Abstract
The impact of cotransfection of mixtures of mutant and wild type (WT) virus on the observed phenotype and replication capacity (RC) in a single-cycle human immunodeficiency virus (HIV) phenotypic assay has been investigated by cotransfecting mutant HIV clones expressing the firefly luciferase expression gene with a WT clone expressing Renilla luciferase. Four mutant constructs with different genotypes displayed <1% RC when transfected alone. Cotransfection of as little as 9% of the WT clone resulted in an 18- to 33-fold increase in the RC of the mutant clones. In addition, the 50% inhibitory concentration (IC50) of lopinavir against seven mutant clones decreased by up to 97% after incremental cotransfection of 9 to 50% of the WT clone. The enhancement of RC and decrease in IC50 for mutant variants following cotransfection with the WT variant appear to be due to complementation rather than genetic recombination. These findings suggest that the RC and susceptibility of plasma isolates from patients who are off therapy or not adherent to treatment, in which WT virus may expand to significant levels, should be interpreted with caution.
Human immunodeficiency virus (HIV) resistance testing methods have been shown to improve the success of antiretroviral therapy in treatment-experienced patients (3, 10, 11, 25, 36). As such, resistance testing is now recommended to help guide the choice of regimens after first- and multiple-drug treatment failure by three consensus panels (the International AIDS Society-USA, the EuroGuideline Group, and the U.S. Department of Health and Human Services) (10, 12, 34). In addition, consideration of resistance testing in primary HIV infection is also recommended (12, 34). Two types of tests can be beneficial: genotypic assays, which document the presence of mutations known to confer decreased drug susceptibility (29), and phenotypic assays, which determine the concentration of an antiretroviral agent that reduces HIV replication by 50% (IC50) in tissue culture (14, 16, 18, 24). Conventional phenotypic assays are labor-intensive, time-consuming, and inefficient because of the requirement for virus isolation from peripheral blood mononuclear cells (18). To overcome this problem, recombinant phenotypic assays, based on direct amplification of the patient's gene of interest (protease [PR] gene and a portion of the reverse transcriptase [RT] gene) from viral RNA in plasma, have been developed (14, 24). These recombinant virus assays are rapid, reproducible, and adaptable to large-scale application through use of robotics. To capture and preserve the PR and RT sequence heterogeneity of the plasma virus, the target cells were cotransfected with either the “pool” of DNA generated from patient plasma together with a linearized HIV genomic vector or with the pool of the resistance testing vectors generated by cloning the pool of PCR-generated DNA into a modified HIV vector that lacks the analogous sequence (14, 24). In both cases, a pool of recombinant viruses were generated from the transfected cells and used for drug susceptibility determination. Then, these population-based approaches measure the drug susceptibility of the viral population, compared to a standard wild-type (WT) virus, represented as the incremental factor of change in IC50 (FC).
Recent modifications of the single-cycle phenotypic assay, employing a standardized transfection of HIV proviral DNA, have also allowed the estimation of the replication capacities (RCs) of patient viruses (20, 24, 27; R. Haubrich, T. Wrin, and N. Hellmann, Abstr. XI Int. HIV Drug Resist. Workshop, abstr. 121, 2002). Both the FC and RC have been shown to correlate with clinical response (3, 10, 11, 25, 36; Haubrich et al., Abstr. XI Int. HIV Drug Resist. Workshop; N. Hellmann, T. Wrin, and M. Bates, Abstr. XI Int. HIV Drug Resist. Workshop, abstr. 63, 2002). However, plasma samples from treatment-experienced patients might contain mixtures of HIV variants with substantially different FCs and RCs, particularly if therapy has been discontinued for a sufficient period of time to allow archival WT strains to expand within the quasispecies (6, 17, 28). Under conditions of the single-cycle assay, cotransfection of different viral variants into the same cell might provide the opportunity for genetic recombination and/or complementation. Since viruses with drug resistance mutations often exhibit reduced RCs compared to standard WT strains (21), the impact of mixed species on RC and FC is uncertain. In one study (24), mixing a mutant strain with 25, 50, and 75% of WT virus reduced the FC of nelfinavir by 70, 88, and 93%, respectively. In each case, the percent decrease in observed FC was greater than that for the cotransfected WT strain, suggesting an interaction between mutant and WT viruses in the cotransfection. The effect of this potential interaction (complementation or recombination) between WT and mutant on the output (FC and RC) of resistance testing assays has not been thoroughly analyzed. The objective of the present study was to quantitate the effect of different proportions of WT virus on the apparent drug susceptibility and RC of mutant virus when both are present in mixtures by using WT and mutant clones that express different reporter genes.
MATERIALS AND METHODS
Plasma sample.
Plasma samples were obtained from PR inhibitor (PI)-experienced individuals receiving lopinavir (LPV)-ritonavir therapy either at baseline or at the time of viral rebound.
Construction of mutant and WT HIV clones.
