Abstract
Hepatitis C virus (HCV) infection is an increasing health problem worldwide. Quantitative assays for HCV viral load are valuable in predicting response to therapy and for following treatment efficacy. Unfortunately, most quantitative tests for HCV RNA are limited by poor sensitivity. We have developed a convenient, highly sensitive real-time reverse transcription-PCR assay for HCV RNA. The assay amplifies a portion of the 5′ untranslated region of HCV, which is then quantitated using the TaqMan 7700 detection system. Extraction of viral RNA for our assay is fully automated with the MagNA Pure LC extraction system (Roche). Our assay has a 100% detection rate for samples containing 50 IU of HCV RNA/ml and is linear up to viral loads of at least 109 IU/ml. The assay detects genotypes 1a, 2a, and 3a with equal efficiency. Quantitative results by our assay correlate well with HCV viral load as determined by the Bayer VERSANT HCV RNA 3.0 bDNA assay. In clinical use, our assay is highly reproducible, with high and low control specimens showing a coefficient of variation for the logarithmic result of 2.8 and 7.0%, respectively. The combination of reproducibility, extreme sensitivity, and ease of performance makes this assay an attractive option for routine HCV viral load testing.
Hepatitis C virus (HCV) is an increasingly important infectious agent in the United States, with an estimated 1.8% of the U.S. population currently infected (1). Only about 20% of infected individuals spontaneously clear the virus, while the rest become chronically infected. Of chronically infected individuals, 10 to 30% will eventually develop cirrhosis or hepatocellular carcinoma. The most effective current therapy for HCV infection includes a combination of pegylated interferon and ribavirin and is associated with an approximately 50 to 60% long-term virological response (3, 6, 12). The response to pegylated interferon and ribavirin therapy is influenced by the genotype of HCV and also by the viral load at the initiation of therapy. For patients with unfavorable genotypes or viral loads, an extended period of therapy may be warranted. Measurement of viral load is also useful in monitoring response to therapy after initiation of treatment.
Several alternatives are available for the determination of HCV viral loads. The comparison of these assays has been facilitated by the recent introduction of the World Health Organization international standard, which expresses viral load in international units (IU) (22). Commercial assays are available which rely on amplification of HCV nucleic acid by reverse transcription-PCR (RT-PCR; Amplicor; Roche) or on amplification of signal from hybridization product (bDNA; Bayer). In general, the available assays have suffered from limitations due to trade-offs between sensitivity and the ability to provide quantitative results. Thus, the most sensitive assays have provided only qualitative results, while assays with the most accurate quantitation have offered limited sensitivity.
We have developed a real-time RT-PCR assay for HCV RNA using the TaqMan 7700 detection system combined with an automated RNA extraction system (MagNA Pure LC; Roche). The assay is extremely sensitive, with a lower limit of detection of less than 50 IU/ml. The assay allows linear and accurate quantitation of HCV loads from less than 50 IU/ml up to at least 109 IU/ml. Because the extraction, amplification, and detection steps have all been automated, our assay is rapid and requires a minimum of technologist hands-on time. This approach should be attractive to laboratories seeking a sensitive, quantitative, and cost-effective method for determination of HCV viral load.
MATERIALS AND METHODS
Patient sample selection.
A total of 486 patient EDTA plasma or serum samples were used for method comparison testing. All samples had originally been submitted to the clinical laboratories for either HCV qualitative or quantitative testing. The initial clinical qualitative testing was done with the COBAS Amplicor 2.0 system (Roche Diagnostics Corp., Indianapolis, Ind.), and the quantitative testing was done with the VERSANT HCV RNA 3.0 bDNA assay (Bayer Diagnostics, Tarrytown, N.J.) according to the manufacturers' protocols. HCV RNA-positive samples were selected to obtain samples with a range of quantitative values spanning the bDNA assay measuring range, with an enhanced number of the less common genotypes, as follows: 1a, n = 107; 1b, n = 68; 2a, n = 44; 2b, n = 48; 3a, n = 60; and 4, n = 2. A small group of samples was selected that had been previously demonstrated to be bDNA negative and Amplicor positive (100 to 800 IU/ml). A second group of samples was selected that had very high levels of HCV above the measuring range of the bDNA assay (>7.7 million IU/ml). For most comparison studies between methods, original sample results from the Amplicor or bDNA assays were compared to results obtained with the real-time assay after sample storage at −70°C for 3 to 24 months. In a few instances when results were discrepant and additional sample was available, either the real-time assay and/or bDNA assay or the Amplicor assay was repeated on the stored sample.
