Abstract
Understanding the relatedness of strains within a bacterial species is essential for monitoring reservoirs of antimicrobial resistance and for epidemiological studies. Pulsed-field gel electrophoresis (PFGE), ribotyping, and multilocus sequence typing are commonly used for this purpose. However, these techniques are either nonquantitative or provide only a limited estimation of strain relatedness. Moreover, they cannot extensively define the genes that constitute an organism. In the present study, 21 oxacillin-resistant Staphylococcus aureus (ORSA) isolates, representing eight major ORSA lineages, and each of the seven strains for which the complete genomic sequence is publicly available were genotyped using a novel GeneChip-based approach. Strains were also subjected to PFGE and ribotyping analysis. GeneChip results provided a higher level of discrimination among isolates than either ribotyping or PFGE, although strain clustering was similar among the three techniques. In addition, GeneChip signal intensity cutoff values were empirically determined to provide extensive data on the genetic composition of each isolate analyzed. Using this technology it was shown that strains could be examined for each element represented on the GeneChip, including virulence factors, antimicrobial resistance determinants, and agr type. These results were validated by PCR, growth on selective media, and detailed in silico analysis of each of the sequenced genomes. Collectively, this work demonstrates that GeneChips provide extensive genotyping information for S. aureus strains and may play a major role in epidemiological studies in the future where correlating genes with particular disease phenotypes is critical.
Monitoring the acquisition and maintenance of genes within bacterial populations is an essential component of understanding the epidemiology of emerging infectious diseases. Accordingly, much effort has been devoted toward developing methods to delineate the relatedness of Staphylococcus aureus strains that are circulating within both health care institutions and community settings.
The techniques that are currently used for strain surveillance include spa typing, ribotyping, pulsed-field gel electrophoresis (PFGE), and multilocus sequence typing (MLST) (2, 3, 8, 20). Although these methods have proven valuable in monitoring strain relatedness, none extensively defines the genes that constitute the organism(s) under investigation. Further evaluation of the genes of interest within an individual isolate can be accomplished by PCR amplification of the particular loci of interest followed by restriction enzyme analysis or sequencing. Vandenesch and colleagues recently combined PFGE, MLST, and PCR for 24 individual virulence genes in a study that was designed to examine the relatedness and genetic composition of 117 community-associated oxacillin-resistant S. aureus (CO-ORSA) isolates from several countries (22). The study demonstrated that despite geographic origin or relatedness of the strains, all CO-ORSA isolates contained the Panton-Valentine leukocidin virulence factor locus. Other virulence factors that were analyzed were not detected within all strains. It is likely that additional genes, which were not subjected to PCR analysis in that study, are also conserved across all CO-ORSA isolates and may play a direct role in the prevalence of these strains within the community.
Recently, Musser and colleagues used a more comprehensive approach to evaluate the relatedness and genomic composition of 36 S. aureus clinical isolates (9). Using a DNA microarray constructed from the genomic sequence of the S. aureus COL strain, a comparison was made among the hybridization patterns of genomic DNA isolated from each isolate. The study identified genomic elements that were present in COL but absent from the strains of interest. Inferences could also be made regarding the relatedness of strains analyzed which, although not extensively evaluated, generally correlated well with more-established methods of determining genetic relationships, such as multilocus enzyme electrophoresis. However, the study was limited to comparing strains of interest with respect to COL open reading frames (ORFs) represented on the microarrays used. As a result, genes that were present within clinical isolates but that were not contained within the COL DNA sequence could not be identified, despite their potential importance in pathogenesis. In addition, many well-studied genes are not present in COL and could not be analyzed, including collagen adhesin (cna), epidermal cell differentiation inhibitor (edin), exfoliative toxins A and B (eta and etb), staphylokinase (sak), Panton-Valentine leukotoxin (luk-PV), and toxic shock syndrome toxin (tst). Similarly, all antimicrobial resistance determinants (except tetracycline resistance), loci for heavy metal resistance, non-type 5 capsule biosynthesis genes, and 27 enterotoxin and exotoxin genes, as well as 315 hypothetical genes from the genomes of S. aureus strain N315 or Mu50, or individual GenBank records could not be interrogated by the microarray used.
In the present study, we have used an Affymetrix GeneChip that represents predicted ORFs from six genetically divergent S. aureus strains and novel GenBank entries to analyze the relatedness of 21 ORSA isolates and each of the strains for which genomic sequencing information is currently available. The 21 isolates represent strains of eight U.S. oxacillin- and methicillin-resistant lineages (15). We compared these results with ribotyping and PFGE results and found that the GeneChip typing method used was more discriminative than conventional approaches. In addition, we established parameters that predict whether individual genes are present within a given strain. These parameters have been validated by PCR analysis, growth on selective media, and DNA sequence analysis. Collectively, these results demonstrate that GeneChips provide an extensive discriminative genotyping procedure and simultaneously provide the ability to identify specific loci that are present or absent within an S. aureus strain.
MATERIALS AND METHODS
Sources of sequence and ORF predictions for design of the Saur2a array.
Genomic sequences and ORF predictions for the S. aureus N315 and Mu50 published genomes were obtained from GenBank. Unpublished genomic sequences for MRSA252 (complete) and MSSA476 (incomplete) were obtained from the Wellcome Trust Sanger Institute (http://www.sanger.ac.uk). The unfinished S. aureus NCTC 8325 sequence was obtained from the University of Oklahoma Advanced Center for Genome Technology (http://www.genome.ou.edu). S. aureus COL genomic sequence and ORF predictions were obtained from The Institute for Genomic Research (http://www.tigr.org). Additional annotated S. aureus records were retrieved from GenBank coding regions. Genomic fragments of the unfinished genomes from strains NCTC 8325 and MSSA476 were approximately ordered and oriented by comparison with the N315 and COL genomes. Prior to running ORF prediction algorithms, the sequence CTAACTAATTAG, which contains translation stop codons in all six reading frames, was placed at the 3′ end of each fragment, so that ORF predictions could be run on the entire sequence without the possibility of predicted genes crossing fragments. For all six genomes, Glimmer 2.02 (5) and GeneMark (10) were used to predict ORFs. Glimmer was performed with default parameters, except that a minimum ORF size of 75 nucleotides was set and start codons ATG, TTG, CTG, and GTG were allowed. Ribosomal binding site location was used to guide the placement of the translation start site (-f option). GeneMark 1.42a was performed with default parameters with the gene model saureus_4.mat (http://opal.biology.gatech.edu/GeneMark/download/saureus_4.mat).