Proviral molecular clones pNL4-3-Fluc and pNL4-3-Rluc were kindly provided by J. He and N. Landau at Indiana University and The Salk Institute, respectively. pNL4-3-Fluc was constructed to contain a firefly luciferase expression gene in the nef region and a frameshift in the envelope (13). A unique XmaI restriction cut site was introduced into pNL4-3-Fluc downstream of the PR coding region to construct the shuttle vector designated pNL4-3-Fluc-x. The pNL4-3-Fluc-x construct was used as the vector for construction of a variety of mutants.
Viral RNA was isolated from 200 μl of plasma with QIAamp RNA extraction kits (QIAGEN, Hilden, Germany) according to the manufacture's instructions. Reverse transcription was performed with Superscript II (Gibco/BRL, Gaithersburg, Md.). A fragment spanning the PR and the p7/p1 and p1/p6 cleavage sites was amplified with the Platinum Taq DNA polymerase high-fidelity system (Invitrogen, Carlsbad, Calif.) with forward and reverse primers containing ApaI and XmaI sites, respectively. After digestion with ApaI and XmaI, the PCR products were inserted into the pNL4-3-Fluc-x vector, in which the ApaI-XmaI fragment had been removed. Ligation reaction products were transformed into competent Escherichia coli (Invitrogen). DNA plasmids purified from each individual colony were sequenced to determine their genotypes. The resulting constructs were designated mutant clones.
Drug susceptibility and RC assays.
Drug susceptibility was determined by a single-cycle assay. To generate recombinant pseudotyped viruses, human embryonic kidney 293 cells were cotransfected with either WT pNL4-3-Fluc-x or a mutant pNL4-3-Fluc-x clone along with an expression vector carrying the vesicular stomatitis virus envelope gene with Lipofectamine PLUS reagent (Invitrogen). The transfection conditions were optimized to achieve >90% transfection efficiency according to the manufacturer's instruction. Three hours after transfection, cells were trypsinized and seeded into 96-well plates containing serial dilutions of LPV. The culture supernatants harvested at 2 days posttransfection were used to infect fresh 293 cells at a concentration of 104 cells/well. To investigate the effect on drug susceptibility of viral mixtures, each mutant clone (expressing firefly luciferase) was mixed with WT pNL4-3-Rluc (expressing Renilla luciferase) at ratios of 1:0.1 (9%), 1:0.5 (33%), and 1:1 (50%). The mixtures of mutant and WT DNA were used for cotransfection as described above. Firefly and Renilla luciferase activities for each sample were sequentially measured with the Dual-Luciferase reporter assay system according to the manufacturer's instructions (Promega Corporation, Madison, Wis.) in order to minimize experimental variations from differences in pipetting volumes, cell lysis efficiency, amount of cells, and assay efficiency. In each experiment, mutant pNL4-3-Fluc-x and WT pNL4-3-Rluc as well as pNL4-3-Fluc-x were transfected alone to serve as controls. Because of the single-replication-cycle format of this assay, the level of luciferase activity in transfected cells is proportional to the amount of HIV DNA transfected; therefore, the luciferase activities from transfected or cotransfected cells were measured for each transfection in order to normalize the transfection efficiency. The percent inhibition was calculated from the formula [1 − (luciferase activity in the presence of drug/luciferase activity in the absence of drug)] × 100. The IC50 was determined by nonlinear regression curve fitting with the Prism program. As controls, the LPV IC50 for the WT was determined following cotransfection with the mutant clones and transfection of the WT pNL4-3-Rluc clone as well as transfection of WT pNL4-3-Fluc-x alone. The FC is the IC50 for the mutant virus/the IC50 for WT pNL4-3-Fluc-x.
The RC for each mutant was calculated by comparing the firefly luciferase activity generated by the mutants to that generated by WT pNL4-3-Fluc-x, after adjusting for minor differences in transfection efficiencies. The RC values were expressed as percentages of that for the WT and reflect the levels of replication for mutant viruses compared to that for the WT control.
Sequence analysis.
The ApaI-SmaI fragment containing the p7/p1 and p1/p6 cleavage sites as well as the PR gene was amplified by RT-PCR from supernatant harvested from cotransfection of mutant and WT constructs. The amplified products were purified and then blunt end ligated into the TA cloning vector (Invitrogen). Miniprep plasmid DNA from individual colonies was purified and then sequenced with an ABI-373 DNA sequencer (Applied Biosystems, Foster City, Calif.).
RESULTS
The impaired RC of mutants is restored by cotransfected WT DNA.