Sample extraction and RNA purification.
Initial studies were performed on samples extracted using the QIAamp viral RNA mini kit (QIAGEN, Valencia, Calif.). Initial sample volume was 140 ml, and elution volume was 60 ml (2.2× concentrated). The majority of samples were extracted with the Roche MagNA Pure LC instrument with the MagNA Pure LC total nucleic acid isolation kit, large volume, according to the manufacturer's instructions. Initial sample volume was 1.0 ml, and elution volume was 80 ml (12.5× concentrated). For all MagNA Pure LC extractions, as an extraction control the GeneAmplimer pAW 109 RNA primer sequencer (Applied Biosystems, Inc., Foster City, Calif.) was doped into the extraction buffer to make a solution containing 10,000 copies/ml. GeneAmplimer pAW 109 RNA primer sequencer is an RNA template transcribed from the plasmid pAW109. The quantity of GeneAmplimer pAW 109 RNA primer sequencer present in each sample at the end of extraction was approximately 600 copies/ml.
RT-PCR assay.
Purified RNA was amplified according to the manufacturer's recommendations with the EZ RT-PCR core reagents (Applied Biosystems) to measure HCV and an internal control.
(i) HCV assay.
Twenty microliters of RNA was added to 30 ml of master mix for the HCV amplification and detection reaction. Each HCV reaction was done in duplicate. The Mn concentration was 3 mM. Each of the three 5′ primers was at a final concentration of 360 nM, and the probes were each at 80 nM. A mixture of four different primers and two probes was used to amplify a region of the 5′ untranslated region (5′UTR) of HCV. Multiple primers and probes were necessary to ensure efficient amplification with all major U.S. genotypes. The primer set was originally designed by Glen Knight at the Lahey Clinic, Burlington, Mass. (unpublished data). The 5′ primers (spanning bp 19 to 41, 20 to 41, and 23 to 41, respectively, relative to the HCV-H strain) were GCGACACTCCACCATAGATCACT, CGACACTCCACCATGAATCACT, and CACTCCGCCATGAAYCACT (where Y = C or T). The 3′ primer (spanning bp 294 to 313 relative to the HCV-H strain) was CACTCGCAAGCACCCTATCA. The probes (spanning bp 256 to 281 and 256 to 280, respectively, relative to the HCV-H strain) were 6-carboxyfluorescein-AGGCCTTTCGCGACCCAACACTACTC-tetramethyl carboxyrhodamine (TAMRA) and 6-carboxyfluorescein-AGGCCTTTCGCAACCCAACGCTACT-tetramethyl carboxyrhodamine.
(ii) Internal control assay (GeneAmplimer pAW 109 RNA primer sequencer).
A single separate reaction well was used to determine extraction quality. Ten microliters of RNA was added to 40 ml of master mix. The 5′ primer GCCTGGGTTCCCTGTTCC and the 3′ primer CGACGTACCCCTGACATGG were each used at a final concentration of 1,080 nM, and the probe VIC-CAGGCCAATGTCTCACCAAGCTCTG-minor groove binder, nonfluorescent quencher was used at a final concentration of 480 nM. The HCV and GeneAmplimer pAW 109 RNA primer sequencer reactions were performed under identical conditions. The amplification reaction was carried out in an ABI 7700 instrument with the following cycles: 50°C for 2 min, 60°C for 30 min, 95°C for 2 min, then repeat cycling of 95°C for 15 s and then 60°C for 1 min for a total of 45 cycles. Analysis was performed with the ABI SDS software. The failure rate for the internal control reaction in clinical use was 0.4%.
Assay standards and controls.
