Abstract
Malassezia yeasts are associated with several dermatological disorders. The conventional identification of Malassezia species by phenotypic methods is complicated and time-consuming, and the results based on culture methods are difficult to interpret. A comparative molecular approach based on the use of three molecular techniques, namely, amplified fragment length polymorphism (AFLP) analysis, sequencing of the internal transcribed spacer, and sequencing of the D1 and D2 domains of the large-subunit ribosomal DNA region, was applied for the identification of Malassezia species. All species could be correctly identified by means of these methods. The results of AFLP analysis and sequencing were in complete agreement with each other. However, some discrepancies were noted when the molecular methods were compared with the phenotypic method of identification. Specific genotypes were distinguished within a collection of Malassezia furfur isolates from Canadian sources. AFLP analysis revealed significant geographical differences between the North American and European M. furfur strains.
The genus Malassezia has received considerable attention in recent years from dermatologists and other clinicians. This group of basidiomycetous yeasts, long known to be the causal agents of pityriasis (tinea) versicolor, is also increasingly being associated with the causation of folliculitis, papillomatosis, and invasive human infections, as well as potential immunogenic triggering of atopic dermatitis, seborrheic dermatitis, and dandruff (6, 9, 11, 14, 18, 25, 26, 29). Malassezia species are listed among the new and emerging yeast pathogens (22, 24).
The genus Malassezia, until recently, was characterized on the basis of rRNA sequences as consisting of seven species, including the lipid-dependent species M. furfur, M. sympodialis, M. globosa, M. obtusa, M. restricta, and M. slooffiae and the lipophilic species M. pachydermatis (12, 13). Recently, two new species have been identified: M. dermatis (39), which was isolated from atopic dermatitis patients, and M. equi (unpublished), a species that was isolated from the skin of horses (32). The latter species has only tentatively been named and still awaits formal description; the present study deals only with the eight described species.
Guillot et al. (17) introduced a physiological system based on lipid assimilation and other phenotypic characteristics for identifying the various Malassezia species. This phenotypic system has been used as the conventional method of identification, though in practice, the test results are not always easy to read. Therefore, several research groups have explored the use of molecular techniques, such as pulsed-field gel electrophoresis (3, 4, 36), randomly amplified polymorphic DNA analysis (1, 3), amplified fragment length polymorphism (AFLP) analysis (40), denaturing gradient gel electrophoresis (40), multilocus enzyme electrophoresis (30), sequencing analysis (37), restriction analysis of PCR amplicons of ribosomal sequences (8, 15, 16, 20, 28), and chitin synthase gene sequence analysis (1, 4, 8, 37), for the identification of Malassezia species. Recently, Gemmer et al. (9) devised a remarkably efficient, novel technique, terminal fragment length polymorphism analysis, for the rapid and reliable identification of Malassezia species. It eliminates the need for strain cultivation in direct investigations of Malassezia populations on skin samples, as well as the need for restriction enzyme digestion of nucleic acids. The technique, however, is not suitable for epidemiological typing, as its ability to show heterogeneity within a species is limited.
The objective of the present study was to facilitate an improved understanding of the epidemiology of Malassezia infections by determining which molecular characterization methodologies could most effectively be used for the identification and strain typing of Malassezia species. It is important to identify the different species correctly, as some studies have given preliminary indications that different Malassezia species may occupy well-defined niches on the human body and may play differential roles in causing various diseases (11, 19, 21). However, these studies have not all been in agreement about which species is prevalent or etiologically important with regard to a specific disease, suggesting that technical improvements in identification methods is critical in producing definitive results.
AFLP analysis is a universally applicable technique that has proven to be useful for the identification and strain typing of microorganisms (2, 5, 34). In this study, we investigated the use of AFLP analysis as well as rapid sequencing of the internal transcribed spacer (ITS) and large-subunit (LSU) regions of the nuclear ribosomal DNA (rDNA) for the identification of the eight currently recognized Malassezia species. We compared the two molecular methods with the conventional method of identification to see if any discrepancies could be observed. The molecular techniques were used to investigate the genetic diversity of Malassezia isolates obtained from a variety of patients in Canada. These patients were mostly from the province of Ontario, a region with a notably diverse population developed through immigration from many areas of the world. In order to understand the extent to which the Ontario isolates might reflect the overall genetic biodiversity of Malassezia spp. from human sources and to ensure that the techniques used were maximally developed as tools for identifying genetic types within Malassezia species, a diverse comparison sample of Malassezia strains from different worldwide geographic regions was chosen from the collection of the Centraalbureau voor Schimmelcultures (CBS; Utrecht, The Netherlands).
MATERIALS AND METHODS
Yeast strains.
One hundred four isolates representing the eight currently recognized Malassezia species were studied. The origins, locations, and hosts of the strains are listed in Table 1. Sixty-five of the strains had originally been isolated and phenotypically identified (17) at the Mycology Laboratory, Ontario Ministry of Health, Toronto, Ontario, Canada, and at Mediprobe Laboratories Inc., London, Ontario, Canada. Thirty-nine isolates were taken from the collection of the CBS. Three strains were received as a gift from Gillian Midgley (London, United Kingdom), and one strain was received from Jan Faergemann (Göteborg, Sweden). Six of the study strains had been isolated from patients residing in Hawaii, South Africa, and Hong Kong. The strains were maintained at 30°C on Leeming and Notman medium, consisting of 1% peptone, 0.5% glucose, 0.01% yeast extract, 0.4% desiccated ox bile, 0.1% (vol/vol) glycerol, 0.5% glycerol monostearate, 0.05% (vol/vol) Tween 60, 1% (vol/vol) high-fat cow's milk, and 1.5% agar in distilled water. The percentages given are for weight per volume unless otherwise specified.
TABLE 1.
