Abstract
Identification of the emerging pathogen Vibrio vulnificus biotype 3 has become a challenge for clinical laboratories in the last few years. In this study, the abilities of five commercial systems to identify this new species have been evaluated for the first time, using a unique collection of strains. Fifty-one well-documented wild strains of V. vulnificus biotype 3 were processed using API 20 NE, GNI+ Vitek 1 cards, ID-GNB Vitek 2 cards, Neg Combo 20 Microscan panels, and NMIC/ID-5 BD Phoenix panels. The numbers of strains identified as V. vulnificus by ID-GNB, NMIC/ID-5, and GNI+ were 50 (98.0%), 46 (90.2%), and 7 (13.7%), respectively. Neg Combo 20 Microscan panels and API 20 NE were unable to identify any of the strains of this emerging pathogen to the species level and mostly misidentifies them as other species of the Vibrionaceae family. Data on the phenotypic pattern of V. vulnificus biotype 3 when processed in all five systems as presented here could help clinical laboratories in identifying this new pathogen.
Since it was first described by Reichelt et al. in 1976 (9), for many years only two biotypes or serovars of the human pathogen Vibrio vulnificus have been recognized (1).
In the midsummer of 1996, we reported the first isolation of a new pathogen, V. vulnificus biotype 3, which causes septicemia and severe soft-tissue infections following contact with fish from artificial freshwater ponds (2). Since then, the identification of this emerging pathogen has become a challenge for clinical laboratories. With the phenotypic behavior of this new biotype being different from that of the V. vulnificus biotypes known so far, most of the commercial systems in use do not include the relevant data in their software, making correct identification of the new bacteria problematic.
In this study, the phenotypic pattern of this new biotype when tested with five commercial systems was defined, and the abilities of the current software of those systems to correctly identify V. vulnificus biotype 3 to the species level have been evaluated for the first time with a unique collection of strains.
MATERIALS AND METHODS
Identification systems.
Fifty-one well-documented V. vulnificus biotype 3 strains isolated during the years 1996 to 1997 were processed by five commercial identification systems: API 20 NE, GNI+ Vitek 1 cards, ID-GNB Vitek 2 cards (BioMerieux, Marcy l-Etoile, France), Neg Combo 20 Microscan panels (Dade Behring Inc.), and NMIC/ID-5 BD Phoenix panels (BD Biosciences).
Given the fact that previous reports have already concluded that the use of API 20 E is inadequate for the identification of V. vulnificus (6), only the API 20 NE system was used in this study. API 20 NE strips were inoculated and processed following the manufacturer's recommendations, and the results were analyzed by using Apilab software, version 3.2.2.
Inoculated Vitek GNI+ cards were processed with a Vitek 1 system, version VTK-R 07.02.
Vitek ID-GNB cards were used with a Vitek 2 system, version VT2-R 02.03.
Microscan Neg Combo 20 panels were inoculated according to the manufacturer's recommendations for halophilic vibrios (0.125 ml of water plus Pluronic plus 0.1 ml of a 0.5 McFarland bacterial suspension in each well) and processed in a Microscan Walkaway 96 apparatus. Results were analyzed by using the Microscan DMS system, version 24.1.
Finally, inoculated NMIC/ID-5 panels were processed with the BD Phoenix system according to the manufacturer's instructions.
Bacterial strains.
Fifty-one bacterial strains tested in the study were isolated from patients during the years 1996 and 1997 in laboratories serving the northern region of Israel, where all cases appear following contact with Tilapia fishes. The strains, which were previously identified at local laboratories, were eventually confirmed by the Central Laboratories of the Israel Ministry of Health in Jerusalem as belonging to V. vulnificus biotype 3 by using an extensive phenotypic workflow proposed by J. J. Farmer from the Centers for Disease Control and Prevention (Atlanta, Ga.).