The HCV standard consisted of the Con1/SG-Neo(I) plasmid, an HCV genotype 1b subgenomic replicon with an NS5a mutation (Apath, St. Louis, Mo.). It was grown and purified to yield stock solutions of DNA plasmid or RNA with concentrations of approximately 1012 copies/ml. DNA amplification was done with the TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.) according to the kit instructions. RNA transcription was done from the linearized DNA plasmid with the T7 MEGAshortscript high-yield transcription kit (Ambion Inc., Austin, Tex.). To determine the concentration of the RNA and DNA standards, dilution curves of each were run in parallel with AcroMetrix nucleic acid panel (NAP) HCV secondary standards (AcroMetrix, Benicia, Calif.) on five separate runs, and the quantities (in international units per milliliter) of RNA and DNA were calculated using the NAP HCV standards. Dilutions of the DNA or RNA were made to produce a standard curve equivalent to patient samples containing about 2 × 109, 2 × 106, 2,000, 200, and 20 IU/ml. All subsequent runs containing patient samples were done using the DNA plasmid as standard. For the GeneAmplimer pAW 109 RNA primer sequencer assay, a three-point standard curve was made from dilutions of the stock, with standard curve points of approximately 100, 1,000, and 10,000 copies/ml. Samples which were less than 25% of the mean GeneAmplimer pAW 109 RNA primer sequencer quantity for each run were considered failed extractions and were repeated.
To monitor assay stability over time, two positive control samples were included with each clinical run. Each consisted of pooled patient serum specimens. The high serum pool control had a mean HCV concentration of 1.17 × 106 IU/ml, and the low serum pool control had a mean of 422 IU/ml.
RESULTS
Performance characteristics of the HCV real-time RT-PCR assay. (i) Selection of standard curve material.
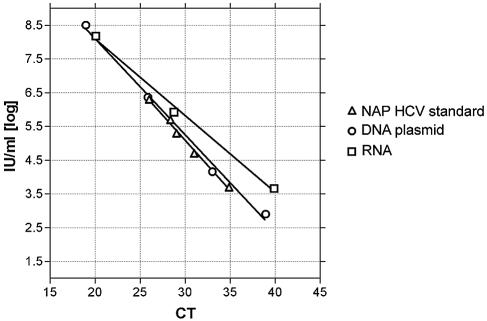
Because of the expense of running the AcroMetrix secondary standards, we produced both RNA and a DNA plasmid containing the appropriate region of the 5′UTR of the virus. Serial dilutions of both plasmids were run in parallel with RNA extracted from the NAP standard curve materials. The DNA plasmid produced a standard curve with a slope more similar to that of the NAP materials than did the RNA (Fig. 1). Based on these results, we determined to use the DNA plasmid as the standard for all subsequent experiments. Assigned quantities for each standard point were determined from means of five different runs, where the DNA quantity (in international units per milliliter) was calculated from the NAP standard curve.
FIG. 1.
Standard curves generated from the commercial NAP standard, plasmid DNA, or in vitro-transcribed RNA. Serial dilutions of plasmid DNA or in vitro-transcribed RNA were run in parallel with serial dilutions of the commercial NAP standard. Shown is the threshold cycle (CT) for the indicated amounts of input nucleic acid.
(ii) Reproducibility of automated RNA extraction.
Several systems are now available that allow automation of the extraction process for viral RNA. Automated extraction is attractive because it increases laboratory throughput and lessens the likelihood of repetitive stress injury. Additionally, automated extractions may be expected to have increased reproducibility compared to manual extractions. We chose to evaluate the MagNA Pure LC automated extraction system (Roche), because it allows extraction of relatively large amounts of specimen (up to 1 ml), which should improve the sensitivity of our assay. We therefore evaluated the reproducibility of automated extractions with the MagNA Pure LC system relative to that of manual extractions with manual QIAGEN columns. Internal control GeneAmplimer pAW 109 RNA primer sequencer RNA was exogenously added to clinical specimens. After extraction, the GeneAmplimer pAW 109 RNA primer sequencer was amplified and detected by real-time RT-PCR. The CV for threshold cycle values (Ct) was very small for both methods, averaging 0.9% for manual extractions and ranging from 0.9 to 1.33% for MagNA Pure LC extractions (Table 1). The CV for the absolute GeneAmplimer pAW 109 RNA primer sequencer quantitative result ranged from 22.6 to 30.4% for the manual extraction and 21.5 to 27.8% for the MagNA Pure LC extraction. Thus, automated extraction of HCV RNA using the MagNA Pure LC system is at least as reproducible as manual extraction by highly experienced technologists.
