 10.1126/science.4023714. [DOI] [PubMed] [Google Scholar]
- Rose G. D., Gierasch L. M., Smith J. A. Turns in peptides and proteins. Adv Protein Chem. 1985;37:1–109. doi: 10.1016/s0065-3233(08)60063-7. [DOI] [PubMed] [Google Scholar]
- Rose G. D. Prediction of chain turns in globular proteins on a hydrophobic basis. Nature. 1978 Apr 13;272(5654):586–590. doi: 10.1038/272586a0. [DOI] [PubMed] [Google Scholar]
- Rose G. D., Roy S. Hydrophobic basis of packing in globular proteins. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4643–4647. doi: 10.1073/pnas.77.8.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg W. S., Terwilliger T. C. Influence of interior packing and hydrophobicity on the stability of a protein. Science. 1989 Jul 7;245(4913):54–57. doi: 10.1126/science.2787053. [DOI] [PubMed] [Google Scholar]
- Shakhnovich E. I., Gutin A. M. Implications of thermodynamics of protein folding for evolution of primary sequences. Nature. 1990 Aug 23;346(6286):773–775. doi: 10.1038/346773a0. [DOI] [PubMed] [Google Scholar]
- Singh J., Thornton J. M. SIRIUS. An automated method for the analysis of the preferred packing arrangements between protein groups. J Mol Biol. 1990 Feb 5;211(3):595–615. doi: 10.1016/0022-2836(90)90268-Q. [DOI] [PubMed] [Google Scholar]
- Sweet R. M., Eisenberg D. Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J Mol Biol. 1983 Dec 25;171(4):479–488. doi: 10.1016/0022-2836(83)90041-4. [DOI] [PubMed] [Google Scholar]
- Taylor J. W., Kaiser E. T. Structure-function analysis of proteins through the design, synthesis, and study of peptide models. Methods Enzymol. 1987;154:473–498. doi: 10.1016/0076-6879(87)54091-5. [DOI] [PubMed] [Google Scholar]
- Thomas K. A., Smith G. M., Thomas T. B., Feldmann R. J. Electronic distributions within protein phenylalanine aromatic rings are reflected by the three-dimensional oxygen atom environments. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4843–4847. doi: 10.1073/pnas.79.16.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warme P. K., Morgan R. S. A survey of amino acid side-chain interactions in 21 proteins. J Mol Biol. 1978 Jan 25;118(3):289–304. doi: 10.1016/0022-2836(78)90229-2. [DOI] [PubMed] [Google Scholar]
- Warme P. K., Morgan R. S. A survey of atomic interactions in 21 proteins. J Mol Biol. 1978 Jan 25;118(3):273–287. doi: 10.1016/0022-2836(78)90228-0. [DOI] [PubMed] [Google Scholar]
- White S. H., Jacobs R. E. Statistical distribution of hydrophobic residues along the length of protein chains. Implications for protein folding and evolution. Biophys J. 1990 Apr;57(4):911–921. doi: 10.1016/S0006-3495(90)82611-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R., Andersson L., Cullis P. M., Southgate C. C. Affinities of amino acid side chains for solvent water. Biochemistry. 1981 Feb 17;20(4):849–855. doi: 10.1021/bi00507a030. [DOI] [PubMed] [Google Scholar]