Several well-conserved genes were divided into three segments so that 5′/3′ signal ratios could be tracked (SA0014[rp1I], SA0352[rpsF], SA0459 [rplY], SA0506[tuf], SA0578[conserved hypothetical], SA0600[conserved hypothetical], SA1129[conserved hypothetical], SA1334[proC], SA1456[aspS], and SA1682[conserved hypothetical]). Several very large genes encoding surface proteins were divided into 5,000-bp segments (SA0173 and homologs in other strains [gramicidin S synthetase], SA1267 and homologs [ebhA], SA1964 and homologs [fmtB], COL-SA0379 [bacteriophage L54a peptidase], COLSA1472 and homologs [putative pathogenicity protein], COL-SA2676 [LPXTG surface protein], and biofilm-associated protein [bap]). Genes known to contain constant and variable domains, such as agrB and agrC, were separated into individual domains.
Clustering.
The ORFS were clustered to identify homologs among the individual strains by using the CAT 4.5 (clustering and alignment tool) software from DoubleTwist (4). To maximize both the discriminatory power of the array and the number of unique probe sets representing each qualifier, the sequence identity threshold for inclusion in an alignment was set at 97%. Each resulting cluster contained a set of highly similar ORFs that were aligned to derive consensus sequences. The clusters were manually curated to correct any obvious errors, such as those in which two adjacent genes merged into a single cluster as a result of a read-through error in a sequence record.
Intergenic regions and probe selection.
Intergenic sequences of >50 bases in length were extracted from the N315 genome by using the published ORF coordinates. Any portion of an intergenic region with >90% identity to an ORF in another genome or in the N315 Glimmer ORF set was excised. Intergenic sequences (both strands) were clustered separately from ORFs. The ORF and intergenic sequences were submitted to Affymetrix (Santa Clara, Calif.) for probe selection. Thirty-four probe pairs were requested for each ORF, and 15 were requested for each intergenic region. The final array contained 7,792 S. aureus qualifiers recognizing 4,380 ORFS, 3,343 intergenic regions, and 69 exogenous control probe sets.
DNA isolation and labeling.
S. aureus strains were grown overnight in brain heart infusion medium in ambient air at 37°C with vigorous aeration. For chromosomal isolation, 1.5 ml of an overnight culture was placed in a 1.5-ml Eppendorf tube and was centrifuged for 5 min at 4°C at high speed in a table-top centrifuge. Supernatants were discarded, and cell pellets were resuspended in an equal volume of ice-cold TE buffer (10 mM Tris, 1 mM EDTA; pH 8.0). Suspensions were then placed in 2-ml lysing matrix tubes (Bio 101, Vista, Calif.). Cells were lysed by shaking in an FP120 reciprocating shaker (Bio 101) two times at 6,000 rpm for 20 s, and cell debris was pelleted by centrifugation at high speed in a table-top centrifuge for 10 min. Chromosomal and plasmid DNA was then purified from the supernatant on a Qiagen DNA tissue easy column (Valencia, Calif.), following the manufacturer's recommendations for bacterial DNA purification. Two micrograms of purified DNA was subjected to electrophoresis on a 0.8% native agarose gel to assess DNA integrity. For DNA labeling, 5 μg of purified DNA was incubated at 90°C for 3 min and then plunged into an ice bath, followed by standard DNA fragmentation and labeling procedures according to the manufacturer's (Affymetrix Inc.) instructions for labeling mRNA for antisense prokaryotic arrays. A 1.5-μg aliquot of labeled DNA was hybridized to a GeneChip and was processed as per the manufacturer's protocol for GeneChip hybridization and washing. GeneChips were scanned as previously described (7). Signal intensities for elements tiled onto each GeneChip were normalized to account for loading errors and differences in labeling efficiencies by dividing each signal intensity by the mean signal intensity for an individual GeneChip. Results were analyzed using GeneSpring version 6.1 (Silicon Genetics) and Spotfire version 7.0.
PCRs.
All PCR assays were performed using Invitrogen's platinum PCR Supermix kits (Carlsbad, Calif.), following the manufacturer's recommendations. Amplification of the cna gene was accomplished using the primers 5′-ACTGGACACATACGTGGACAGGATT and 5′-TTTTCCTGTTGCTTTTCCATCTTGA. Primers for PCR amplification of the srtA gene included 5′-AGCAGCAAGCTAAACCTCAAATTCC and 5′-AAGATTTTACGTTTTTCCCAAACGC. PCR amplification and typing of the agr locus was accomplished following the procedure described in reference 14.
Ribotyping and PFGE.
Strains were subjected to PFGE as previously described (15). Ribotyping was performed using the RiboPrinter system (Qualicon, Wilmington, Del.) according to the manufacturer's instructions. Each strain was analyzed using two restriction enzymes, EcoRI and PvuII. Computer-generated riboprints for each strain were assigned to an EcoRI or PvuII ribogroup by the software and then visually inspected for correct assignment into ribogroups. Individual ribotypes were assigned to a strain based on identity of ribogroups for both restriction enzymes.
RESULTS
The availability of six S. aureus genomic sequences allowed the development of a second-generation Affymetrix GeneChip that was designed to monitor a more expansive repertoire of S. aureus loci than previously measurable by microarrays. Based on these sequences, an Affymetrix GeneChip (Saur2a) was developed that represents (i) consensus sequences for genes that are conserved in all strains analyzed, (ii) alleles, which we have defined as any gene that contains less than 97% nucleotide identity to another gene, (iii) genes that are unique to an individual strain, (iv) unique GenBank entries, and (v) N315 intergenic regions that are greater than 50 nucleotides in length. In the present study we used this GeneChip to develop a method to monitor strain relatedness and genetic composition of each of the sequenced S. aureus strains and 21 clinical isolates.