Malassezia isolates included in the study
| Isolatea | Origin | Location | AFLP subtypeb | Host | ITS accession no. | LSU accession no. | % Similarity of ITS sequences | % Similarity of LSU sequences |
|---|---|---|---|---|---|---|---|---|
| M. furfur | ||||||||
| CBS 1878, NT of Pityro- sporum ovale* | Dandruff | Unknown | 2 | Human | AY387100 | AY387196 | 100 | 100 |
| EL8 = CBS 9365 | Skin | France | 2 | Elephant | AY387101 | AY387197 | 100 | 100 |
| CBS 4172 | Skin | Sweden | 2 | Elk | AY387102 | AY387198 | 100 | 100 |
| CBS 7969 | Skin | France | 2 | Elephant | AY387103 | AY387199 | 100 | 100 |
| WF8 = CBS 9371 | Arm of tinea versicolor patient | Hawaii | 2 | Human | AY387104 | AY387200 | 100 | 100 |
| 98F8480 = CBS 9376 | Lesion on skin | Canada | 1 | Human | AY387123 | AY387219 | 100 | 100 |
| 2ATB1 | Back | Canada | 1 | Human | AY387124 | AY387220 | 98.62 | 99.64 |
| SWB1 = CBS 9371 | Back | Canada | 1 | Human | AY387125 | AY387221 | 98.62 | 99.64 |
| TPB1 = CBS 9372 | Back | Canada | 1 | Human | AY387126 | AY387222 | 98.62 | 99.64 |
| SOS3 = CBS 9369 | Scalp | Canada | 1 | Human | AY387127 | AY387223 | 98.62 | 99.64 |
| CBS 7982 | Skin of ear from healthy subject | France | 1 | Human | AY387128 | AY387224 | 98.62 | 99.64 |
| 4SJCMC1 = CBS 9374 | Chest | Canada | 1 | Human | AY387131 | AY387227 | 98.62 | 99.64 |
| PM312 | Urine of neonate | Germany | 4 | Human | AY387118 | AY387214 | 100 | 100 |
| PM314 | Tracheal secretion | Germany | 4 | Human | AY387119 | AY387215 | 100 | 100 |
| PM316 | Throat of neonate | Germany | 4 | Human | AY387120 | AY387216 | 100 | 100 |
| JLPK23 | Catheter, blood | France | 4 | Human | AY387121 | AY387217 | 99.59 | 100 |
| JLPK18 | Catheter, liver recipient, blood | France | 4 | Human | AY387122 | AY387218 | 99.59 | 100 |
| GM420 = CBS 9367 | Unknown | United Kingdom | 5 | Human | AY387105 | AY387201 | 100 | 100 |
| JLPK13 | Urine | France | 5 | Human | AY387106 | AY387202 | 99.59 | 100 |
| CBS 7860 | Skin of ear from neonate | United Kingdom | 5 | Human | AY387114 | AY387210 | 99.59 | 100 |
| CBS 7867 | Skin of ear from neonate | United Kingdom | 5 | Human | AY387115 | AY387211 | 99.59 | 100 |
| CBS 7710 | Skin | The Netherlands | 5 | Human | AY387117 | AY387213 | 100 | 99.64 |
| HK8 = CBS 9366 | Skin | Hong Kong | 6 | Human | AY387107 | AY387203 | 98.9 | 100 |
| NCF3 = CBS 9364 | Forehead | Canada | 6 | Human | AY387108 | AY387204 | 98.9 | 100 |
| NCB3 = CBS 9368 | Back | Canada | 6 | Human | AY387109 | AY387205 | 98.9 | 100 |
| NCC2 = CBS 9375 | Chest | Canada | 6 | Human | AY387110 | AY387206 | 98.9 | 100 |
| MF9 | Forehead | Canada | 6 | Human | AY387111 | AY387207 | 100 | 100 |
| NCF2 = CBS 9373 | Forehead | Canada | 6 | Human | AY387112 | AY387208 | 98.9 | 99.82 |
| CBS 6000 | Dandruff | India | 7 | Human | AY387113 | AY387209 | 100 | 99.64 |
| CBS 7985 | Wing of ostrich | France | 8 | Ostrich | AY387129 | AY387225 | 98.62 | 99.82 |
| CBS 7984 | Ear of healthy elephant | France | 8 | Elephant | AY387130 | AY387226 | 98.48 | 99.82 |
| M. globosa | ||||||||
| CBS 7966 T* | Pityriasis versicolor | United Kingdom | Human | AY387132 | AY387228 | 100 | 100 | |
| CBS 7874 | Dandruff | United Kingdom | Human | AY387133 | AY387229 | 92.9 | 99.82 | |
| HK10 | Skin | Hong Kong | Human | AY387134 | AY387230 | 95.49 | 100 | |
| MPS3 | Scalp | Canada | Human | AY387135 | AY387231 | 94.98 | 99.82 | |
| CBC1 | Chest | Canada | Human | AY387136 | AY387232 | 95.37 | 99.82 | |
| M. obtusa | ||||||||
| CBS 7876 T, GM 215* | Skin | United Kingdom | Human | AY387137 | AY387233 | 100 | 100 | |
| CBS 7968 | Atopic dermatitis | United Kingdom | Human | AY387138 | AY387234 | 100 | 100 | |
| M. pachydermatis | ||||||||
| CBS 1879, NT of M. pachy- dermatis* | Skin | France | Pig | AY387139 | AY387235 | 100 | 100 | |
| CBS 1919 | Ulcerated ear of dog | Europe | Dog | AY387140 | AY387236 | 100 | 100 | |
| CBS 1885 | Ear of dog with otitis extema | Sweden | Dog | AY387141 | AY387237 | 95.27 | 99.46 | |
| CBS 1884 | Ear of dog with otitis extema | Sweden | Dog | AY387142 | AY387238 | 100 | 100 | |
| M. restricta | ||||||||
| CBS 7877, T* | Healthy skin | United Kingdom | Human | AY387143 | AY387239 | 100 | 100 | |
| CBS 8747 | Healthy scalp | Canada | Human | AY387144 | AY387240 | 99.85 | 100 | |
| CBS 7991 | Normal skin | United Kingdom | Human | AY387145 | AY387241 | 90.32 | 99.46 | |
| M. slooffiae | ||||||||
| CBS 7956 T, JG 554* | Healthy ear | France | Pig | AY387146 | AY387242 | 100 | 100 | |
| CBS 7973 | Pityriasis versicolor | France | Human | AY387147 | AY387243 | 99.25 | 100 | |
| RBF2 | Forehead | Canada | Human | AY387148 | AY387244 | 99.11 | 100 | |
| CBS 7975 | Dandruff | United Kingdom | Human | AY387149 | AY387245 | 98.96 | 100 | |
| CBS 7875 | Dandruff | United Kingdom | Human | AY387150 | AY387246 | 99.23 | 87.84 | |
| CBS 7972 | Pityriasis versicolor | France | Human | AY387151 | AY387247 | 99.25 | 100 | |
| CBS 7971 | Scalp | United Kingdom | Human | AY387152 | AY387248 | 99.7 | 99.82 | |
| TV1 | Back | Canada | Human | AY387153 | AY387249 | 98.94 | 100 | |
| AWC3 | Chest | Canada | Human | AY387154 | AY387250 | 99.7 | 99.82 | |
| GM150 | Unknown | United Kingdom | Human | AY387155 | AY387251 | 99.49 | 99.82 | |
| SF2 | Skin | South Africa | Human | AY387156 | AY387252 | 99.1 | 99.82 | |
| M. sympodialis | ||||||||