To assess the effect of cotransfection of WT HIV on the RC of mutant strains, we compared the RCs of 11 mutant clones after transfection alone or after cotransfection with WT pNL4-3-Rluc at ratios of mutant to WT of 10:1 (9% of WT), 2:1 (33% of WT), and 1:1 (50% of WT). WT pNL4-3-Rluc and WT pNL4-3-Fluc-x were included as controls. The genotypes of the 11 mutant clones are shown in Table 1. Each mutant clone contains one or two primary mutations and a number of secondary PI resistance mutations. Prior to the cotransfection experiments, the transfection efficiencies were optimized to achieve greater than 90% efficiency. Luciferase activity, measured 48 h posttransfection, correlated well with the levels of p24 production from transfected cells (data not shown). Under optimized transfection conditions, the difference in transfection efficiency between different reactions was minor (<30%; data not shown). For 95% of the determinations, the values of RC from replicate assays differed by less than twofold. The values of standard deviations of RC were <30% of the means for 80% of the determinations.
TABLE 1.
Genotype of the mutant clones
| Mutant clone | Mutations in PR |
|---|---|
| 1 | L10I, L24I, E35D, M36I, M46L, I54V, L63P, I64V, V82A |
| 2 | L10F, L24I, E35D, M36V, N37S, R41K, M46I, I54V, D60E, Q61E, I62V, L63P, I64V, V82A |
| 3 | L10F, I15T, E35D, M36V, N37S, R41K, I54V, D60E, Q61E, I62V, L63P, I64V, V82A |
| 4 | L10V, V32I, N37S, M46I, I47V, I62V, L63P, V77I, P79S, Q92K, I93L, C95F |
| 5 | L10F, V32I, E35D, M36I, M46L, I54V, L63P, I64V, V82A |
| 6 | L10I, E35D, N37D, L63P, A71V, T74P, I84V, L90M, I93L |
| 7 | L10I, E35D, N37D, M46I, I54V, L63P, A71V, T74P, I84V, L90M, I93L |
| 8 | L10I, V32I, M46I, I47A, I62V, L63P, V77I, Q92K, I93L, C95F |
| 9 | L10I, G48V, I54V, L63P, A71V, I72M, V77I, V82A, L90M, I93L |
| 10 | L10I, V32F, G48V, I54V, V56K, L63P, A71V, I72M, V77I, V82A, L90M, I93L |
| 11 | L10V, I15V, G16E, K20R, E35D, M36I, R41K, M46I, I50V, I54V, K55R, R57K, Q61G, I64L, A71V, I72R, V82A, L89I, L90M, Q92K |
Mutant clones 1 to 4 displayed low RCs (<1% of WT) when transfected alone. In contrast, cotransfection with as little as 9% of the WT clone increased the RCs of these clones by 18- to 33-fold (Table 2). Cotransfection with 33 and 50% of WT further enhanced the RCs of those four mutant clones to at least 10% and up to 76% of WT, respectively (incremental increase of 68- to 690-fold). In contrast, the other seven mutants (mutant clones 5 to 11) displayed only modestly decreased RCs compared to the WT clone (15 to 68%) when transfected alone. The RCs of those seven mutants did not change significantly upon cotransfection with up to 50% of WT (one- to fourfold). Similarly, there was no significant change in the RC of WT virus after cotransfection with up to 67% mutants, as indicated by Renilla luciferase activity (data not shown).
TABLE 2.
Effect of cotransfection with WT virus on the RCs of mutant viruses
| Mutant clone | Mean RC (%) ± SDa (fold increase in RCb) at indicated % of WT DNA cotransfected
|
|||
|---|---|---|---|---|
| 0% | 9% | 33% | 50% | |
| 1 | 0.07 ± 0.008 | 1.6 ± 0.5 (22) | 10.9 ± 1.4 (155) | 22.1 ± 6.3 (315) |
| 2 | 0.08 ± 0.05 | 1.7 ± 0.7 (21) | 43.7 ± 10.4 (546) | 55.2 ± 4.2 (690) |
| 3 | 0.16 ± 0.6 | 5.3 ± 0.7 (33) | 48.2 ± 2.8 (301) | 64.3 ± 11.2 (402) |
| 4 | 0.7 ± 0.4 | 12.3 ± 4.7 (18) | 47.7 ± 4.1 (68) | 75.9 ± 8.1 (108) |
| 5 | 14.7 ± 3.0 | 19.8 ± 3.6 (1) | 46.8 ± 5.8 (3) | 57.0 ± 5.5 (4) |
| 6 | 67.6 ± 4.9 | 83.5 ± 18.0 (1) | 146.8 ± 2.4 (2) | 160 ± 5.5 (2) |
| 7 | 32.5 ± 8.5 | 32.9 ± 7.3 (1) | 34.3 ± 7.6 (1) | 50.2 ± 1.6 (2) |
| 8 | 32.7 ± 0.3 | 41.8 ± 3.4 (1) | 52.5 ± 1.1 (2) | 56.0 ± 10.1 (2) |
| 9 | 47.9 ± 4.8 | 47.2 ± 4.8 (1) | 57.3 ± 2.4 (1) | 97.4 ± 5.5 (2) |
| 10 | 20.4 ± 5.6 | 21.5 ± 1.9 (1) | 27.3 ± 7.7 (1) | 22.6 ± 0.3 (1) |
| 11 | 38.6 ± 13.2 | 31.6 ± 1.8 (1) | 44.1 ± 2.6 (1) | 40.5 ± 5.1 (1) |
The relative RC was determined by single-cycle assay as described in Materials and Methods. RC percentages are calculated from the formula: (mean luciferase activity of the test samples/mean luciferase activity of WT pNL4-3-Fluc-x from six replicates for each experiment) × 100. Values were derived from two or three separate experiments.