Analysis of sequenced strains.
Prior to using the Saur2a array to evaluate the genetic composition and relatedness of clinical isolates, we determined the accuracy with which the array monitored the genes that constitute each of the six strains that were used in its development. We also analyzed S. aureus strain MW2, whose sequence was published after Saur2a was developed (1). An initial analysis was performed using Affymetrix Microarray Suite 5.0 algorithms, which provide detection calls (present, absent, or marginal) for RNA transcripts. Although these algorithms have been optimized for RNA analysis, they initially served to analyze DNA purified from each of the sequenced strains. DNA from each strain was labeled and hybridized to a Saur2a array. The signal intensity for each qualifier (predicted ORF or intergenic region) was measured. Affymetrix algorithms determined a detection call for each qualifier. Results are shown in Table 1.
TABLE 1.
Analysis of sequenced S. aureus strains
| Straina | Predicted presentb | Called presentc | Called marginal or absentd | Incorrectly called presente |
|---|---|---|---|---|
| COL (laboratory 1) | 5,335 | 5,330 | 5 | ND |
| COL (laboratory 2) | 5,335 | 5,330 | 5 | ND |
| COL (laboratory 3) | 5,335 | 5,330 | 5 | 1,207 |
| COL (repository) | 5,335 | 5,084 | 251 | ND |
| COL (type 1) | 5,335 | 5,329 | 6 | ND |
| COL (type 2) | 5,335 | 5,105 | 239 | ND |
| MRSA252 | 4,397 | 4,387 | 10 | 1,838 |
| MSSA476 | 5,242 | 5,236 | 6 | 1,277 |
| Mu50 | 6,412 | 6,407 | 5 | 464 |
| MW2 | 5,208 | 5,202 | 6 | 1,268 |
| N315 | 6,339 | 6,331 | 8 | 424 |
| NCTC 8325 | 5,366 | 5,359 | 7 | 1,102 |
Sequenced S. aureus strains, with the source referenced in parentheses.
Number of genes expected to be present based on DNA sequence.
Number of genes determined to be present (called present) by GeneChip Microarray Suite 5.0 analysis.
Number of genes determined to be not present by microarray analysis.
Number of qualifiers that were not contained within the genomic sequence of a given sequenced strain but were incorrectly identified to be present based on Affymetrix algorithms. ND, not determined.
For the sequenced strains, a qualifier was expected to be present if at least 70% of the qualifier's perfect-match oligonucleotides were contained within the genome being analyzed. The 70% matching requirement is one of the criteria Affymetrix software requires for an RNA transcript to be considered present. For each of the seven strains tested, there were between 5 and 10 discrepancies between the expected and actual present-detection calls, resulting in a 0.1 to 0.2% error rate. Despite this low error rate, there were two immediate concerns with the data. Firstly, 251 genes (4.7%) that were expected to be present in strain COL [COL (repository)] were determined to be absent by Affymetrix algorithms, raising concern either that (i) the GeneChip was not properly constructed, (ii) the Affymetrix approach used to make present or absent calls was not appropriate for DNA samples on Saur2a GeneChips; and/or (iii) the strain analyzed was not the sequenced COL strain. Secondly, the false-positive error rate (genes that were known to be absent but incorrectly determined to be present) was between 5.0 and 23.5% for each of the sequenced strains. This latter issue is addressed in more detail in subsequent sections.
The COL strain used was obtained from a publicly available strain repository. Upon further investigation, the strain produced two colony morphologies following extended incubation on nutrient-rich agar plates. Each colony type was purified and was subjected to microarray analysis (Table 1). One colony type (COL type 1) demonstrated an error rate of 0.1%, was resistant to tetracycline, and matched the profiles of COL strains obtained from three independent laboratories. The second colony type (COL type 2) demonstrated an error rate of 4.5% and was tetracycline susceptible. These results suggested that the initial strain analyzed was contaminated with at least one additional S. aureus strain, which was likely to be the cause of major discrepancies between the expected and observed results. Scientists at the strain stock center confirmed that the repository stock was contaminated with another S. aureus strain. (The strain stock center subsequently notified investigators to whom contaminated stocks had been shipped.)
Although Affymetrix algorithms accurately detected genes known to constitute each of the strains under investigation, between 424 and 1,838 other genes were erroneously identified to be present in each strain (Table 1). Based on sequencing data, these genes were known to be missing from the genomic sequence of a given strain but were called present by Affymetrix standards. This finding indicated that Affymetrix algorithms tended to overestimate the number of genes that were present within a DNA sample. As a result, we set out to redefine parameters that would allow more accurate call determinations to be made for all genes of an organism under investigation.
Calculating present and absent call determination values.
The goal was to evaluate each sequenced strain independently and empirically identify signal intensity cutoff values to define whether a gene was present or absent within a given strain. A present call cutoff value was set so that 90% of the qualifiers known to be present in a given strain would have signal intensities above the designated value. Similarly, a second cutoff was to be defined so that 90% of the qualifiers expected to be absent would have signal intensities below this value.
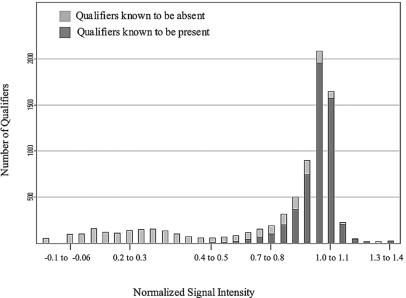
The initial step in defining the appropriate cutoff values was identifying all the genes expected to be present in a given sequenced strain by using the 70% matching requirement described above. Next, GeneChip data for that strain were normalized to account for differences in labeling efficiencies. Normalization was accomplished by dividing the log-transformed raw signal intensity of each qualifier by the mean signal for the entire chip. For each strain, the distribution of the normalized values for the qualifiers expected to be present was examined, and the cutoff value that defined 90% of the signals was determined. This value was termed the adjusted present call determination value. Similarly, an adjusted absent call determination value was calculated based on the distribution of normalized signal values for the qualifiers expected to be absent. Qualifiers demonstrating intermediate signal intensities (between the absent and present call determinates) were considered undeterminable. The distribution of expected present and absent qualifiers for strain NCTC 8325 is shown in Fig. 1. All other sequenced strains demonstrated a similar distribution (results not shown).