TABLE 1.
Extraction efficiency and yield for a spiked internal control (GeneAmplimer pAW 109 RNA primer sequencer)
| Extraction method | No. of runs, (no. of samples) | CV of actual Ct values (%) | CV of quantitative result (%) | Yield (%) |
|---|---|---|---|---|
| Manual Qiagen column | 2 (10 each) | 0.9 | 22.4 | 60.4 |
| 0.9 | 30.4 | |||
| MagNA Pure instrument | 2 (32 and 24) | 1.3 | 27.8 | 59.2 |
| 0.9 | 21.5 |
(iii) Sensitivity.
We next sought to determine the sensitivity of our assay for the detection of very low levels of HCV RNA. The assay routinely detects better than 99% of the time the 28-IU/ml DNA standard, run in duplicate in each assay. However, since this standard is DNA, it is not taken through the extraction process or the RT step. To establish the sensitivity of patient samples taken through the entire test process, we did testing in two ways. First, we ran the NAP 100 and the 95-IU/ml sample pool in duplicate (76 wells) over 38 runs. All 76 wells were positive. Second, we used the NAP 50 standard, the 95-IU/ml sample pool, and dilutions of the 95-IU/ml sample pool to 71 and 47.5 IU/ml. All four sample types were extracted in multiples and run by RT-PCR in multiples to determine the positive rate. All assays (6 for the NAP 50 and 12 each for the 95-, 71-, and 47.5-IU/ml samples) were positive. Thus, our assay is extremely sensitive with a 100% detection rate down to at least 50 IU/ml.
(iv) Extraction linearity at high viral loads.
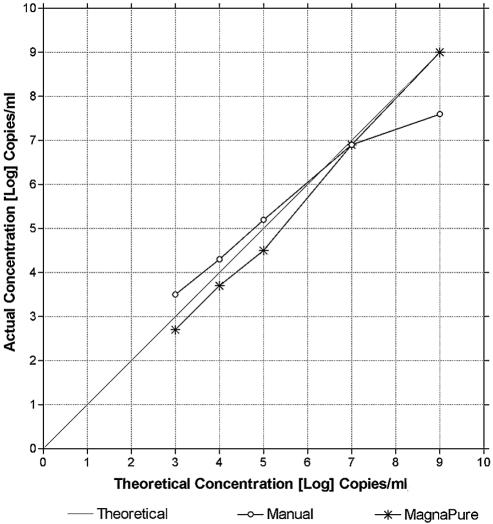
Some extraction methods show loss of linearity at high amounts of input nucleic acid, as the binding capacity of the capture matrix is exceeded. Since the levels of HCV RNA in infected patients can vary widely (by 9 logs or more), we evaluated the linearity of our extraction procedures through this dynamic range. Dilutions of the RNA standard into normal serum (lyophilized normal human serum; Bio-Rad, Hercules, Calif.) were made, and then each dilution was extracted with either the MagNA Pure or the manual QIAGEN method. Similar extraction efficiencies were seen at the lower concentrations, but at the high end the manual method gave a lower yield than the MagNA pure method (Fig. 2). Overall, the MagNA Pure LC extraction system showed very good linearity throughout the tested range (102 to 109 IU/ml). The manual extraction in general also performed well, except for an approximately 1.5 log underestimation of viral quantity at the upper end, i.e., 109 IU/ml of input HCV RNA (Fig. 2). We conclude that the MagNA Pure LC system shows good linearity of extraction efficiency throughout the clinically relevant measuring range, and it may be superior to manual extraction with the QIAGEN QIAamp viral RNA mini kit at high levels of input HCV RNA.