Eventually, all the strains were confirmed as V. vulnificus at the Nuffield Department of Clinical Laboratory Sciences of John Radcliffe Hospital, University of Oxford (Oxford, United Kingdom), using PCR for the cytotoxin-hemolysin (CTH) gene, as described by Brauns et al. (4). The DNeasy kit (QIAGEN GmbH, Hilden, Germany) was used to extract DNA, and the protocol for gram-negative bacteria was followed. Briefly, several colonies from a single clone of each bacterial culture were picked off into phosphate-buffered solution and centrifuged at 7,500 rpm (5,000 × g) for 10 min. The cell pellet was resuspended in 180 μl of tissue lysis buffer, and then 20 μl of proteinase K (10 mg/ml) was added and the sample was incubated at 55°C until the tissue was completely lysed. Next, 200 μl of lysis buffer was added, and incubation was at 70°C for 10 min. The DNA in the clear viscous lysates was precipitated with ethanol (95% [vol/vol]) and added to DNeasy mini-columns. Ethanol (70% [vol/vol])-based buffers (AW1 and AW2) were added sequentially to the columns and centrifuged at 8,000 rpm (6,000 × g). The supernatants were discarded, and the DNA was resuspended in sterile water and used for amplification.
PCR amplification was carried out with previously described oligonucleotide primers (4). Each 50-μl amplification reaction mixture comprised 10 ng of chromosomal DNA, 100 pmol of each PCR primer (MWG Biotech, Ebersberg, Germany), 10× PCR buffer with 1.5 mM MgCl2 (QIAGEN GmbH), 0.5 U of Taq DNA polymerase (QIAGEN GmbH), and 1.6 mM deoxynucleoside triphosphates (ABgene, Epsom, United Kingdom). The reaction conditions were denaturation at 94°C for 1 min, primer annealing at 68°C for 1 min, and extension at 72°C for 1 min, for 30 cycles.
A volume (5 μl) of PCR-amplified DNA was separated on a 0.8% agarose gel containing 0.5 μg of ethidium bromide ml−1. The gel was run in Tris-borate-EDTA buffer at 120 V for 30 min with a 100-bp DNA ladder (ABgene, Epsom, United Kingdom) as a size standard. DNA fragments in the gel were visualized under a UV transilluminator (Gel Doc 1000; Bio-Rad Laboratories Ltd., Bio-Rad House, Hemel Hempstead, United Kingdom).
In addition, two control strains were also tested with all systems but NMIC/ID-5 BD-Phoenix panels: V. vulnificus biotype 1 (CDC strain 9028A95) and V. vulnificus biotype 2 (L. Hoi, Denmark; strain 96-7-138), both from an Israel Ministry of Health strain collection.
RESULTS
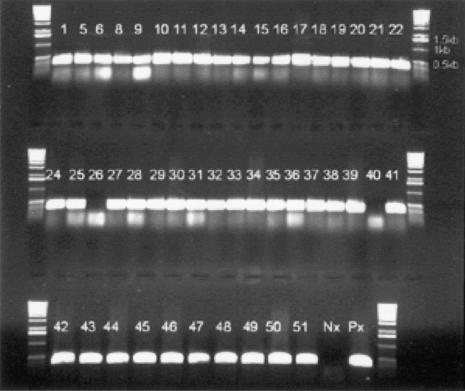
The results by PCR for 48 of the strains are shown in Fig. 1. Samples 2, 3, and 4 were tested separately and were also positive for the CTH gene. Samples 26 and 40, shown as negative for the CTH gene, gave a positive reaction when retested using an annealing temperature of 60°C.
FIG. 1.
PCR results for the CTH gene. Px, positive control strain; Nx, negative control strain.
The numbers of strains correctly identified by the different systems are shown in Table 1. The numbers of isolates misidentified as other Vibrionaceae or non-Vibrio species or not identified at all are also shown in the same table.
TABLE 1.
Percentages of strains correctly identified as V. vulnificus by all systems
| System | % (No.) of correct IDa | % (No.) of strains misidentified as:
|
% (no.) of strains not identified | |
|---|---|---|---|---|
| Other Vibrio | Not Vibrio | |||
| API 20 NE | 0 (0) | 100 (51) | 0 (0) | 0 (0) |
| Neg Combo 20/Microscan | 0 (0) | 100 (51) | 0 (0) | 0 (0) |
| GNI+/Vitek 1 | 17.6 (9) | 21.6 (11) | 60.8 (31) | 0 (0) |
| ID-GNB/Vitek 2 | 98.0 (50) | 0 (0) | 0 (0) | 2.0 (1) |
| NMIC/ID-5/BD Phoenix | 90.2 (46) | 3.9 (2) | 3.9 (2) | 2.0 (1) |
ID, identifications.