FIG. 1.
Distribution of strain NCTC 8325 normalized signal intensity values for all qualifiers represented on Saur2a. Light grey indicates qualifiers with <70% perfect-match probe matches to strain NCTC 8325 chromosomal DNA. Dark grey indicates qualifiers with ≥70% perfect-match probe matches to the strain NCTC 8325 genome.
Adjusted present and absent call determination values were 0.894 ± 0.01 and 0.981 ± 0.01, respectively, for all sequenced strains. As shown in Table 2, based on these call determination values an average of 80.8% qualifiers known to be present within a strain were correctly identified, 10% were incorrectly called absent, and 9.9% were undeterminable. A total of 84.6% of the qualifiers that were known to be absent from a strain were correctly called absent, 10% were incorrectly called present, and 5.7% were undeterminable. In each case, intergenic regions accounted for the majority of the erroneous call determinations. For instance, a total of 774 qualifiers were incorrectly determined to be present or absent based on adjusted call determination values for strain NCTC 8325. Of these 774 erroneous calls, 637 (82%) were intergenic regions (data not shown). As a result, 96.9% of the 4,380 ORFs represented on the Saur2a array were correctly determined for strain NCTC 8325 by using adjusted call determinations. Similar values were observed for all other sequenced strains. Analyses with cutoff values other than 90% and with the exclusion of intergenic regions were also examined but were less accurate in predicting the presence or absence of genes within sequenced strains (data not shown).
TABLE 2.
Adjusted call determinations for sequenced S. aureus strains
| Strain | No. of qualifiers
|
|||||
|---|---|---|---|---|---|---|
| <0.70 fraction matchedb
|
≥0.70 fraction matchedc
|
|||||
| Absent | No call | Present | Absent | No call | Present | |
| COL (laboratory 1) | 1,973 (82.6) | 176 (7.4) | 239 (10.0) | 534 (10.0) | 693 (13.0) | 4,108 (77.0) |
| COL (laboratory 2) | 1,970 (82.5) | 179 (7.5) | 239 (10.0) | 534 (10.0) | 598 (11.2) | 4,203 (78.8) |
| COL (laboratory 3) | 1,956 (81.9) | 193 (8.1) | 239 (10.0) | 534 (10.0) | 745 (14.0) | 4,056 (76.0) |
| COL (repository) | 1,955 (81.9) | 194 (8.1) | 239 (5.0) | 534 (10.0) | 685 (12.8) | 4,116 (77.2) |
| MRSA252 | 2,713 (81.6) | 280 (8.4) | 333 (10.0) | 440 (10.0) | 467 (10.6) | 3,490 (79.4) |
| MSSA476 | 2,034 (82.0) | 198 (8.0) | 249 (10.0) | 525 (10.0) | 745 (14.2) | 3,972 (75.8) |
| Mu50 | 1,185 (90.4) | 0 (0.0) | 126 (9.6) | 574 (9.0) | 0 (0.0) | 5,838 (91.1) |
| MW2 | 2,023 (80.4) | 239 (9.5) | 253 (10.1) | 522 (10.0) | 1,025 (19.7) | 3,661 (70.3) |
| N315 | 1,264 (91.3) | 0 (0.0) | 120 (8.7) | 442 (7.0) | 0 (0.0) | 5,897 (93.0) |
| NCTC 8325 | 1,896 (80.4) | 225 (9.6) | 236 (10.0) | 538 (10.0) | 937 (17.5) | 3,891 (72.5) |
Adjusted call determinations-for Saur2a qualifiers containing ≥70% or <70% perfect-match probe sets to chromosomal sequences of each sequenced strain.
Values are numbers of qualifiers that would actually be determined to be absent, undeterminable, or present based on strain-specific adjusted call cutoffs. Values in parentheses are percentages.
The <0.70 fraction included an analysis of all qualifiers that contained less than 70% perfect-match probe sets to chromosomal DNA for each sequenced strain.
A similar analysis was performed for qualifiers with ≥70% perfect-match probe sets that represent a gene within each sequenced strain.
Despite their strain divergence (described below), present and absent call determination values were very similar for each of the seven sequenced strains. These adjusted values appeared to accurately predict the presence or absence of ∼6,000 qualifiers for each strain. To test the reproducibility of the adjusted call determination method, DNA isolation, GeneChip hybridization, and analysis of the adjusted detection calls were repeated for strains COL (two times) and Mu50 (one time). As expected, an average of 6,410 qualifiers (83.2%) were accurately determined, and 736 qualifiers (9.5%) were incorrectly identified as present or absent (data not shown). These results suggested that the adjusted determination calls allowed for reproducible results among a diverse strain set.
The accuracy of the adjusted call method was analyzed in a number of ways. First, comparisons were made between the Affymetrix and adjusted call determinations for 315 genes that are known to be missing from COL but represented on the Saur2a array (data not shown). COL strains from three independent laboratories were analyzed. Affymetrix software erroneously identified between 28 and 46 genes to be present in the three strains (8.8 to 14.6% false-positive rate), whereas 10 to 15 genes (3.0 to 4.7%) were undeterminable. In contrast, adjusted present call determinations indicated that one gene was incorrectly identified to be present in each sample (0.3% false-positive rate) and either none or one gene (<0.3%) was considered undetermined for each replicate. Comparisons of genes that are known to be present in COL and represented on the array demonstrated a slight advantage to using Affymetrix algorithms (0.1% false-negative rate) compared to adjusted call values (1.2% false-negative rate). To further evaluate the two processes, GeneChip analysis was performed on 21 clinical isolates that were obtained from the Centers for Disease Control and Prevention (CDC), using both Affymetrix and adjusted present and absent call determination values of 0.894 and 0.981, respectively.