FIG. 2.
Comparison of extraction linearity for HCV RNA with the QIAGEN manual method versus the MagNA Pure LC automated extraction system. The RNA standard was serially diluted and then extracted by either the QIAGEN manual method or the MagNA Pure LC automated extraction system. Shown is the theoretical quantity based on the dilution factor (x axis) versus the actual real-time RT-PCR result for each dilution (y axis).
(v) Effect of viral genotype on assay linearity.
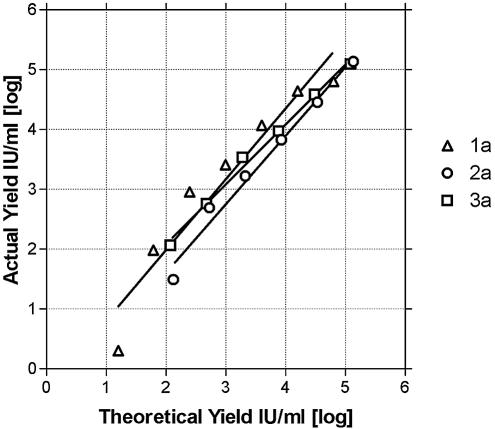
HCV has multiple genotypes, and the RNA sequences of these genotypes can vary in the 5′UTR amplified in our assay. To allow detection of these various sequences, we used a cocktail of three 5′ primers, one 3′ primer, and two TaqMan probes for our assay. We therefore sought to determine whether our assay would in fact detect the HCV genotypes relevant to our patient population and whether these genotypes would amplify with similar efficiencies, thus allowing accurate quantitation. To address this issue, three patient samples, one each of the 1a, 2a, and 3a genotypes, each of which had >5,000,000 IU/ml HCV, were serially 40-fold diluted into normal serum, and then each dilution was extracted with the MagNA Pure system. For all three genotypes, the experimental result closely matched the expected result throughout the measured range (Fig. 3), and the slope of the regression line was very similar for all three genotypes (R2 = 1.19, 1.14, and 0.99). We conclude that our assay detects genotypes 1a, 2a, and 3a with equal efficiency and allows accurate quantitation of viral load.
FIG. 3.
Linearity of yield with RNA dilutions isolated from genotype 1a, 2a, and 3a samples. A sample from each genotype was serially diluted, extracted using the MagNA Pure LC instrument, and subjected to real-time RT-PCR. Shown is the theoretical quantity based on the dilution factor (x axis) versus the actual real-time RT-PCR result from each dilution (y axis).
Clinical performance of HCV real-time RT-PCR. (i) Comparison of real-time RT-PCR versus bDNA for HCV.
We next evaluated the performance of our real-time RT-PCR assay by using actual clinical specimens submitted for HCV RNA testing. A total of 486 patient samples previously tested with Bayer VERSANT HCV RNA 3.0 or Roche Amplicor were analyzed, including negative samples and a range of positive samples with an approximately equal mix of genotypes 1a, 1b, 2a, 2b, and 3a. The samples were extracted either manually or by using the MagNA Pure LC instrument. A total of 325 samples positive for HCV RNA by bDNA were tested, of which 323 were positive with our assay (Table 2). Of the 157 samples reported as negative by the ultrasensitive (but qualitative) Amplicor assay, all were negative by our real-time RT-PCR assay as well. Four specimens were available that were bDNA negative but positive by the qualitative Amplicor assay, presumably because they were near or below the lower limit of detection of the bDNA assay (280, 350, 821, and 2,083 IU/ml). All four of these were clearly positive by our real-time RT-PCR assay. Two samples were weakly positive by bDNA (1,564 and 7,787 IU/ml) but negative by our real-time RT-PCR assay. The bDNA 3.0 assay has been reported to have a specificity of 95.9 to 98.2% (4, 16, 26), and so given that we tested 325 bDNA-positive specimens, several false positives would be expected. Unfortunately, insufficient material was available for further studies on these discrepant samples.
TABLE 2.