ID-GNB, NMIC/ID5, and GNI+ correctly identified 50 (98.0%), 46 (90.2%), and 9 (17.6%) strains, respectively. Microscan Neg Combo 20 and API 20 NE were unable to correctly identify any of the strains of this emerging pathogen.
For the single strain that ID-GNB was unable to identify, the Vitek 2 system proposed Plesiomonas shigelloides and V. vulnificus with the same probability rates and asked for further tests.
From the five strains not identified by NMIC/ID5 as V. vulnificus, only two were misidentified by the system as other Vibrio spp.
GNI+ misidentified 10 strains as other Vibrio spp. Thirty-one strains, all of them presenting the same phenotypic pattern, were misidentified as P. shigelloides (75% probability), with V. vulnificus as the second choice (19% probability).
Microscan Neg Combo 20 misidentified all the strains as other Vibrionaceae, most of them as Vibrio mimicus and Vibrio parahaemolyticus.
Finally, API 20 NE also misidentified all the strains as other Vibrionaceae, most of them as Vibrio alginolyticus.
Tables 2 to 6 show the phenotypic profiles presented by all 51 strains of biotype 3 tested with the API 20 NE, GNI+, ID-GNB, NMIC/ID5, and Microscan Neg Combo 20 systems, respectively. In addition, Tables 2, 3, 4, and 6 show the results for the control strains of biotypes 1 and 2 with the respective systems. Table 2 also shows the expected result for V. vulnificus with API 20 NE as it appears in the kit insert.
TABLE 2.
API 20 NE profile of the 51 strains of biotype 3a
| Substrate | % (No.) of positive biotype 3 strains | Result with control strain
|
Expected resultc (% positive) | |
|---|---|---|---|---|
| Biotype 1 | Biotype 2 | |||
| Potassium nitrate | 100 (51) | + | + | 100 |
| Tryptophan | 100 (51) | + | + | 95 |
| Glucose | 0 (0) | + | + | 95 |
| Arginine | 0 (0) | − | − | 0 |
| Urea | 0 (0) | − | − | 1 |
| Esculin | 0 (0) | − | − | 95 |
| Gelatin | 100 (51) | + | + | 99 |
| PNPG | 0 (0) | + | + | 99 |
| Glucose | 73 (37) | − | − | 9 |
| Arabinose | 0 (0) | − | − | 0 |
| Mannose | 61 (31) | − | − | 10 |
| Mannitol | 0 (0) | − | + | 9 |
| N-Acetyl-glucosamine | 6 (3) | − | − | 1 |
| Maltose | 100 (51) | − | + | 6 |
| Gluconate | 61 (31) | − | + | 28 |
| Caprate | 0 (0) | − | − | 0 |
| Adipate | 0 (0) | − | − | 0 |
| Malate | 89 (45) | + | + | 95 |
| Citrate | 33 (17) | − | + | 91 |
| Phenyl-acetate | 0 (0) | − | − | 0 |
| Oxidaseb | 100 (51) | + | + | 100 |
Substrates that give a positive result with all 51 strains are boldfaced. PNPG, p-nitrophenyl-β-d-glucoside.
External test.
Expected result for V. vulnificus according to the insert chart provided by the manufacturer.
TABLE 6.
Profile of the 51 strains of biotype 3 in Microscan system, using Neg Combo 20 panelsa
| Substrate | % (No.) of positive strains | Result for control strain
|
|
|---|---|---|---|
| Biotype 1 | Biotype 2 | ||
| Glucose | 100 (51) | + | + |
| Raffinose | 0 (0) | − | − |
| Inositol | 0 (0) | − | − |
| Urea | 0 (0) | − | − |
| Lysine | 0 (0) | − | − |
| Tryptophan deaminase | 0 (0) | − | − |
| Citrate | 14 (7) | − | − |
| Colistin (4 mcg/ml) (gr.) | 100 (51) | + | − |
| Sucrose | 0 (0) | − | − |
| Rhamnose | 0 (0) | − | − |
| Adonitol | 0 (0) | − | − |
| H2S | 0 (0) | − | − |
| Arginine | 0 (0) | − | − |
| Esculine | 0 (0) | − | − |
| Malonate | 0 (0) | − | − |
| Cephalotin (8 mcg/ml) (gr.) | 14 (7) | − | + |
| Sornitol | 0 (0) | − | − |
| Arabinose | 0 (0) | − | − |
| Mellobiose | 0 (0) | − | − |
| Indol | 100 (51) | − | − |
| Omithine | 0 (0) | − | − |
| VP | 0 (0) | − | − |
| ONPG | 0 (0) | − | + |
Substrates that give a positive result with all 51 strains are boldfaced. gr., growth; VP, Voges-Proskeuer; ONPG, o-nitrophenyl-β-d-galactopyranoside.