Values are RCs of mutants in mixture/RCs of mutant transfected alone.
The apparent phenotypes of mutants are decreased by cotransfection with WT DNA.
We also investigated the effect of cotransfection of WT HIV on the phenotypic susceptibility (FC) of mutant clones. Prior to this study, validation of this phenotypic assay was performed. The susceptibility of WT pNL4-3-Fluc to LPV was determined in four independent experiments, and each experiment includes triplicate wells. The IC50 values for LPV derived from four independent experiments were 5.0-, 5.5-, 7.4-, and 7.8-fold, respectively. We also tested other PIs (ritonavir, indinavir, nelfinavir, and saquinavir) in this assay; the IC50 values obtained with this assay correlated well with the results published in the literature (data not shown). The variations among different experiments for at least 95% of the determinations were less than threefold.
Since mutant clones 1 to 4 displayed insufficient RC for phenotypic evaluation, these experiments were limited to mutant clones 5 to 11. The reductions in LPV susceptibility of the seven mutants after transfection alone ranged from 21- to 277-fold, compared to the WT standard (Table 3). Upon cotransfection with 9% WT DNA, the FCs for six of seven mutants decreased by approximately one-half (range, 41 to 63%). Cotransfection of 33 and 50% WT DNA substantially decreased the FCs of all mutants (72 to 97%). In all cases, the percent decrease in FC for the mutant was greater than the percentage for the cotransfected WT strain, suggesting that FC values for mutant strains are underestimated when mixtures of mutant and WT DNA are cotransfected. In contrast, the susceptibility of the WT clone was not significantly affected by cotransfection with up to 50 to 67% of the each of four mutant clones (Table 4). In each case, a twofold or less change in IC50 after cotransfection with the mutant was observed.
TABLE 3.
Effect of cotransfection with WT clone on the phenotypes of mutant viruses
| Mutant clone | Mean FC for LPV compared to WT ± SDa (% decrease in FC of the mutant upon cotransfection with WTb) at indicated % of WT DNA cotransfected
|
|||
|---|---|---|---|---|
| 0% | 9% | 33% | 50% | |
| 5 | 25.8 ± 4.1 | 13.5 ± 1.3 (48) | 7.2 ± 1.2 (72) | 6.7 ± 0.9 (74) |
| 6 | 21.2 ± 7.2 | 7.9 ± 1.2 (63) | 4.5 ± 1.0 (79) | 2.8 ± 0.6 (97) |
| 7 | 66.2 ± 21.1 | 29.6 ± 10.4 (55) | 6.1 ± 1.2 (91) | 3.0 ± 2.1 (95) |
| 8 | 276.9 ± 27.4 | 140.9 ± 20.8 (49) | 29.5 ± 6.6 (89) | 9.0 ± 2.4 (97) |
| 9 | 35.8 ± 11.8 | 21.1 ± 0.36 (41) | 7.2 ± 0.7 (80) | 3.1 ± 0.9 (91) |
| 10 | 54.5 ± 6.6 | 24.4 ± 3.8 (55) | 5.0 ± 1.7 (91) | 3.0 ± 0.4 (95) |
| 11 | 150.1 ± 17.1 | 132.6 ± 37.0 (11) | 31.3 ± 2.8 (79) | 16.7 ± 3.5 (89) |
Values were determined from the formula IC50 of mutant either alone or in mixture/IC50 of WT pNL4- 3-Fluc. The mean changes in IC50 were derived from two or three independent experiments, and each experiment contains triplicates.
Values are calculated from the formula (FC of mutant in mixture/FC of mutant alone) × 100.
TABLE 4.
Effect of cotransfection of mutant clones on the LPV susceptibility of WT virus
| Mutant clone cotransfected | IC50 for WT pNL4-3-Rluc of LPV (μM) at indicated % of mutant DNA cotransfected
|
|
|---|---|---|
| 50% | 67% | |
| 4 | 0.011 | 0.021 |
| 8 | 0.013 | 0.013 |
| 9 | 0.014 | 0.024 |
| 10 | 0.017 | 0.025 |
| pNL4-3-Fluc-x (WT) | 0.014 | 0.013 |
The interaction between WT and mutant occurs in cotransfected cells.