| CBS 7222 T, EG 604* | Human ear | United States | Human | AY387157 | AY387253 | 100 | 100 | |
| WBC2 | Chest | Canada | Human | AY387158 | AY387254 | 100 | 100 | |
| 2SMC3 | Chest | Canada | Human | AY387159 | AY387255 | 99.52 | 100 | |
| ZHC1 | Chest | Canada | Human | AY387160 | AY387256 | 100 | 100 | |
| VMB3 | Back | Canada | Human | AY387161 | AY387257 | 100 | 100 | |
| 98F9925 | Lesion on trunk | Canada | Human | AY387162 | AY387258 | 99.52 | 100 | |
| WF39 | Patient with folliculitis | Hawaii | Human | AY387163 | AY387259 | 99.52 | 100 | |
| LNC1 | Chest | Canada | Human | AY387164 | AY387260 | 99.52 | 100 | |
| ZHC3 | Chest | Canada | Human | AY387165 | AY387261 | 99.52 | 100 | |
| 3SKB3 | Back | Canada | Human | AY387166 | AY387262 | 99.84 | 100 | |
| RBB1 | Back | Canada | Human | AY387167 | AY387263 | 99.52 | 100 | |
| 2MBB1 | Back | Canada | Human | AY387168 | AY387264 | 100 | 100 | |
| SSB2 | Back | Canada | Human | AY387169 | AY387265 | 99.52 | 100 | |
| 2CC3 | Chest | Canada | Human | AY387170 | AY387266 | 100 | 100 | |
| 98F8139 | Lesion on skin | Canada | Human | AY387171 | AY387267 | 100 | 100 | |
| LMB3 | Back | Canada | Human | AY387172 | AY387268 | 99.52 | 100 | |
| RPC1 | Chest | Canada | Human | AY387173 | AY387269 | 99.52 | 100 | |
| RBC1 | Chest | Canada | Human | AY387174 | AY387270 | 100 | 100 | |
| KEB1 | Back | Canada | Human | AY387175 | AY387271 | 99.52 | 100 | |
| LGB3 | Back | Canada | Human | AY387176 | AY387272 | 99.52 | 100 | |
| RGC2 | Chest | Canada | Human | AY387177 | AY387273 | 100 | 100 | |
| DKC1 | Chest | Canada | Human | AY387178 | AY387274 | 100 | 100 | |
| RPB2 | Back | Canada | Human | AY387179 | AY387275 | 99.52 | 100 | |
| JF05 | Back of healthy 23-year-old female | Sweden | Human | AY387180 | AY387276 | 100 | 100 | |
| WF42 | Back of patient with folliculitis | Hawaii | Human | AY387181 | AY387277 | 100 | 100 | |
| WBB2 | Back | Canada | Human | AY387182 | AY387278 | 100 | 100 | |
| 3SKB1 | Back | Canada | Human | AY387183 | AY387279 | 99.52 | 100 | |
| AEB2 | Back | Canada | Human | AY387184 | AY387280 | 99.84 | 100 | |
| BMB3 | Back | Canada | Human | AY387185 | AY387281 | 100 | 100 | |
| CBS 7979 | Skin | United Kingdom | Human | AY387186 | AY387282 | 99.52 | 100 | |
| CBS 7978 | Pityriasis versicolor | United Kingdom | Human | AY387187 | AY387283 | 99.52 | 100 | |
| ZHB2 | Back | Canada | Human | AY387188 | AY387284 | 100 | 100 | |
| CWB1 | Back | Canada | Human | AY387189 | AY387285 | 99.52 | 100 | |
| GM323 | Unknown | United Kingdom | Human | AY387190 | AY387286 | 100 | 100 | |
| RBC3 | Chest | Canada | Human | AY387190 | AY387287 | 100 | 100 | |
| CWB2 | Back | Canada | Human | AY387192 | AY387288 | 99.52 | 100 | |
| GMB1 | Back | Canada | Human | AY387193 | AY387289 | 100 | 100 | |
| JSB2 | Back | Canada | Human | AY387194 | AY387290 | 99.52 | 100 | |
| 98F | Lesion on skin | Canada | Human | AY387195 | AY387291 | 100 | 100 | |
| CBS 7865 | Skin | United Kingdom | Human | AY387116 | AY387212 | 99.59 | 100 | |
| M. dermatis | ||||||||
| CBS 9145 | Atopic dermatitis | Japan | Human | AB070360 | AB070365 | 100 | 100 | |
| CBS 9169 | Atopic dermatitis | Japan | Human | AB070356 | AB070361 | 100 | 100 | |
| CBS 9170 | Atopic dermatitis | Japan | Human | AB070358 | AB070363 | 100 | 100 | |
| Unknown species | ||||||||
| ISB2 | Back | Canada | Human | Unknown | Unknown | Unknown | Unknown | |
| LNS2 | Scalp | Canada | Human | Unknown | Unknown | Unknown | Unknown | |
| KHS2 | Scalp | Canada | Human | Unknown | Unknown | Unknown | Unknown | |
| BSB1 | Back | Canada | Human | Unknown | Unknown | Unknown | Unknown | |
| TPF2 | Forehead | Canada | Human | Unknown | Unknown | Unknown | Unknown | |
| DRC2 | Chest | Canada | Human | Unknown | Unknown | Unknown | Unknown | |
| 4GMC1 | Chest | Canada | Human | Unknown | Unknown | Unknown | Unknown |
Asterisks indicate the type strains used for determining similarity values.
AFLP subtypes are shown for M. furfur only.
DNA extraction.
DNA was extracted according to the cetyltrimethylammonium bromide (CTAB) method (33) from 4- to 5-day-old cultures. Briefly, 3 loopfuls of yeast growth was transferred to a 1.5-ml microcentrifuge tube containing about 100 mg of sterile sand and 800 μl of CTAB buffer (33). Cells were disrupted mechanically for approximately 1 min by using a pestle. The mixture was vortexed and incubated for 4 h at 65°C. The suspension was centrifuged for 30 min at 18,300 × g rpm at 4°C, and 700 μl of the supernatant was transferred to a fresh microcentrifuge tube. Subsequently, 700 μl of chloroform-isoamyl alcohol (24:1 by volume) was added, and the solution was shaken vigorously. The solution was centrifuged at 18,300 × g for 20 min at 4°C, and 500 μl of the supernatant was transferred to a fresh tube. To this, 500 μl of chloroform-isoamyl alcohol was added, and the suspension was centrifuged again at 200 × g for 10 min at 4°C. From this suspension, 350 μl of the aqueous layer was taken and mixed with 150 μl of CTAB buffer. To this, 300 μl of ice-cold isopropanol (kept at −20°C) was added, and the DNA was precipitated by centrifuging at 18,300 × g for 10 min at 4°C. The pellet obtained was washed with cold 70% ethanol. After being dried, the pellet was suspended in 100 μl of sterile water plus 4 μl of RNase (10 mg/ml) (USB Corp., Cleveland, Ohio). The samples were stored at −20°C.