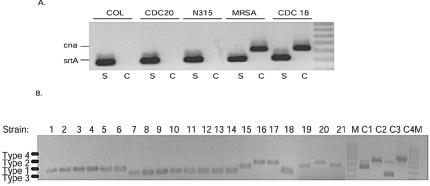
Table 3 compares Affymetrix and adjusted call determinations for the genes encoding collagen adhesion (cna) and sortase (srtA) virulence factors. Both Affymetrix and adjusted call determinations indicated that the gene encoding sortase (srtA) was present in every strain analyzed. These results were validated by PCR detection. However, discrepancies were observed between the two detection methods when analyzing the gene encoding collagen adhesion. Affymetrix algorithms determined that cna is present in both isolate 20 and the sequenced strain Mu50. cna was also considered present in two of three COL samples tested, and it was undeterminable in the third sample (laboratory 3) and also in strain N315. In contrast, adjusted call determination parameters indicated that cna was absent in each of these strains (isolate 20, Mu50, N315, and each of the COL samples). In each case, PCR analysis, as well as sequence analysis of Mu50, N315, and COL demonstrated that the adjusted determinations were correct and that cna is not present in these strains, further validating the accuracy of the methodology (Table 3; Fig. 2A).
TABLE 3.
Comparison of Affymetrix, adjusted present calls, and PCR detection for sequenced and clinical isolates
| Strain |
srtA
|
cna
|
agr type
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Raw | Adjusted | PCR | Raw | Adjusted | PCR | Raw | Adjusted | PCR | |
| CDC 1 | + | + | + | − | − | − | 1 | 1 | 1 |
| CDC 2 | + | + | + | − | − | − | 1 | 1 | 1 |
| CDC 3 | + | + | + | − | − | − | 1 | 1 | 1 |
| CDC 4 | + | + | + | − | − | − | 1 | 1 | 1 |
| CDC 5 | + | + | + | − | − | − | 1 | 1 | 1 |
| CDC 6 | + | + | + | − | − | − | 1 | 1 | 1 |
| CDC 7 | + | + | + | + | + | + | 3 | 3 | 3 |
| CDC 8 | + | + | + | + | + | + | 3 | 3 | 3 |
| CDC 9 | + | + | + | + | + | + | 3 | 3 | 3 |
| CDC 10 | + | + | + | + | + | + | 3 | 3 | 3 |
| CDC 11 | + | + | + | + | + | + | 3 | 3 | 3 |
| CDC 12 | + | + | + | + | + | + | 3 | 3 | 3 |
| CDC 13 | + | + | + | + | + | + | 3 | 3 | 3 |
| CDC 14 | + | + | + | + | + | + | 3 | 3 | 3 |
| CDC 15 | + | + | + | + | + | + | 1 | 1 | 1 |
| CDC 16 | + | + | + | − | − | − | 2 | 2 | 2 |
| CDC 17 | + | + | + | − | − | − | 2 | 2 | 2 |
| CDC 18 | + | + | + | + | + | + | 3 or 4 | 3 | 3 |
| CDC 19 | + | + | + | − | − | − | 1 or 4 | 1 | 1 |
| CDC 20 | + | + | + | + | − | − | 2 | 2 | 2 |
| CDC 21 | + | + | + | − | − | − | 1 | 1 | 1 |
| COL (lab 1) | + | + | + | + | − | − | 1 | 1 | 1 |
| COL (lab 2) | + | + | + | + | − | − | 1 | 1 | 1 |
| COL (lab 3) | + | + | + | +/− | − | − | 1 | 1 | 1 |
| MRSA252 | + | + | + | + | + | + | 3 | 3 | 3 |
| MSSA476 | + | + | + | + | + | + | 3 | 3 | 3 |
| Mu50 | + | + | + | + | − | − | 2 | 2 | 2 |
| MW2 | + | + | + | + | + | + | 3 | 3 | 3 |
| N315 | + | + | + | +/− | − | − | 2 | 2 | 2 |
| NCTC 8325 | + | + | + | − | − | − | 1 or 4 | 1 | 1 |
Genes were present (+) or absent (−) based on Affymetrix algorithms (raw), adjusted call determinations, and PCR detection. agr type determinations are also shown (1 to 4).
FIG. 2.
PCR detection of cna, srtA, and agr type. PCR analysis was performed for cna and srtA and to distinguish agr alleles for all strains in this study. In each case, PCR results validated adjusted call predictions. (A) PCR detection of srtA (S) and cna (C) for several strains in which there were discrepancies between Affymetrix and adjusted call determinations. Positive (MRSA252) and negative (COL, laboratory 1) control strains are also shown. (B) Multiplex PCR detection of S. aureus agr alleles, as described in reference 14. CDC strains tested are indicated across the top. M, molecular marker; C1 to C4, controls for agr types 1 to 4, respectively.
Next, the two procedures were used to evaluate allelic differences between strains. The agr locus encodes members of a two-component regulatory system that, in part, modulates virulence factor expression in S. aureus. Ji et al. demonstrated that polymorphisms within the locus define at least four agr groups (12). Because the Saur2a contains alleles of the genes constituting the agr locus, we investigated the accuracy of the microarray for determining agr types. Although Affymetrix algorithms provided ambiguous results for three of the strains analyzed, applying adjusted determination values resulted in accurate agr typing of each strain, as confirmed by PCR analysis (Table 3; Fig. 2B).
Use of GeneChips to monitor strain relatedness.