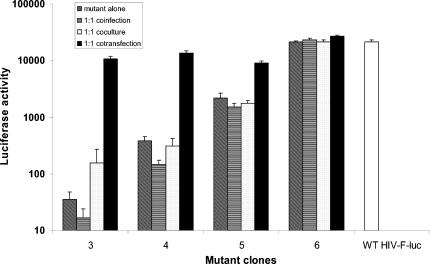
To investigate the mechanism of the observed interaction between mutant and WT viruses in the single-cycle assay, we performed coculture, coinfection, and cotransfection experiments comparing the WT and two mutant clones with poor RCs (clones 3 and 4), as well as two with higher RCs (clones 5 and 6). In contrast to the results of cotransfection experiments, coculture of cells containing individually transfected mutants and WT at a ratio of 1:1 did not affect the RCs of the mutants, as indicated by firefly luciferase activity (Fig. 1). Similarly, upon coinfection of cells with a 1:1 ratio of mutants and WT viruses harvested from individually transfected cells the RCs of the mutant clones did not change significantly from those observed upon infection without WT virus (Fig. 1). These results indicate that the complementation of the mutant RC by WT HIV occurs primarily in cotransfected cells when the mutant and WT DNA coexist in the same cells.
FIG. 1.
Relative replication capacity was determined by measuring the single-cycle growth of recombinant viruses containing a luciferase reporter gene. Mutant alone, WT pNL4-3-Rluc and mutant DNAs individually transfected into 293 cells. Equivalent amounts of WT pNL4-3-Fluc-x or mutant viruses harvested from the individual transfected cells were used to infect fresh 293 cells. Coinfection was performed by infecting fresh 293 cells with a 1:1 mixture of mutants and WT viruses that were harvested from individually transfected cells 48 h posttransfection. Coculture was done by mixing cells containing individually transfected mutants and WT at a ratio of 1:1 3 h after transfection and then coculturing for 48 h. Cotransfection was performed by transfecting a 1:1 mixture of WT and mutant DNA. Luciferase activity was quantified 48 h postinfection, and relative virus growth was normalized to that of its parental WT pNL4-3-Fluc-x. The results were derived from a single experiment with six replicates.
No evidence of recombination in cotransfected cells.
To address whether the enhancement of RCs and decrease in FCs of the mutant clones following cotransfection with the WT clone are due to genetic recombination, sequences of 16 to 20 individual clones from the supernatants harvested from the cotransfection of mutant clones 4 and 6 with WT were determined. As shown in Table 5, the ratios of mutant to WT sequences corresponded to the relative amounts of mutant and WT clones used for cotransfection (1:1, 2:1, and 10:1, respectively). No recombination was observed, suggesting that the interaction of mutant and WT viruses in the above single-cycle replication assays is not due to recombination.
TABLE 5.
Frequencies of mutant and WT sequences detected in supernatant from the cotransfection
| Mutant clone cotransfected | % of WT cotransfected | No. of positive clones of (% of total)
|
||
|---|---|---|---|---|
| Mutant | WT | Total | ||
| 4 | 50 | 9 (45) | 11 (55) | 20 |
| 33 | 14 (73) | 5 (27) | 19 | |
| 9 | 20 (100) | 0 (0) | 20 | |
| 6 | 50 | 8 (45) | 10 (55) | 18 |
| 33 | 11 (68) | 5 (32) | 16 | |
| 9 | 16 (84) | 3 (16) | 19 | |
DISCUSSION
In this study, we analyzed the effect of mixtures of viral strains on the apparent RCs and FCs of HIV molecular clones. We observed that even relatively small amounts of WT virus within a viral population could significantly enhance the apparent RCs and decrease the FCs of mutant strains. Cotransfection of the mutant strain with as little as 9% WT clone resulted in a significant increase in the RCs of all four mutant strains that had low RCs when transfected alone. Cotransfection with 33 and 50% WT strain further increased the RC to a level close to that for the WT. In contrast, we observed that, when mutant clones with only modestly reduced RCs were cotransfected with WT virus, the RC did not change significantly. Furthermore, the RC of the WT virus was not affected by cotransfection with mutants with low RC.
Similarly, cotransfection of as little as 9% WT clone decreased the FCs of mutants by up to 63%. Similar results were obtained in a previous study in which the apparent FC of mutant virus decreased by 70 to 93% upon mixing with 25 to 75% WT virus (24). The agreement between these two studies suggests that (i) the assay used in this study is reliable and (ii) the observed effects of virus mixtures on the RC and FC in the present study are likely to be observed in any recombinant phenotypic assay employing transfection of mixed populations.