AFLP analysis.
AFLP analysis was performed according to the manufacturer's instructions in the AFLP microbial fingerprinting protocol (PE Biosystems, Nieuwerkerk aan de IJssel, The Netherlands), with some modifications. Restriction and ligation were performed simultaneously on 10 ng of genomic DNA by using 1 U of MseI, 5 U of EcoRI, and 3 U of T4 DNA ligase (Biolabs, Westburg, The Netherlands). The sequences of the primers EcoRI and MseI were 5′-GACTGCGTACCAATTCAC-3′ and 5′-GATGAGTCCTGAGTAAC-3′, respectively. The adaptors used were EcoRI (5′-CTCGTAGACTGCGTACC-3′, forward; 3′-CATCTGACGCATGGTTAA-5′, reverse) and MseI (5′-GACGATGAGTCCTGAG-3′, forward; 3′-CTACTCAGGACTCAT-5′, reverse). The reaction was allowed to take place in a total volume of 5.5 μl with the following constituents: a 0.36 μM concentration of the EcoRI adaptor and a 3.64 μM concentration of the MseI adaptor from the AFLP microbial fingerprinting kit (PE Biosystems), 0.1 M NaCl, 0.91 mM Tris-HCl (pH 7.8), 0.18 mM MgCl2, 0.18 mM dithiothreitol, 18 μM ATP, and 91.36 μg of bovine serum albumin ml−1. The restriction ligation mixture was incubated for 2 h at 37°C and later diluted by adding 25 μl of sterile double-distilled water. The first PCR was performed with two preselective primers (EcoRI core sequence and MseI core sequence) and the AFLP amplification core mix from the AFLP microbial fingerprinting kit, according to the manufacturer's manual, under the following conditions: 2 min at 72°C, followed by 20 cycles of 20 s at 94°C, 30 s at 56°C, and 2 min at 72°C each. The PCR product was diluted by adding 25 μl of sterile double-distilled water. The second PCR used more-selective primers, EcoRI-A FAM and MseI-G. The conditions were 2 min at 94°C; 10 cycles consisting of 20 s at 94°C, 30 s at 66°C (decreasing 1°C every step of the cycle), and 2 min at 72°C; and then 25 cycles consisting of 20 s at 94°C, 30 s at 56°C, and 2 min at 72°C. The samples were prepared for acrylamide capillary electrophoresis with the following loading mix: 2.0 μl of selective amplification product, 24 μl of deionized formamide, and 1 μl of GeneScan-500 labeled with 6-carboxy-X-rhodamine (Applied Biosystems, Foster City, Calif.) as an internal size standard. After incubation for 5 min at 95°C, the samples were run on an ABI 310 genetic analyzer for 30 min each. Data were analyzed with the Bionumerics software package (version 2.5; Applied Maths, Kortrijk, Belgium), by using (i) Pearson correlation based on similarities of the densitometric curves and (ii) the unweighted pair group method with arithmetic means analysis. The statistical reliability of the clusters was confirmed by using the cophenetic values, which calculate the correlation between the calculated similarities and the dendrogram-derived similarities.
PCR amplification and sequencing of ITS and LSU regions.
The ITS I and ITS II regions and the D1 and D2 domains of the LSU region were amplified with primers V9 (5′-TGCGTTGATTACGTCCCTGC) and RLR3R (5′-GGTCCGTGTTTCAAGAC). The PCR conditions were 5 min at 94°C; 35 cycles of 45 s at 94°C, 40 s at 56°C, and 2 min at 72°C; followed by chilling at 4°C. The PCR products were purified by using GFX columns (Amersham Pharmacia Biotech Inc., Roosendaal, The Netherlands) and visualized on electrophoresis gel after ethidium bromide staining. The rDNA was sequenced with the BigDye terminator cycle sequencing kit (Applied Biosystems) and analyzed on an ABI Prism 3700 sequencer (Applied Biosystems) by using the standard conditions recommended by the vendor. The primers used in the sequence reaction were ITS4 (5′-TCCTCCGCTTATTGATATGC) and ITS5 (5′-GGAAGTAAAAGTCGTAACAAGG) for analysis of the ITS region and NL1 (5′-GCATATCAATAAGCGGAGGAAAAG) and RLR3R (5′-GGTCCGTGTTTCAAGAC) for analysis of the D1 and D2 domains of 26S rDNA. The PCR conditions were as follows: 25 PCR cycles of 96°C for 10 s (denaturation), 50°C for 5 s (annealing), and 60°C for 4 min (extension). The sequencing products were purified with Sephadex (Amersham Pharmacia). Sequences were assembled and edited with Seqman II software (DNAStar Inc., Madison, Wis.) and aligned with Megalign (DNAStar). The sequences were visually corrected. Phylogenetic analysis was performed by using PAUP (version 4) parsimony analysis, random stepwise addition, and tree bisection-reconnection.
Statistical analysis.
Likelihood ratios were computed by using SPSS for Macintosh (release 11.0.2, 2003; SPSS, Chicago, Ill.).
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study have been deposited in GenBank. The GenBank numbers for D1 and D2 domain and ITS sequences are listed in Table 1.
RESULTS
AFLP analysis. (i) Identification of species.