Summary of patient results with TaqMan RT-PCR versus bDNA and Amplicor
| Comparison test(s) and result(s) | No. of patients tested by Taqman that were:
|
|
|---|---|---|
| Positive | Negative | |
| bDNA positive | 323 | 2 |
| Amplicor negative | 0 | 157 |
| bDNA negative, Roche Amplicor positive | 4 | |
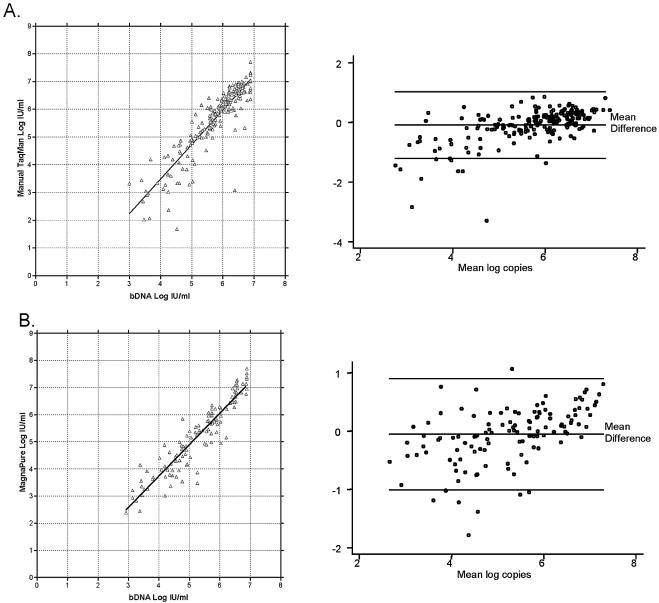
We then compared the levels of HCV RNA determined by our assay with the HCV RNA levels determined by bDNA. In general our real-time RT-PCR assay agreed well with the bDNA assay, whether performed on specimens extracted manually (Fig. 4A) (n = 199; slope = 1.16; R2 = 0.808) or using the MagNA Pure LC system (Fig. 4B) (n = 124; slope = 1.08; R2 = 0.871). Approximately 77% of specimens showed agreement of viral load within 0.5 log, as determined by the real-time RT-PCR versus bDNA methods, and approximately 95% agreed within 1 log. We also identified nine genotype 4 samples and three genotype 6 samples which had been previously evaluated by our genotyping assay. These genotypes are rare in our patient population, and only two of the samples (both genotype 4) had sufficient volume to perform both RT-PCR and bDNA. All 12 genotype 4 or 6 samples were amplified and detected by our RT-PCR assay. For the two samples that could be tested by both methods, the RT-PCR and bDNA assays agreed within 0.4 to 0.9 log. Thus, our assay provides quantitative results for HCV RNA comparable to that of the bDNA assay, but with the advantage of sensitivity to 50 IU/ml or less, compared to 615 IU/ml for the bDNA assay.
FIG. 4.
Comparison of quantitation of HCV RNA in clinical specimens by real-time RT-PCR versus bDNA. (A) A total of 199 clinical specimens previously found to be HCV positive by bDNA were subjected to manual QIAGEN RNA extraction followed by real-time RT-PCR. Shown in the left panel is the comparison of the viral quantitation by each method. The right panel is an agreement plot, with the x axis representing the quantity of virus by RT-PCR and the y axis representing the difference in quantity by the two methods [(log copy by RT-PCR) − (log copy by bDNA)]. Reference lines represent the mean difference and 95% agreement limits. (B) A total of 124 clinical specimens previously found to be HCV positive by bDNA were subjected to automated RNA extraction using the MagNA Pure LC instrument, followed by real-time RT-PCR. Shown in the left panel is a comparison of the viral quantitation by each method. The right panel is an agreement plot, with the x axis representing the quantity of virus by RT-PCR and the y axis representing the difference in quantity by the two methods [(log copy by RT-PCR) − (log copy by bDNA)]. Reference lines represent the mean difference and 95% agreement limits.
(ii) Reproducibility of real-time RT-PCR in clinical use.