The present results indicate that cotransfected WT virus can complement the replication defect of mutant strains, presumably through an intracellular interaction. The construction of WT and mutant clones expressing different luciferase reporters allows quantitation of the effect of this complementation. Since no recombinant clones were observed upon sequence analysis of the viruses harvested from the cotransfection, genetic recombination is unlikely to be responsible for the effect. Lack of evidence of recombination is in agreement with the nature of single-cycle assays and the short period of viral replication (4 days total). Our results also indicate that complementation occurs only following cotransfection, not during coculture or coinfection, suggesting that the mechanism of the complementation requires the presence of both mutant and WT DNA in the same cell. Under coculture or coinfection conditions, the probability of coexistence of WT and mutant strains within the same cell is low (8, 26, 30). The fact that the complementation between WT and mutant occurs only in cotransfected cells indicates that this event may have no significant impact on the RCs and drug susceptibilities of mutants in vivo because it requires superinfection, which could occur in vivo but is not common. The reduced RC of PI-resistant virus is likely in many cases to be attributable to reduced efficiency in the cleavage of the Gag and Gag-Pol polyproteins by a mutant HIV PR, through decreased substrate binding and cleavage and/or impaired dimerization (4, 20, 27, 35). Indeed, some of the mutations selected during PI therapy apparently contribute to resistance by enhancing proteolytic efficiency and RC (21, 22, 23). Furthermore, mutations in the p1/p7 and NC/p1 substrate sites can also partially alleviate the reduced RC of PI-resistant virus (5, 20). Our results suggest that, following cotransfection of WT and mutant HIV DNA, independent transcription and translation produce both mutant and WT Gag-Pol polyproteins. The coexistence of WT and mutant PR may complement the defective catalytic efficiency of mutant PR. Gag and Pol proteins processed by WT PR would be expected to nonselectively package HIV RNA derived from either pNL4-3-Rluc (WT) or the mutant clone, leading to an artificial increase in the number of infectious “mutant” particles (those containing mutant RNA and leading to the expression of firefly luciferase). A similar mechanism is likely to produce the decrease in the FCs of the mutants. The reduced susceptibility of PI-resistant virus is generally attributable to the loss in affinity for the binding of a PI to the mutant HIV PR active site (4, 7, 32). In cells cotransfected with WT and mutant strains, a significant amount of total (WT plus mutant) PR activity is inhibited at drug concentrations substantially lower than the IC50 of the pure mutant strain (WT PR is preferentially inhibited). If proteolytic processing is rate-determining for replication (15, 19, 31), the lowered total PR activity (due to smaller amounts of active PR and reduced processing efficiency of mutant PR, particularly for WT Gag and Gag-Pol) would be expected to lower the observed IC50 of the mutant (firefly luciferase activity). In contrast, the presence of mutant PR in the cotransfected cells would presumably have little impact on the RC of the WT strain because sufficient WT PR is present to complete processing. Furthermore, if the WT PR is responsible for the majority of processing, the effect of cotransfecting mutant PR is expected to have minimal impact on the FC of the WT strain. Further studies are needed for fully understanding the mechanism of the above observations.
These observations have implications for the interpretation of RC and susceptibility during the clinical management of patients by phenotypic resistance testing assays. Since the cotransfection of even relatively small amounts of WT virus (that may not be detectable by population sequencing methods) may significantly increase RC and reduce FC, these parameters should be interpreted with caution in the following groups of patients: (i) patients who were previously heavily treated and are currently off therapy or not adherent to treatment, in which WT virus or mutant variants with better fitness may expand to significant levels in the population (9, 17), and (ii) treatment-naive patients whose resistant mutants are just emerging and are in competition with WT virus (1, 33). The present study was limited to the investigation of mixing WT and mutant strains; the effect of mixing divergent mutants has not been assessed. The present study is also limited by the use of individual clones and is not amenable to the more-complex viral mixtures that are likely to be present in vivo (9, 17). Furthermore, the present study is limited to the investigation of the effect of cotransfection of a mixture on the phenotypic susceptibility of mutants to LPV. It is speculated that this apparent decrease in resistance may be also seen with other PIs. However, it is not clear if this would be the case for RT inhibitors (RTIs). The effect of cotransfection of mixtures on the susceptibility to other PIs and RTIs should be further studied. Nonetheless, our results suggest that complementation between viral populations can occur with samples from patient plasma. Since the effect of complementation is not assessable to current commercial HIV resistance testing methodology, the interpretation of testing results should be always performed in the context of other clinical data and treatment history.
In summary, because of the unique cotransfection step inherent to single-cycle HIV resistance assays, even relatively small amounts of WT virus within a viral population can significantly impact the apparent RCs and phenotypes of mutant strains. This effect appears to be due to complementation rather than genetic recombination. The RCs and susceptibilities of plasma isolates should be interpreted with caution for patients who are off therapy or not adherent to treatment.
Acknowledgments
We are grateful to Johnny He and Nathaniel R. Landau for providing pNL4-3-Fluc and pNL4-3-Rluc constructs and to Scott C Brun, William Kohlbrenner, and Shing Chang for supporting this study.