TABLE 3.
Profiles of the 51 strains of biotype 3 in Vitek 1 using GNI+ cardsa
| Substrate | % (No.) of positive biotype 3 strains | Result for control strain
|
|
|---|---|---|---|
| Biotype 1 | Biotype 2 | ||
| DP-300 | 0 (0) | − | − |
| Glucose (ox.) | 100 (51) | + | + |
| Acetamide | 0 (0) | − | − |
| Esculine | 0 (0) | − | − |
| Plant indican | 0 (0) | − | − |
| Urea | 0 (0) | − | − |
| Citrate | 0 (0) | − | − |
| Malonate | 0 (0) | − | − |
| Tryptophan | 0 (0) | − | − |
| Polymyxin B | 0 (0) | − | − |
| Lactose | 0 (0) | − | − |
| Maltose | 100 (51) | + | + |
| Mannitol | 0 (0) | + | + |
| Xylose | 0 (0) | − | − |
| Raffinose | 0 (0) | − | − |
| Sorbitol | 0 (0) | − | − |
| Sucrose | 0 (0) | − | − |
| Inositol | 0 (0) | − | − |
| Adonitol | 0 (0) | − | − |
| p-Coumaric | 0 (0) | − | − |
| H2S | 0 (0) | − | − |
| ONPG | 0 (0) | − | − |
| Rhamnose | 0 (0) | − | − |
| l-arabinose | 0 (0) | − | − |
| Glucose (ferm.) | 100 (51) | + | + |
| Arginine | 0 (0) | − | − |
| Lysine | 78 (40) | − | + |
| Omithine | 63 (32) | − | − |
Substrates that give a positive result with all 51 strains are boldfaced. ox., oxidation; ferm., fermentation; ONPG, o-nitrophenyl-β-d-galactopyranoside.
TABLE 4.
Profile of the 51 strains of biotype 3 in Vitek 2, using ID-GNB cardsa
| Substrate | % (No.) of positive biotype 3 strains | Result with control strain
|
|
|---|---|---|---|
| B1 | B2 | ||
| Adonitol | 0 (0) | − | − |
| l-arabinose | 0 (0) | − | − |
| d-cellobiose | 0 (0) | − | + |
| d-galacturonate | 0 (0) | − | − |
| d-glucose | 100 (51) | + | + |
| Glucose-1-phosp. | 100 (51) | + | + |
| d-glucuronate | 96 (49) | − | + |
| Myoinositol | 0 (0) | − | − |
| 5-Keto-d-gluconate | 0 (0) | − | − |
| d-maltose | 98 (50) | + | + |
| d-mannitol | 0 (0) | − | + |
| d-melibiose | 0 (0) | − | − |
| Palatinose | 0 (0) | − | − |
| d-raffinose | 0 (0) | − | − |
| l-rhamnose | 0 (0) | − | − |
| Saccharose/sucrose | 0 (0) | − | − |
| d-sorbitol | 0 (0) | − | + |
| d-trehalose | 100 (51) | + | + |
| Lysine | 100 (51) | + | + |
| Decarboxylase | |||
| Omithine decarbox. | 71 (36) | − | − |
| Urease | 0 (0) | − | − |
| Malonate | 0 (0) | − | − |
| Tryptophane deaminase | 0 (0) | − | − |
| Alpha-arabinosidase | 0 (0) | − | + |
| Alpha-galactosidase | 0 (0) | − | + |
| Alpha-glutamate | 0 (0) | − | − |
| Beta-cellobiosidase | 16 (8) | − | − |
| Beta-galactosidase | 0 (0) | − | + |
| Beta-glucosidase | 0 (0) | − | − |
| Beta-glucuronidase | 0 (0) | − | − |
| Beta-mannosidase | 100 (51) | + | + |
| Beta-N-acetyl-gluc. | 100 (51) | + | + |
| Beta-N-acetyl-galac. | 0 (0) | + | + |
| Beta-xylosidase | 0 (0) | − | − |
| Glu-Gly-Arg-arylamidase | 0 (0) | − | − |
| Gamma-glutamyl-trans. | 0 (0) | − | − |
| l-lysine-arylamidase | 100 (51) | + | + |
| Phosphatase | 0 (0) | − | − |
| l-proline-arylamidase | 100 (51) | + | + |
| l-pyrrolidonyl-arylam. | 0 (0) | − | − |
| CBZ-Arg-arylamidase | 0 (0) | − | − |
Substrates that give a positive result with all 51 strains are boldfaced. phosp., phosphatase; decarbox., decarboxylase; gluc., glucosidase; galac., galactosidase; trans., transferase; arylam., arylamidase; CBZ, carbobenzyloxy; B1, V. vulnificus biotype 1; B2, V. vulnificus biotype 2.