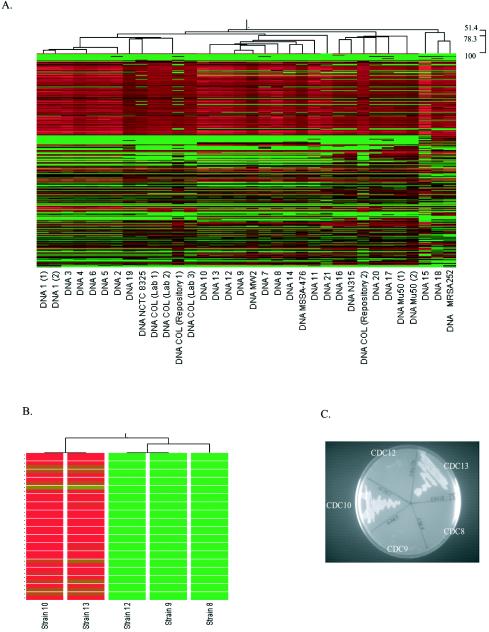
In addition to simultaneously providing an ability to obtain gene-by-gene information for a strain under investigation, the relatedness of each strain analyzed could also be determined with Saur2a. This was accomplished by using hierarchical clustering to develop a dendrogram that compared the normalized signal intensity of each qualifier for a given strain to the signal intensity of the same qualifier across all strains analyzed (Fig. 3A). Using this approach, strains that have similar signal intensities for all qualifiers are positioned closer together on the dendrogram than strains with divergent genomic compositions (differing signal intensities for the same qualifiers).
FIG. 3.
GeneChip-based genotyping. (A) Saur2a-derived dendrogram (top) with heat map (beneath) for all qualifiers that were analyzed in each strain. The dendrogram illustrates the relatedness of each strain based on the signal intensity of each qualifier across all strains. Within the heat map, each qualifier (total, 7,723) is shown vertically for each strain. Red indicates a high signal intensity; green indicates a low signal intensity. The order of qualifiers is identical for all strains. Scanning horizontally identifies qualifiers that have high signal intensity (red) in some strains but low intensities (green) in others. (B) Dendrogram of CDC strains 10, 13, 12, 9, and 8, which were all considered to be identical strains by both ribotyping and PFGE. The heat map illustrates 36 qualifiers (horizontally) that are considered present in strains 10 and 13 but absent in other strains, based on adjusted call determinations. (C) Growth characteristics of CDC strains 10, 13, 12, 9, and 8 on kanamycin-containing agar plates.
The data were validated by several observations. First, as shown in Fig. 3A, strains 1, 10 and 13 (both are the same strain), COL, and Mu50 were independently tested multiple times, and replicates were considered more closely related than other strains analyzed. Isolates 10 and 13 are the same strain; they were included twice to serve as a control for this analysis. Second, in silico comparisons demonstrated that, among sequenced strains, (i) MW2 is most closely related to MSSA476, (ii) Mu50 is closely related to N315 and moderately related to MRSA252, and (iii) COL is closely related to NCTC 8325. Each of these relationships was detected in the dendrogram (Fig. 3A). Finally, both ribotyping and PFGE clustering nicely agreed with the dendrogram derived from the GeneChip data (Table 4).
TABLE 4.
Ribotyping, GeneChip, and PFGE genotyping results
| Strain | Genotyping resulta
|
||
|---|---|---|---|
| GeneChip | Ribotype | PFGE | |
| CDC 1 | 1.1 | XII | USA300 (0.0114) |
| CDC 3 | 1.1 | XII | USA300 (0.0114) |
| CDC 4 | 1.1 | XII | USA300 (0.0114) |
| CDC 6 | 1.1 | XII | USA300 (0.0114) |
| CDC 5 | 1.1 | XII | USA300 (0.0114) |
| CDC 2 | 1.2 | XII | USA300 (0.0114) |
| CDC 19 | 1.3 | XII | USA500 TYPE (0.0004) |
| NCTC 8325 | 1.4 | XIII | ND |
| COL (lab 1) | 1.5 | IX | ND |
| COL (lab 2) | 1.5 | ND | ND |
| COL (repository 1) | 1.5 | ND | ND |
| COL (lab 3) | 1.5 | ND | ND |
| CDC 10 | 2.1 | XI | USA400 (0.0051) |
| CDC 13 | 2.1 | XI | USA400 (0.0051) |
| CDC 12 | 2.2 | XI | USA400 (0.0051) |
| CDC 9 | 2.2 | XI | USA400 (0.0051) |
| MW2 | 2.3 | XI | ND |
| CDC 7 | 2.4 | IV | USA400 (0.0199) |
| CDC 8 | 2.5 | XI | USA400 (0.0051) |
| CDC 14 | 2.6 | X | USA400 (0.0172) |
| MSSA476 | 2.7 | XI | ND |
| CDC 11 | 2.8 | XI | USA400 (0.0080) |
| CDC 21 | 2.9 | VI | USA700 TYPE (0.0097) |
| CDC 16 | 3.1 | V | USA100/800 |
| N315 | 3.2 | ND | ND |
| COL (repository 2) | 3.3 | ND | ND |
| CDC 20 | 3.4 | II | USA600 TYPE |
| CDC 17 | 3.5 | VII | USA100-B (0.0022) |
| Mu50 (1) | 3.6 | ND | ND |
| Mu50 (2) | 3.6 | ND | ND |
| CDC 15 | 4.1 | III | USA600 (0.0121) |
| CDC 18 | 4.2 | VIII | USA200 TYPE |
| MRSA252 | 4.3 | I | ND |
Ribotyping, GeneChip, and PFGE results are shown for each strain. Strains were observed to fit into four major clusters (1 to 4) by GeneChip analysis (Fig. 3A). Individual strains within each of these clusters were further distinguished. For example, GeneChip profiles 2.2 and 2.3 are different strains within cluster number 2. Strains with the same profile numbers are identical. Ribotyping results distinguished strains as belonging to one of 12 different ribogroups (I to XII). PFGE results demonstrated that strains belonged to eight different groups (USA100 to USA800; 80% identity cutoff). Numbers in parentheses represent the strain's identification number. Strains with the same identification number are considered identical. ND, not determined.