REFERENCES
- 1.Bi, X., H. Gatanaga, S. Ida, K. Tsuchiya, S. Matsuoka-Aizawa, S. Kimura, and S. Oka. 2003. Emergence of protease inhibitor resistance-associated mutations in plasma HIV-1 precedes that in proviruses of peripheral blood mononuclear cells by more than a year. J. Acquir. Immune Defic. Syndr. 34:1-6. [DOI] [PubMed] [Google Scholar]
- 2.Call, S. A., M. S. Saag, A. O. Westfall, J. L. Raper, S. V. Pham, J. M. Tolson, N. S. Hellmann, G. A. Cloud, and V. A. Johnson. 2001. Phenotypic drug susceptibility testing predicts long-term virologic suppression better than treatment history in patients with human immunodeficiency virus infection. J. Infect. Dis. 183:401-408. [DOI] [PubMed] [Google Scholar]
- 3.Cohen, C. J., S. Hunt, M. Sension, C. Farthing, M. Conant, S. Jacobson, J. Nadler, W. Verbiest, K. Hertogs, M. Ames, A. R. Rinehart, and N. M. Graham. 2002. A randomized trial assessing the impact of phenotypic resistance testing on antiretroviral therapy. AIDS 16:579-588. [DOI] [PubMed] [Google Scholar]
- 4.Croteau, G., L. Doyon, D. Thibeault, G. McKercher, L. Pilote, and D. Lamarre. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frost, S. D., M. Nijhuis, R. Schuurman, C. A. Boucher, and A. J. Brown. 2000. Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: the relative roles of drift and selection. J. Virol. 74:6262-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldblum, A. 1990. Modulation of the affinity of aspartic proteases by the mutated residues in active site models. FEBS Lett. 261:241-244. [DOI] [PubMed] [Google Scholar]
- 8.Gonzales, M. J., E. Delwart, S. Y. Rhee, R. Tsui, A. R. Zolopa, J. Taylor, and R. W. Shafer. 2003. Lack of detectable human immunodeficiency virus type 1 superinfection during 1072 person-years of observation. J. Infect. Dis. 188:397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hance, A. J., V. Lemiale, J. Izopet, D. Lecossier, V. Joly, P. Massip, F. Mammano, D. Descamps, F. Brun-Vezinet, and F. Clavel. 2001. Changes in human immunodeficiency virus type 1 populations after treatment interruption in patients failing antiretroviral therapy. J. Virol. 75:6410-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanna, G. J., and R. T. D'Aquila. 2001. Clinical use of genotypic and phenotypic drug resistance testing to monitor antiretroviral chemotherapy. Clin. Infect. Dis. 32:774-782. [DOI] [PubMed] [Google Scholar]
- 11.Harrigan, P. R., K. Hertogs, W. Verbiest, R. Pauwels, B. Larder, S. Kemp, S. Bloor, B. Yip, R. Hogg, C. Alexander, and J. S. Montaner. 1999. Baseline HIV drug resistance profile predicts response to ritonavir-saquinavir protease inhibitor therapy in a community setting. AIDS 13:1863-1871. [DOI] [PubMed] [Google Scholar]
- 12.Haubrich, R., and L. Demeter. 2001. International perspectives on antiretroviral resistance. Clinical utility of resistance testing: retrospective and prospective data supporting use and current recommendations. J. Acquir. Immune Defic. Syndr. 26(Suppl. 1):S51-S59. [DOI] [PubMed] [Google Scholar]
- 13.He, J., and N. R. Landau. 1995. Use of a novel human immunodeficiency virus type 1 reporter virus expressing human placental alkaline phosphatase to detect an alternative viral receptor. J. Virol. 69:4587-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hertogs, K., M. P. de Bethune, V. Miller, T. Ivens, P. Schel, A. Van Cauwenberge, C. Van Den Eynde, V. Van Gerwen, H. Azijn, M. Van Houtte, F. Peeters, S. Staszewski, M. Conant, S. Bloor, S. Kemp, B. Larder, and R. Pauwels. 1998. A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency virus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother. 42:269-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iga, M., Z. Matsuda, A. Okayama, W. Sugiura, S. Hashida, K. Morishita, Y. Nagai, and H. Tsubouchi. 2002. Rapid phenotypic assay for human immunodeficiency virus type 1 protease using in vitro translation. J. Virol. Methods 106:25-37. [DOI] [PubMed] [Google Scholar]
- 17.Izopet, J., C. Souyris, A. Hance, K. Sandres-Saune, M. Alvarez, C. Pasquier, F. Clavel, J. Puel, and P. Massip. 2002. Evolution of human immunodeficiency virus type 1 populations after resumption of therapy following treatment interruption and shift in resistance genotype. J. Infect. Dis. 185:1506-1510. [DOI] [PubMed] [Google Scholar]
- 18.Japour, A. J., D. L. Mayers, V. A. Johnson, D. R. Kuritzkes, L. A. Beckett, J. M. Arduino, J. Lane, R. J. Black, P. S. Reichelderfer, R. T. D'Aquila, C. S. Crumpacker, the RV-43 Study Group, and the AIDS Clinical Trials Group Virology Committee Resistance Working Group. 1993. Standardized peripheral blood mononuclear cell culture assay for determination of drug susceptibilities of clinical human immunodeficiency virus type 1 isolates. Antimicrob. Agents Chemother. 37:1095-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jardine, D. K., D. P. Tyssen, and C. J. Birch. 2000. Effect of protease inhibitors on HIV-1 maturation and infectivity. Antiviral Res. 45:59-68. [DOI] [PubMed] [Google Scholar]