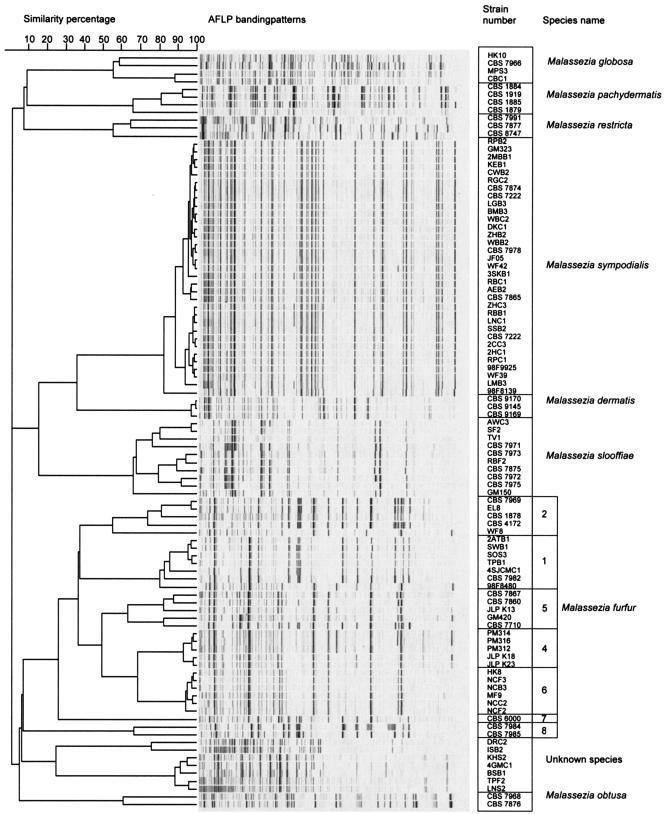
The dendrogram obtained by unweighted pair group method with arithmetic means analysis is shown in Fig. 1. The eight Malassezia species could clearly be distinguished in the AFLP-based tree (Fig. 1). The tree comprised nine main groups, or clusters, and within those clusters, those subtending M. furfur, M. slooffiae, and M. globosa had distinct phylogenetic substructures. Genotypes were assigned to each cluster as shown in Fig. 1. The cophenetic and similarity values for various groups are shown in Table 2.
FIG. 1.
AFLP analysis banding patterns of Malassezia species. Unknown species, strains not associated with any recognized Malassezia taxon.
TABLE 2.
Cophenetic and similarity values for various clusters of Malassezia spp. based on comparisons with type strain of each species
| Species and cluster no. | No. of strains | % Similarity value | Cophenetic value |
|---|---|---|---|
| M. furfur | 29 | 26.49 | 85 |
| Cluster 1 | 4 | 74.15 | 94 |
| Cluster 2 | 1 | 55.27 | 90 |
| Cluster 3 | 7 | 80.52 | 83 |
| Cluster 4 | 5 | 63.37 | 93 |
| Cluster 5 | 5 | 93.07 | 95 |
| Cluster 6 | 6 | 94.37 | 84 |
| Cluster 7 | 1 | 26.49 | 85 |
| M. sympodialis | 33 | 82.39 | 79 |
| Cluster 1 | 21 | 74.88 | 78 |
| Cluster 2 | 11 | 82.39 | 79 |
| Cluster 3 | 1 | 82.39 | 79 |
| M. dermatis | 3 | 97.33 | 90 |
| M. globosa | 4 | 55.15 | 98 |
| M. pachydermatis | 4 | 65.81 | 94 |
| M. restricta | 3 | 55.41 | 92 |
| M. slooffiae | 10 | 66.33 | 74 |
| Cluster 1 | 4 | 76.37 | 93 |
| Cluster 2 | 5 | 79.65 | 84 |
| Cluster 3 | 1 | 66.33 | 74 |
| M. obtusa | 2 | 61.19 | 100 |
| Unknown species | |||
| Cluster 1 | 2 | 75.95 | 100 |
| Cluster 2 | 5 | 88.83 | 66 |
Some interesting observations were made for the M. furfur cluster. AFLP patterns revealed that considerable genetic diversity exists within this species. Eight well-distinguished subtypes were revealed. Strains from subtype 3 of this species as delineated by Theelen et al. (40) were not included in the present study, but this subtype designation has been retained to avoid confusion. The distribution of the 31 M. furfur isolates studied across the subtypes discussed here was relatively even, a circumstance which, unfortunately, limited the sample number per subtype, thus making analysis problematic. Nonetheless, it can be noted that just two subtypes, types 2 and 8, contained all five zoonotic isolates tested, including strains from elephant, elk, and ostrich. Subtype 2 also contained two isolates from human skin. Subtypes 1 and 6 contained the majority (13 of 17) of the skin (including scalp) isolates obtained from nonneonatal humans. Two skin isolates from the ears of neonates, however, were found to belong to subtype 5, which also contained one isolate from nonneonatal skin. Subtype 4 appeared remarkable in that all five isolates that were available were from internal body sites, catheter sites, or mucosae rather than from healthy skin; two of these isolates were recorded as coming from neonates. Subtype 5, apart from containing the two neonatal ear skin isolates previously mentioned, contained the only other internal body site isolate examined, an isolate from urine. Of the isolates obtained in connection with hospitals (including neonatal wards) and/or systemic disease, then, all belonged to subtypes 4 and 5, and at least 8 of 10 isolates in these subtypes were derived from such sources. The source of subtype 5 isolate GM 420/CBS 9367 has not been traced.
Because many of the subtype 1 and 6 isolates were from the trunk (back and chest), we tested whether these genotypes might be significantly associated with this body site. Isolate numbers permitted statistical testing for the significance of differences in M. furfur subtypes among isolates from the trunk, the head (including the scalp and ear, whether neonatal or not), and internal sites or sites of apparent systemic invasion (including catheter sites). In this isolate breakdown, differences among AFLP types were not significant (likelihood ratio, 13.039; P = 0.221).
The delineation of possible host differences among the subtypes was rendered problematic by the superimposed geographic differences among the isolates. Subtypes 1 and 6 contained mainly isolates from Ontario (11 of 13 total isolates), but the majority of isolates from adult skin (11 of 17) also came from the Ontario sample. The subtype 4 isolates from internal sites as well as the internal and neonatal isolates of subtype 5 were from Western Europe (Germany, France, The United Kingdom, and The Netherlands), but that region was the source of only one of the nonneonatal human skin isolates available for study. That isolate belongs to subtype 5. Statistical testing of AFLP type distributions between North America and Europe showed that the differences seen were significant (P < 0.001; likelihood ratio, 23.689); however, it was not possible to concomitantly test whether the differences observed were truly attributable to geographic factors rather than to the accompanying differences in host body sites. One M. furfur isolate, CBS 6000, clustered separately and was designated type 7. It was from human dandruff sampled in India. The only other Asian isolate in the sample was from adult skin sampled in Hong Kong. It belonged to subtype 6.
M. sympodialis formed a coherent AFLP cluster. The majority of strains were from the backs or chests of healthy subjects from Ontario. Some of the M. sympodialis strains studied were from patients who had skin lesions. The lesional and nonlesional isolates showed no genetic differences based on their AFLP types.
Tight clustering was seen among M. sympodialis and M. dermatis isolates; but in contrast, the clusters of M. pachydermatis, M. globosa, M. restricta, and M. slooffiae were relatively loosely structured. The low internal similarity values (Table 1) seen within each of these four species support the impression that they are genetically diverse species.