Despite the similarity between the three genotyping approaches, GeneChip results appeared to be the most discriminative. For instance, ribotyping data indicated that seven strains fit into ribogroup XII and eight strains belonged to ribogroup XI. As shown in Table 4, both PFGE and Saur2a-based typing further distinguished members of each ribogroup into subgroups. In the case of ribogroup XII, PFGE and GeneChip analysis further distinguished strains into identical subgroups. However, five strains from ribogroup XI were considered identical by PFGE (isolates 8, 9, 10, 12, and 13) but were further distinguished as three separate strains by Saur2a (Table 4; Fig. 3A and B). To determine which typing method provided more accurate results, adjusted call determinations were compared for all qualifiers across these five strains. As shown in Fig. 3B, 36 genes including the antimicrobial resistance determinants ermA, bleO, and aadA were considered to be present in strains 10 and 13 but absent from strains 9, 12, and 8. To determine if these GeneChip predictions were correct, strains were tested for growth on antibiotic-containing agar plates. Strains 10 and 13 formed colonies on plates containing kanamycin, whereas isolates 8, 9, and 12 did not, confirming that the five strains are not identical in genetic composition (Fig. 3C). In addition, adjusted detection call predictions indicated that 31 genes were present in strains 9 and 12 but absent from strains 10 and 13 (data not shown). Collectively, these results suggested that GeneChip-based genotyping was more discriminative than both ribotyping and PFGE.
DISCUSSION
We have previously reported results of studies that used a first-generation S. aureus oligonucleotide array (7, 17, 21). That array was based on an incomplete version of the S. aureus strain COL DNA sequence and, as a result, could not evaluate 15% of the genes within the COL genome. Although the array was adequate for use with strains closely related to COL, publication of the completed S. aureus N315 and Mu50 genomes, combined with the availability of four additional genomes, provided strong motivation for developing a second-generation microarray that could monitor a wider repertoire of genes in divergent strains (13). In the present study, we have used this S. aureus Affymetrix oligonucleotide array to determine the relatedness and genetic composition of a series of S. aureus strains. In doing so, we developed an approach that simultaneously evaluates relationships among strains and provides present or absent determinations for every gene that is represented on the microarray.
This technology is expected to provide novel information about S. aureus pathogenesis, antimicrobial resistance, and vaccine tolerance. For example, studies such as those performed by Vandenesch and colleagues that demonstrated that the Panton-Valentine leukocidin virulence factor is present in every CO-ORSA strain they tested can now be extended to identify whether these genes are also present in health care institution-associated strains. It is likely that such a study will be helpful in defining whether a subset of genes can distinguish community-associated from nosocomial ORSA strains. Defining the entire repertoire of genes that are conserved across diverse CO-ORSA strains may also clarify how the proteins that they encode influence the prevalence of ORSA within the community.
Several genes have previously been linked to a particular type of S. aureus infection, such as tst with toxic shock syndrome and exofoliative toxins with scalded-skin syndrome. It is anticipated that this technology will also provide the ability to associate subsets of S. aureus genes with particular types of infections. Moreover, because the GeneChip used contains alleles of many genes, the potential exists to associate a particular phenotype with a gene allele. Studies evaluating agr types have previously demonstrated that allelic types do influence pathogenesis and, thus, their identification is important for epidemiological studies. Most clinical isolates are agr group 1. agr group 3 has been associated with community-associated methicillin-resistant S. aureus, group 2 has been linked to intermediate glycopeptide resistance, and group 4 has been associated with exfoliative toxin-producing strains (6, 11, 16, 18, 19, 23). Because adjusted call predictions accurately determined the agr type of each strain analyzed in this study, it is likely that this technology could be used to analyze the association of a specific agr type(s), and other genes or alleles, with disease-causing strains.
Although the technology described herein globally evaluates the genetic composition of an organism, there are currently limitations to the GeneChip approach. Each qualifier contains an average of 34 probes which collectively monitor the presence or absence of a qualifier. Due to the microarray design stringency, in some instances probes are not distributed equally across a gene. In addition, gaps exist between probes for most genes represented on the GeneChip, and as a result not every nucleotide of a coding sequence is interrogated. Therefore, results should be considered an estimation of whether a particular gene is present or absent within a sample. The same limitation would occur with standard PCR-based microarrays, which are less stringent and would erroneously consider a gene to be present if a fragment of a gene or if homologous sequences hybridized to a qualifier. However, the stringency of the GeneChip probe design provides an additional layer of information that allows genes and promoter regions to be monitored for point mutations and deletions (unpublished data). It is unlikely that small deletions or point mutations within a gene would easily be detected with PCR-based microarrays.
Because Saur2a arrays measure over 7,700 qualifiers, it is not surprising that the array has considerable potential for assessing strain relatedness. Importantly, the described approach was robust enough to identify replicates of four very divergent strains. This indicates that the technology can allow for one to determine whether a group of similar strains under investigation are clonal or slightly divergent in genetic composition. This distinction is a critical aspect of monitoring strain outbreaks.
This technology is also likely to be powerful for analyzing the acquisition of antimicrobial resistance determinants and may provide a means to evaluate whether other genetic determinants confer a predisposition, or contribute to, the development of resistance. McDougal and colleagues have recently shown that oxacillin-resistant strains from the United States belong to eight major lineages (15). The present study describes the use of strains from each of these lineages to develop a novel GeneChip-based method of interrogating S. aureus strains. We currently are using the described methodology to further characterize each lineage.
In most cases MLST, ribotyping, and PFGE provide the level of discrimination needed to monitor strains circulating throughout the community and health care environments. These techniques are more rapid, do not require extensive analysis, and can be accomplished at a fraction of the cost associated with microarrays. However, none of these methods also allows one to simultaneously define the genes that constitute the organism(s) under investigation on a genome scale. In addition to the uses described above, we envision the approach developed here to be helpful in characterizing isolates within the same ribo-, MLST, or PFGE group, or in studies where further characterization is needed.