- 20.Maguire, M. F., R. Guinea, P. Griffin, S. Macmanus, R. C. Elston, J. Wolfram, N. Richards, M. H. Hanlon, D. J. Porter, T. Wrin, N. Parkin, M. Tisdale, E. Furfine, C. Petropoulos, B. W. Snowden, and J. P. Kleim. 2002. Changes in human immunodeficiency virus type 1 Gag at positions L449 and P453 are linked to I50V protease mutants in vivo and cause reduction of sensitivity to amprenavir and improved viral fitness in vitro. J. Virol. 76:7398-7406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mammano, F., V. Trouplin, V. Zennou, and F. Clavel. 2000. Retracing the evolutionary pathways of human immunodeficiency virus type 1 resistance to protease inhibitors: virus fitness in the absence and in the presence of drug. J. Virol. 74:8524-8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menzo, S., A. Monachetti, C. Balotta, S. Corvasce, S. Rusconi, S. Paolucci, F. Baldanti, P. Bagnarelli, and M. Clementi. 2003. Processivity and drug-dependence of HIV-1 protease: determinants of viral fitness in variants resistant to protease inhibitors. AIDS 17:663-671. [DOI] [PubMed] [Google Scholar]
- 23.Nijhuis, M., R. Schuurman, D. de Jong, J. Erickson, E. Gustchina, J. Albert, P. Schipper, S. Gulnik, and C. A. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 24.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piketty, C., E. Race, P. Castiel, L. Belec, G. Peytavin, A. Si-Mohamed, G. Gonzalez-Canali, L. Weiss, F. Clavel, and M. D. Kazatchkine. 1999. Efficacy of a five-drug combination including ritonavir, saquinavir and efavirenz in patients who failed on a conventional triple-drug regimen: phenotypic resistance to protease inhibitors predicts outcome of therapy. AIDS 13:F71-F77. [DOI] [PubMed] [Google Scholar]
- 26.Potash, M. J., and D. J. Volsky. 1998. Viral interference in HIV-1 infected cells. Rev. Med. Virol. 8:203-211. [DOI] [PubMed] [Google Scholar]
- 27.Prado, J. G., T. Wrin, J. Beauchaine, L. Ruiz, C. J. Petropoulos, S. D. Frost, B. Clotet, R. T. D'Aquila, and J. Martinez-Picado. 2002. Amprenavir-resistant HIV-1 exhibits lopinavir cross-resistance and reduced replication capacity. AIDS 16:1009-1017. [DOI] [PubMed] [Google Scholar]
- 28.Rayner, M. M., B. Cordova, and D. A. Jackson. 1997. Population dynamics studies of wild-type and drug-resistant mutant HIV in mixed infections. Virology 236:85-94. [DOI] [PubMed] [Google Scholar]
- 29.Shafer, R. W. 2002. Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin. Microbiol. Rev. 15:247-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taddeo, B., M. Federico, F. Titti, G. B. Rossi, and P. Verani. 1993. Homologous superinfection of both producer and nonproducer HIV-infected cells is blocked at a late retrotranscription step. Virology 194:441-452. [DOI] [PubMed] [Google Scholar]
- 31.Tessmer, U., and H. G. Krausslich. 1998. Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6* protein is essential for efficient Gag polyprotein processing and viral infectivity. J. Virol. 72:3459-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Todd, M. J., I. Luque, A. Velazquez-Campoy, and E. Freire. 2000. Thermodynamic basis of resistance to HIV-1 protease inhibition: calorimetric analysis of the V82F/I84V active site resistant mutant. Biochemistry 39:11876-11883. [DOI] [PubMed] [Google Scholar]
- 33.Vasudevachari, M. B., Y. M. Zhang, H. Imamichi, T. Imamichi, J. Falloon, and N. P. Salzman. 1996. Emergence of protease inhibitor resistance mutations in human immunodeficiency virus type 1 isolates from patients and rapid screening procedure for their detection. Antimicrob. Agents Chemother. 40:2535-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson, J. W. 2003. Update on antiretroviral drug resistance testing: combining laboratory technology with patient care. AIDS (Reading) 13:25-30, 35-38. [PubMed] [Google Scholar]
- 35.Xie, D., S. Gulnik, E. Gustchina, B. Yu, W. Shao, W. Qoronfleh, A. Nathan, and J. W. Erickson. 1999. Drug resistance mutations can effect dimer stability of HIV-1 protease at neutral pH. Protein Sci. 8:1702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zolopa, A. R., R. W. Shafer, A. Warford, J. G. Montoya, P. Hsu, D. Katzenstein, T. C. Merigan, and B. Efron. 1999. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann. Intern. Med. 131:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]