(ii) Putative new species.
There were some strains that fell into a cluster that was not associated with any recognized Malassezia taxon (Fig. 1). These strains appear to belong to a species that is as yet undescribed.
LSU rDNA D1 and D2 domain and ITS sequence analysis.
Sequence analyses of the LSU and ITS regions resulted in eight well-separated, distinct groups representing the eight different Malassezia spp. The D1 and D2 domain and ITS sequences were in the range of 550 and 600 bp, respectively. The nucleotide sequences determined in this study have been deposited in the CBS and GenBank databases, which may be further consulted for identification purposes (www.cbs.knaw.nl/databases/index.htm and www.ncbi.nlm.nih.gov). The phylogenetic trees based on the ITS and LSU sequences were in complete agreement with each other. Some species turned out to be genetically heterogenous with respect to the D1 and D2 domain and ITS sequences. This was true in particular for M. furfur (D1, D2, and ITS), M. sympodialis (ITS), M. pachydermatis (D1, D2, and ITS), M. globosa (D1, D2, and ITS), and M. slooffiae (D1, D2, and ITS). Phylogenetically, M. furfur and M. obtusa appear to be sister species. A sister species relationship is also found with M. restricta and M. globosa. In both trees, the phylogenetic origin of M. slooffiae was unclear.
Comparison of physiological and molecular methods.
The results of the two molecular methods used in our study were concordant. However, some discrepancies were observed when the AFLP analysis and sequencing results were compared with the results of physiologically based identification. One isolate (98F) that had previously been identified as M. furfur was reidentified as M. sympodialis, while three isolates (VMB3, GMB1, and WF39) that had previously been identified as M. globosa were reidentified as M. sympodialis. One isolate (JSB2) that was formerly identified as M. slooffiae was reidentified as M. sympodialis, and one (SF2) that was formerly identified as M. sympodialis was recognized as M. slooffiae. Two isolates (GM420 and 4SJCMC1) which had previously been identified as M. pachydermatis and M. globosa, respectively, were reidentified as M. furfur. Overall, a misidentification rate of 13.8% was observed.
DISCUSSION
Increased interest in gaining a better understanding of the epidemiology of Malassezia infections has led to the development of epidemiological applications for several molecular typing methods that are able to differentiate Malassezia isolates (1, 3, 4, 9, 15-17, 20, 28, 36, 38, 40). The recent identification of two new Malassezia species, M. dermatis (39) and M. equi (32), has further substantiated the need for molecularly specific techniques to distinguish Malassezia species. Furthermore, linking the epidemiological and genotyping data will improve our understanding of the Malassezia species.
Theelen et al. (40) established AFLP analysis as a useful discriminatory technique for Malassezia species identification. AFLP analysis has the capacity to assay a much greater number of loci for polymorphisms than are surveyed by other PCR-based techniques (2) because it is based on the ligation of known sequences (adaptors) to a wide range of restriction fragments; these adaptors then function as targets for PCR primers (2, 34). The use of an internal size standard with every sample for normalization purposes greatly enhances the reproducibility of the results (5). Unlike results from most other PCR-based methods, which analyze only a part of the genome, the banding patterns in AFLP analysis illustrate a broad-ranging subsample of the whole genome and are much easier to analyze than those resulting from restriction fragment length polymorphism and random amplified polymorphic DNA analysis. Another advantage of AFLP analysis is that the patterns can be stored in an accessible general database for future comparison and species identification of additional Malassezia isolates.
Sequencing of the two most varied domains, D1 and D2, of the 26S rDNA has proved useful at both the phylogenetic and taxonomic levels, as the region is sufficiently varied to allow distinction between species, in particular basidiomycetes (27, 35). On the other hand, sequencing of the ITS region has also been found to be useful in discriminating closely related species (10, 23). In the case of Malassezia spp., Gupta et al. (20) observed that PCR-restriction fragment length polymorphism analysis of the ITS region was sufficient to resolve the differences between the physiologically similar species M. furfur, M. sympodialis, and M. slooffiae. Further, sequence diversity within various species has been observed, which suggests the presence of several genotypes within the species (11, 37). In keeping with these observations, we decided to sequence both of these regions and also to add AFLP analysis to further refine the precision of genotypic clustering in this group and to facilitate accurate species identification.
Following up on the studies of Theelen et al. (40), and testing numerous new clinical isolates, we confirmed that the AFLP analysis yields high-quality fingerprints and is species specific in typing Malassezia isolates, though some species contain multiple subtypes. We were able to characterize the major genotypes, including several novel ones, that were present in the Malassezia isolates from Ontario. AFLP clusters seen in M. furfur in our study suggested the existence of some previously undisclosed specificities. For example, our M. furfur genotype 1 isolates came mainly from sites on the chest and back, a remarkable finding given that many of the other Malassezia isolates screened came from body sites, such as the exposed areas of the face, that are only subtly physiologically differentiated from the skin of the trunk. In addition, subtypes 1 and 6 appeared to be especially associated with nonneonatal skin, a habitat in which populations of M. furfur isolates might be expected, to some extent, to have attained an ecological balance among themselves and with other species (i.e., to be living in a way approximating the ecological k-strategy or “climax community”) (7). On the other hand, subtypes 4 and 5 were obtained mostly from sites of internal invasion in immunocompromised patients or from neonatal skin, i.e., from classic newly available sites potentially allowing aggressive, ruderal-type (ecological r-strategy) invasion. Human skin may support such strongly ruderal genotypes in relatively small numbers except in sites of skin disturbance, where they may have an advantage. (M. furfur, as a whole, with its relatively rapid growth and minimally fastidious character in vitro, may already constitute one of the more ruderal components of the spectrum of skin Malassezia species, as would be predicted from its regular involvement in nosocomial infection.)
We could not rule out the possibility that the distributional differences between the subtype 1 and 6 group and the subtype 4 and 5 group reflected not ecology but rather a difference in geographic distribution or a sampling artifact simulating such a difference. Subtypes 1 and 6 were mainly from the Ontario sample, consisting mainly of isolates from adult skin, and subtypes 4 and 5 were mainly from Western Europe, where available M. furfur isolates came mostly from in-hospital studies. It appears highly unlikely that Ontario could have strongly geographically specific Malassezia populations in contradistinction to those of Western Europe, since most Ontarians are of western European origin within their own lifetime or within a small number of generations. Without extensive follow-up sampling, however, the underlying causality of the distributional differences established among subtypes 1, 4, 5, and 6 cannot be discerned. In particular, a sample of hospital-associated isolates from Ontario and skin isolates from adult western Europeans would be of value. A relatively large sample of Malassezia isolates of all species, however, would have to be processed in order to capture sufficient M. furfur isolates to resolve the questions raised by the present study. Development of selective isolation media for individual Malassezia species would be of value.