DISCUSSION
In the last few years, the routine use of molecular biology techniques such as nested PCR (7) has been playing a more important role in clinical microbiology laboratories in developed countries. However, this is not the situation in the majority of the clinical laboratories worldwide, especially in nondeveloped countries. Thus, the use of commercial phenotype-based identification systems will continue to be the mainstream in the next years.
In this study, the abilities of five widely used phenotype-based commercial systems to identify the emerging pathogen V. vulnificus biotype 3 were evaluated.
ID-GNB with Vitek 2 and NMIC/ID5 with BD Phoenix proved to be the best systems for correctly identifying this new biotype as V. vulnificus (98.0 and 90.2% of strains correctly identified, respectively). These two systems offer a wide range of phenotypic reactions, many of them enzymatic, and a considerable number of them were positive with the strains tested. Microscan Neg Combo 20, API 20 NE, and GNI+ with Vitek 1, however, offer a significantly smaller number of reactions, and the number of positive tests was minimal with the tested strains. However, the majority of the strains presented a consistent phenotypic pattern with each of the systems. Thus, clinical laboratories using these systems could tentatively identify strains showing the phenotype presented here as that of V. vulnificus or at least suspect the presence of this pathogen.
So far, this emerging pathogen has been reported only in Israel (2, 3, 5, 8).
However, since commercial systems may misidentify it as another member of the Vibrionaceae family, it is not unlikely that the presence of this pathogen in other countries could be underestimated. Thus, clinical microbiology laboratories should be aware of this possibility and must meticulously check the correct identification of any vibrio isolated from clinical sources.
TABLE 5.
Profile of the 51 strains of biotype 3 in BD-Phoenix, using NMIC/ID-5 cardsa
| Substrate | % (No.) of positive strains |
|---|---|
| Phenylalanine-AMC | 100 (51) |
| 4MU-NAG | 100 (51) |
| Glutamic acid-AMC | 0 (0) |
| Tryptophan-AMC | 86 (44) |
| PYR-AMC | 0 (0) |
| Proline-AMC | 100 (51) |
| Arginine-AMC | 100 (51) |
| Arginine-arginine-AMC | 100 (51) |
| Glycine-AMC | 100 (51) |
| Leucine-AMC | 100 (51) |
| Lysine-alanine-AMC | 100 (51) |
| Glutaryl-glycine-arginine-AMC | 6 (3) |
| Glycine-proline-AMC | 33 (17) |
| Colistin | 0 (0) |
| Polymyxin B | 0 (0) |
| Mannitol | 0 (0) |
| Citrate | 6 (3) |
| Acetate | 6 (3) |
| Adonitol | 0 (0) |
| Malonate | 6 (3) |
| Ketoglutaric acid | 0 (0) |
| Tiglic acid | 4 (2) |
| Proline-NA | 100 (51) |
| Gamma-l-glutamyl-NA | 4 (2) |
| Bis(PNP) phosphate | 100 (51) |
| PNP-β-d-glucoside | 10 (5) |
| Allose | 0 (0) |
| N-acetyl-galactosamine | 4 (2) |
| N-acetyl-glucosamine | 12 (6) |
| Sorbitol | 4 (2) |
| Sucrose | 2 (1) |
| Galacturonic acid | 0 (0) |
| Maltulose | 0 (0) |
| Rhamnose | 0 (0) |
| Gentiobiose | 6 (3) |
| Dextrose | 10 (5) |
| Galactose | 0 (0) |
| Fructose | 8 (4) |
| Gluconic acid | 4 (2) |
| Melibiose | 0 (0) |
| Arabinose | 0 (0) |
| Methyl-β-glucoside | 2 (1) |
| Ornithine | 0 (0) |
| Urea | 8 (4) |
| Esculin | 6 (3) |
Substrates that give a positive result with all 51 strains are boldfaced; AMC, fluorescent coumarin derivative; 4MU-NAG, 4-methylumbelliferone-N-acetyl-bd-glucosaminide; PYR, pyrrolidonyl-α-naphthylamide; NA, p-nitroaniline; PNP, p-nitrophenyl.