ADDENDUM IN PROOF
After submission of this paper, the completed genomes of MRSA252 and MSSA476 were reported (M. T. G. Holden, E. J. Feil, J. A. Lindsay, S. J. Peacock, N. P. J. Day, M.C. Enright, T. J. Foster, C. E. Moore, L. Hurst, R. Atkin, A. Barron, N. Bason, S. D. Bentley, C. Chillingworth, T. Chillingworth, C. Churcher, L. Clark, C. Corton, A. Cronin, J. Doggett, L. Dowd, T. Feltwell, Z. Hance, B. Harris, H. Hauser, S. Holroyd, K. Jagels, K. D. James, N. Lennard, A. Line, R. Mayes, S. Moule, K. Mungall, D. Ormond, M. A. Quail, E. Rabbinowitsch, K. Rutherford, M. Sanders, S. Sharp, M. Simmonds, K. Stevens, S. Whitehead, B. G. Barrell, B. G. Spratt, and J. Parkhill, Proc. Natl. Acad. Sci. 101:9786-9791, 2004).
Acknowledgments
We thank Steven Gill for sharing the complete COL genomic sequence prior to its release in the TIGR Comprehensive Microbial Resource database. Additionally, we thank Stephen Olmsted for helpful suggestions to the manuscript.
REFERENCES
- 1.Baba, T., F. Takeuchi, M. Kuroda, H. Yuzawa, K. Aoki, A. Oguchi, Y. Nagai, N. Iwama, K. Asano, T. Naimi, H. Kuroda, L. Cui, K. Yamamoto, and K. Hiramatsu. 2002. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet 359:1819-1827. [DOI] [PubMed] [Google Scholar]
- 2.Bannerman, T. L., G. A. Hancock, F. C. Tenover, and J. M. Miller. 1995. Pulsed-field gel electrophoresis as a replacement for bacteriophage typing of Staphylococcus aureus. J. Clin. Microbiol. 33:551-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumberg, H. M., D. Rimland, J. A. Kiehlbauch, P. M. Terry, and I. K. Wachsmuth. 1992. Epidemiologic typing of Staphylococcus aureus by DNA restriction fragment length polymorphisms of rRNA genes: elucidation of the clonal nature of a group of bacteriophage-nontypeable, ciprofloxacin-resistant, methicillin-susceptible S. aureus isolates. J. Clin. Microbiol. 30:362-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chou, A., and J. Burke. 1999. CRAWview: for viewing splicing variation, gene families, and polymorphism in clusters of ESTs and full-length sequences. Bioinformatics 15:376-381. [DOI] [PubMed] [Google Scholar]
- 5.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dufour, P., Y. Gillet, M. Bes, G. Lina, F. Vandenesch, D. Floret, J. Etienne, and H. Richet. 2002. Community-acquired methicillin-resistant Staphylococcus aureus infections in France: emergence of a single clone that produces Panton-Valentine leukocidin. Clin. Infect. Dis. 35:819-824. [DOI] [PubMed] [Google Scholar]
- 7.Dunman, P. M., E. Murphy, S. Haney, D. Palacios, G. Tucker-Kellogg, S. Wu, E. L. Brown, R. J. Zagursky, D. Shlaes, and S. J. Projan. 2001. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J. Bacteriol. 183:7341-7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgerald, J. R., D. E. Sturdevant, S. M. Mackie, S. R. Gill, and J. M. Musser. 2001. Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc. Natl. Acad. Sci. USA 98:8821-8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes, W. S., and M. Borodovsky. 1998. How to interpret an anonymous bacterial genome: machine learning approach to gene identification. Genome Res. 8:1154-1171. [DOI] [PubMed] [Google Scholar]
- 11.Jarraud, S., G. J. Lyon, A. M. Figueiredo, L. Gerard, F. Vandenesch, J. Etienne, T. W. Muir, and R. P. Novick. 2000. Exfoliatin-producing strains define a fourth agr specificity group in Staphylococcus aureus. J. Bacteriol. 182:6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ji, G., R. Beavis, and R. P. Novick. 1997. Bacterial interference caused by autoinducing peptide variants. Science 276:2027-2030. [DOI] [PubMed] [Google Scholar]
- 13.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 14.Lina, G., F. Boutite, A. Tristan, M. Bes, J. Etienne, and F. Vandenesch. 2003. Bacterial competition for human nasal cavity colonization: role of staphylococcal agr alleles. Appl. Environ. Microbiol. 69:18-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McDougal, L. K., C. D. Steward, G. E. Kilgore, J. M. Chaitram, S. K. McAllister, and F. Tenover. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moore, P. C., and J. A. Lindsay. 2001. Genetic variation among hospital isolates of methicillin-sensitive Staphylococcus aureus: evidence for horizontal transfer of virulence genes. J. Clin. Microbiol. 39:2760-2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Said-Salim, B., P. M. Dunman, F. M. McAleese, D. Macapagal, E. Murphy, P. J. McNamara, S. Arvidson, T. J. Foster, S. J. Projan, and B. N. Kreiswirth. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., R. P. Novick, L. Venkataraman, C. Wennersten, P. C. DeGirolami, M. J. Schwaber, and H. S. Gold. 2003. Staphylococcus aureus accessory gene regulator (agr) group II: is there a relationship to the development of intermediate-level glycopeptide resistance? J. Infect. Dis. 187:929-938. [DOI] [PubMed] [Google Scholar]
- 19.Sakoulas, G., G. M. Eliopoulos, R. C. Moellering, Jr., C. Wennersten, L. Venkataraman, R. P. Novick, and H. S. Gold. 2002. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob. Agents Chemother. 46:1492-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shopsin, B., M. Gomez, S. O. Montgomery, D. H. Smith, M. Waddington, D. E. Dodge, D. A. Bost, M. Riehman, S. Naidich, and B. N. Kreiswirth. 1999. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J. Clin. Microbiol. 37:3556-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Utaida, S., P. M. Dunman, D. Macapagal, E. Murphy, S. J. Projan, V. K. Singh, R. K. Jayaswal, and B. J. Wilkinson. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719-2732. [DOI] [PubMed] [Google Scholar]
- 22.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Leeuwen, W., W. van Nieuwenhuizen, C. Gijzen, H. Verbrugh, and A. van Belkum. 2000. Population studies of methicillin-resistant and -sensitive Staphylococcus aureus strains reveal a lack of variability in the agrD gene, encoding a staphylococcal autoinducer peptide. J. Bacteriol. 182:5721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]