The disclosure of the intriguing differences among human-associated subtypes, as well as the finding of two genotypes strongly associated with animal sources, dramatically illustrates the power of AFLP analysis in allowing us to discover potentially important epidemiological patterns within Malassezia species. AFLP analysis was also able to discriminate Malassezia strains that could not be identified by physiological tests (Fig. 1). As mentioned above, it is probable that these strains belong to an as-yet-undescribed species. Further investigation is in progress.
The discrepancies observed in our study between phenotypic and molecular methods of identification were comparable to those observed by Makimura et al. (28), who used ITS1 sequencing as a molecular identification standard. They observed that out of 46 clinical isolates that had formerly been identified as M. furfur by employing the conventional phenotypic approach, 22 were M. sympodialis and 5 were M. slooffiae. It is worth mentioning, however, that misidentification may not be the only potential source of such findings. Malassezia, as a lipophile, is relatively hydrophobic, and reliable single-cell cultures may be relatively hard to make. Clumping occurs in water, especially if oil from the medium is present, unless relatively vigorous efforts are made. It appears that cultures of slow-growing Malassezia spp. may conceal relatively inactive inocula of certain faster-growing species and that when the cultures senesce, the contaminating inocula are activated in a way that causes the slow-growing species to be overgrown. Nakabayashi et al. (31) observed that M. globosa, which had been detected in a primary culture, disappeared in several experiments and that only M. sympodialis remained in the culture. More than one Malassezia species may be present in a clinical sample or even in an apparent single colony on a contact plate, necessitating considerable effort to ensure that each Malassezia colony submitted for identification is in a pure state. In some situations, pharmacological selection pressure derived from topical drugs or hygienic materials may also influence the isolation of Malassezia spp. from clinical samples. Trace quantities of inhibitors carried over into primary cultures may result in the growth of some species being partially repressed until later subcultures attenuate the inhibitor levels. More research is required to find out whether the discrepancies in identification are due to technological error in the physiological methods or to sampling artifacts as mentioned above.
REFERENCES
- 1.Aizawa, T., R. Kano, Y. Nakamura, S. Watanabe, and A. Hasegawa. 1999. Molecular heterogeneity in clinical isolates of Malassezia pachydermatis from dogs. Vet. Microbiol. 70:67-75. [DOI] [PubMed] [Google Scholar]
- 2.Blears, M. J., S. A. De Grandis, H. Lee, and J. T. Trevors. 1998. Amplified fragment length polymorphism (AFLP): a review of the procedure and its applications. J. Ind. Microbiol. Biotechnol. 21:99-114. [Google Scholar]
- 3.Boekhout, T., and R. W. Bosboom. 1994. Karyotyping of Malassezia yeasts: taxonomic and epidemiological implications. Syst. Appl. Microbiol. 17:146-153. [Google Scholar]
- 4.Boekhout, T., M. Kamp, and E. Gueho. 1998. Molecular typing of Malassezia species with PFGE and RAPD. Med. Mycol. 36:365-372. [DOI] [PubMed] [Google Scholar]
- 5.Borst, A., B. Theelen, E. Reinders, T. Boekhout, A. C. Fluit, and P. H. M. Savelkoul. 2003. Use of amplified fragment length polymorphism analysis to identify medically important Candida spp., including C. dubliniensis. J. Clin. Microbiol. 41:1357-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, H., H. Miller, N. Watkins, M. Arduino, D. Ashford, S. Midgley, R. Aguero, R. Pinto-Powell, C. F. von Reyn, W. Edwards, M. McNeil, and W. Jarvis. 1998. An epidemic of Malassezia pachydermatis in an intensive care nursery associated with colonization of health care workers' pet dogs. N. Engl. J. Med. 338:706-711. [DOI] [PubMed] [Google Scholar]
- 7.Cooke, R. C., and J. M. Whipps. 1993. Ecophysiology of fungi, p. 3-20. Blackwell Scientific Publications Ltd., Oxford, United Kingdom.
- 8.Gaitanis, G., A. Velegraki, E. Frangoulis, A. Mitroussia, A. Tsigonia, A. Tzimogianni, A. Katsambas, and N. J. Legakis. 2002. Identification of Malassezia species from patient skin scales by PCR-RFLP. Clin. Microbiol. Infect. 8:162-173. [DOI] [PubMed] [Google Scholar]
- 9.Gemmer, C. M., Y. M. DeAngelis, B. Theelen, T. Boekhout, and T. L. Dawson, Jr. 2002. Fast, noninvasive method for molecular detection and differentiation of Malassezia yeast species on human skin and application of the method to dandruff microbiology. J. Clin. Microbiol. 40:3350-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graser, Y., M. El Fari, R. Vilgalys, F. A. Kuijpers, G. S. de Hoog, W. Presber, and H. J. Tietz. 1999. Phylogeny and taxonomy of the family Arthrodermataceae (dermatophytes) using sequence analysis of the ribosomal ITS region. Med. Mycol. 37:315-330. [PubMed] [Google Scholar]
- 11.Gueho, E., T. Boekhout, H. R. Ashbee, J. Guillot, A. Van Belkum, and J. Faergemann. 1998. The role of Malassezia species in the ecology of human skin and as pathogens. Med. Mycol. 36:220-229. [PubMed] [Google Scholar]
- 12.Gueho, E., and S. Meyer. 1989. A reevaluation of the genus Malassezia by means of genomic comparison. Antonie Leeuwenhoek 55:245-251. [DOI] [PubMed] [Google Scholar]
- 13.Gueho, E., G. Midgley, and J. Guillot. 1996. The genus Malassezia with the description of four new species. Antonie Leeuwenhoek 69:337-355. [DOI] [PubMed] [Google Scholar]
- 14.Gueho, E., R. B. Simmons, W. R. Pruitt, S. A. Meyer, and D. G. Ahearn. 1987. Association of Malassezia pachydermatis with systemic infections of humans. J. Clin. Microbiol. 25:1789-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillot, J., M. Deville, F. Berthelemy, F. Provost, and E. Gueho. 2000. A single PCR-restriction endonuclease analysis for rapid identification of Malassezia species. Lett. Appl. Microbiol. 31:400-403. [DOI] [PubMed] [Google Scholar]