Acknowledgments
We thank Bactlab Ltd., Caesarea Industrial Park, Israel, for supplying the NMIC/ID5 BD Phoenix panels and Ilex Ltd., Roch-Ha’ayin, Israel, for supplying the GNI+ and ID-GNB Vitek cards.
We also thank the technical staff of the Microbiology Laboratory of Ha'Emek Medical Center, the Microbiology Laboratory of Maccabi Health Services in Haifa, Israel, the Microbiology Laboratory of Assaf Harofeh Medical Center, Zerifin, Israel, and the Molecular Microbiology Laboratory of John Radcliffe Hospital, University of Oxford, Oxford, United Kingdom.
REFERENCES
- 1.Biosca, E. G., C. Amaro, J. L. Larsen, and K. Pedersen. 1997. Phenotypic and genotypic characterization of Vibrio vulnificus: proposal for the substitution of the subspecific taxon biotype for serovar. Appl. Environ. Microbiol. 63:1460-1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisharat, N., V. Agmon, R. Finkelstein, R. Raz, G. Ben-Dror, L. Lerner, S. Soboh, R. Colodner, D. Cameron, D. Wykstra, D. Swerdlow, and J. J. Farmer. 1999. Clinical, epidemiological and microbiological features of a Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet 354:1421-1424. [DOI] [PubMed] [Google Scholar]
- 3.Bisharat, N., and R. Raz. 1996. Vibrio infection in Israel due to changes in fish marketing. Lancet 348:1585-1586. [DOI] [PubMed] [Google Scholar]
- 4.Brauns, L. A., M. C. Hudson, and J. D. Oliver. 1991. Use of the polymerase chain reaction in detection of culturable and nonculturable Vibrio vulnificus cells. Appl. Environ. Microbiol. 57:2651-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colodner, R., B. Chazan, J. Kopelowitz, Y. Keness, and R. Raz. 2002. Unusual portal of entry of Vibrio vulnificus: evidence of its prolonged survival on the skin. Clin. Infect. Dis. 34:714-715. [DOI] [PubMed] [Google Scholar]
- 6.Dalsgaard, A., I. Dalsgaard, L. Hoi, and J. L. Larsen. 1996. Comparison of a commercial biochemical kit and an oligonucleotide probe for identification of environmental isolates of Vibrio vulnificus. Lett. Appl. Microbiol. 22:184-188. [DOI] [PubMed] [Google Scholar]
- 7.Lee, S. E., S. Y. Kim, S. J. Kim, H. S. Kim, J. H. Shin, S. H. Choi, S. S. Chung, and J. H. Rhee. 1998. Direct identification of Vibrio vulnificus in clinical specimens by nested PCR. J. Clin. Microbiol. 36:2887-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nudelman, A., G. Edelson, A. Linden, and R. Raz. 1997. Infection by Vibrio vulnificus after a prick from the spine of a Tilapia. Harefuah 133:444-445. (In Hebrew). [PubMed]
- 9.Reichelt, J. L., P. Baumann, and L. Baumann. 1976. Study of genetic relationships among marine species of the genera Beneckea and Photobacterium by means of in vitro DNA/DNA hybridization. Arch. Microbiol. 110:101-120. [DOI] [PubMed] [Google Scholar]