- 16.Guillot, J., and E. Gueho. 1995. The diversity of Malassezia yeasts confirmed by rRNA sequence and nuclear DNA comparisons. Antonie Leeuwenhoek 67:297-314. [DOI] [PubMed] [Google Scholar]
- 17.Guillot, J., E. Gueho, M. Lesourad, G. Midgley, B. Chevrier, and B. Dupont. 1996. Identification of Malassezia species. A practical approach. J. Mycol. Med. 6:103-110. [Google Scholar]
- 18.Gupta, A. K., R. Batra, R. Bluhm, T. Boekhout, and T. L. Dawson. Skin diseases associated with Malassezia species. J. Am. Acad. Dermatol, in press. [DOI] [PubMed]
- 19.Gupta, A. K., Y. Kohli, J. Faergemann, and R. C. Summerbell. 2001. Epidemiology of Malassezia yeasts associated with pityriasis versicolor in Ontario, Canada. Med. Mycol. 39:199-206. [DOI] [PubMed] [Google Scholar]
- 20.Gupta, A. K., Y. Kohli, and R. C. Summerbell. 2000. Molecular differentiation of seven Malassezia species. J. Clin. Microbiol. 38:1869-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta, A. K., Y. Kohli, R. C. Summerbell, and J. Faergemann. 2001. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med. Mycol. 39:243-251. [DOI] [PubMed] [Google Scholar]
- 22.Hazen, K. C. 1995. New and emerging yeast pathogens. Clin. Microbiol. Rev. 8:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwen, P. C., S. H. Hinrichs, and M. E. Rupp. 2002. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med. Mycol. 40:87-109. [DOI] [PubMed] [Google Scholar]
- 24.Jahagirdar, B. N., and V. A. Morrison. 2002. Emerging fungal pathogens in patients with hematologic malignancies and marrow/stem-cell transplant recipients. Semin. Respir. Infect. 17:113-120. [DOI] [PubMed] [Google Scholar]
- 25.Kieffer, M., I. M. Bergbrant, J. Faergemann, G. B. Jemec, V. Ottevanger, P. Stahl Skov, and E. Svejgaard. 1990. Immune reactions to Pityrosporum ovale in adult patients with atopic and seborrheic dermatitis. J. Am. Acad. Dermatol. 22:739-742. [DOI] [PubMed] [Google Scholar]
- 26.Kwon-Chung, K. J., and J. E. Bennet. 1992. Infections caused by Malassezia species (tinea versicolor, pityriasis versicolor, dermatomycosis furfuracea, tinea flava, “liver spots”), p. 170-182. In K. J. Kwon-Chung and J. E. Bennet (ed.), Medical mycology. Lea & Febiger, Malvern, Pa.
- 27.Leclerc, M. C., H. Philippe, and E. Gueho. 1994. Phylogeny of dermatophytes and dimorphic fungi based large subunit ribosomal RNA sequence comparisons. J. Med. Vet. Mycol. 32:331-341. [DOI] [PubMed] [Google Scholar]
- 28.Makimura, K., Y. Tamura, M. Kudo, K. Uchida, H. Saito, and H. Yamaguchi. 2000. Species identification and strain typing of Malassezia species stock strains and clinical isolates based on the DNA sequences of nuclear ribosomal internal transcribed spacer 1 regions. J. Med. Microbiol. 49:29-35. [DOI] [PubMed] [Google Scholar]
- 29.Marcon, M. J., and D. A. Powell. 1992. Human infections due to Malassezia spp. Clin. Microbiol. Rev. 5:101-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Midreuil, F., J. Guillot, E. Gueho, F. Renaud, M. Mallie, and J. M. Bastide. 1999. Genetic diversity in the yeast species Malassezia pachydermatis analysed by multilocus enzyme electrophoresis. Int. J. Syst. Bacteriol. 49:1287-1294. [DOI] [PubMed] [Google Scholar]
- 31.Nakabayashi, A., Y. Sei, and J. Guillot. 2000. Identification of Malassezia species isolated from patients with seborrheic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med. Mycol. 38:337-341. [DOI] [PubMed] [Google Scholar]
- 32.Nell, A., S. A. James, C. J. Bond, B. Hunt, and M. E. Herrtage. 2002. Identification and distribution of a novel Malassezia species yeast on normal equine skin. Vet. Rec. 150:395-398. [DOI] [PubMed] [Google Scholar]
- 33.O'Donnel, K., E. Cigelnik, N. S. Weber, and J. M. Trappe. 1997. Phylogenetic relationships among ascomycetous truffles and the true and false morels inferred from 18S and 28S ribosomal DNA sequence analysis. Mycologia 89:48-65. [Google Scholar]
- 34.Savelkoul, P. H. M., H. J. M. Aarts, J. de Haas, L. Dijkshoorn, B. Duim, M. Otsen, J. L. W. Rademaker, L. Schouls, and J. A. Lenestra. 1999. Amplified-fragment length polymorphism analysis: the state of an art. J. Clin. Microbiol. 37:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scorzetti, G., J. W. Fell, A. Fonseca, and A. S. Tallman. 2002. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res. 2:495-517. [DOI] [PubMed] [Google Scholar]
- 36.Senczek, D., U. Siesenop, and K. Bohm. 1999. Characterization of Malassezia species by means of phenotypic characteristics and detection of electrophoretic karyotypes by pulsed field gel electrophoresis (PFGE). Mycoses 42:409-414. [DOI] [PubMed] [Google Scholar]
- 37.Sugita, T., M. Kodama, M. Saito, I. Tomonobu, Y. Kato, R. Tsuboi, and A. Nishikawa. 2003. Sequence diversity of the intergenic spacer region of the rRNA gene of Malassezia globosa colonizing the skin of patients with atopic dermatitis and healthy individuals. J. Clin. Microbiol. 41:3022-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugita, T., H. Suto, T. Unno, R. Tsuboi, H. Ogawa, T. Shinoda, and A. Nishikawa. 2001. Molecular analysis of Malassezia microflora on the skin of atopic dermatitis patients and healthy subjects. J. Clin. Microbiol. 39:3486-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugita, T., M. Takashima, T. Shinoda, H. Suto, T. Unno, R. Tsuboi, H. Ogawa, and A. Nishikawa. 2002. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J. Clin. Microbiol. 40:1363-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Theelen, B., M. Silvestri, E. Gueho, A. Van Belkum, and T. Boekhout. 2001. Identification and typing of Malassezia yeasts using amplified fragment length polymorphism (AFLP), random amplified polymorphic DNA (RAPD) and denaturing gradient gel electrophoresis (DGGE). FEMS Yeast Res. 1:79-86. [DOI] [PubMed] [Google Scholar]