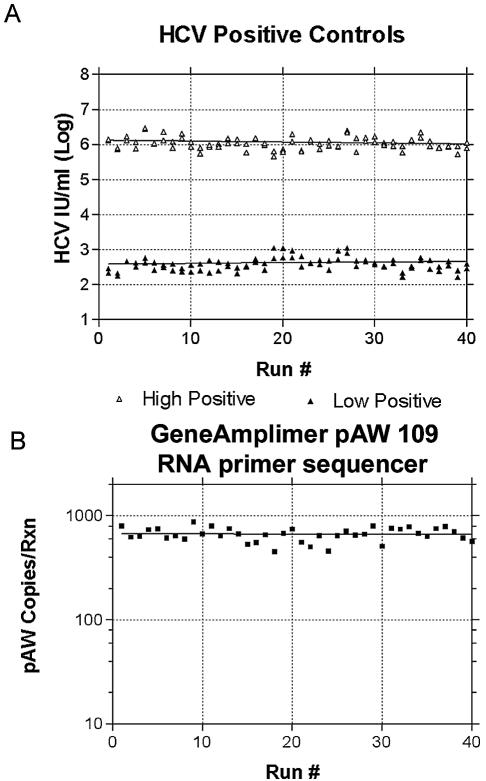
In 40 consecutive runs containing clinical patients, run by two different technologists, the high serum pool control (mean, 1.17 × 106) had a CV for the logarithmic value of 2.8% (the CV of the non-log-transformed value was 43.3%), and the low serum pool control (mean, 422) had a CV for the logarithmic value of 7.0% (the CV of the non-log-transformed value was 48.3%) (Fig. 5A). The GeneAmplimer pAW 109 RNA primer sequencer internal control (mean, 664 copies/ml) had a CV for the logarithmic value of 2.4% (the CV of the non-log-transformed value was 14.9%) (Fig. 5B). Out of 1,280 extractions on the 40 runs, there were 5 failed extractions in which the GeneAmplimer pAW 109 RNA primer sequencer result was less than 25% of the GeneAmplimer pAW 109 RNA primer sequencer mean for that run, for an overall failure rate of 0.4%. Based on these results, the real-time RT-PCR assay was demonstrated to be extremely reproducible during routine use in the clinical laboratory.
FIG. 5.
Reproducibility of real-time RT-PCR for HCV RNA in clinical use. Shown are the real-time RT-PCR results for the high and low positive controls (A) and the GeneAmplimer pAW 109 RNA primer sequencer internal control (B) over 40 consecutive clinical runs.
DISCUSSION
Testing for HCV RNA is an important part of the clinical care for HCV-infected patients. RT-PCR testing is indicated as follow-up to the detection of antibody directed to HCV, both as a confirmatory test and to differentiate active viremia from resolved infections. Quantitative determination of viral load is an important part of the pretreatment evaluation of patients with HCV infection. Low pretreatment HCV RNA levels are a predictor of sustained response to therapy (21, 24). An early viral response, defined as a minimum 2-log decrease in viral load during the first 12 to 24 weeks of treatment, is predictive of a sustained virologic response. Patients who fail to achieve an early viral response by 24 weeks have only a small chance of achieving a sustained viral response even if treated for 1 year (2). Treatment may be discontinued for such patients, or alternative therapies may be considered. Most patients who are HCV RNA negative 6 months after completion of therapy will remain negative indefinitely (11).
Before the development of our real-time RT-PCR assay, our laboratory used the Bayer Versant bDNA 3.0 assay for quantitation of HCV RNA. This assay shows good intra- and interassay CVs (26). However, the bDNA assay is limited by relatively poor sensitivity, with a lower limit of detection of 615 IU/ml. Therefore, we previously used the Roche Amplicor version 2.0 system for detection of low-level viremia. This assay has a much better lower limit of detection (100 IU/ml), but it does not provide quantitative results. While the combination of the bDNA and Amplicor assays allowed us to detect specimens containing 100 or more IU of HCV RNA/ml and to quantitate those above 615 IU/ml, it was relatively inefficient for us to perform, since any specimens testing negative by bDNA would need follow-up testing by the Amplicor assay. For those specimens subsequently positive by the Amplicor assay, the result would be delayed until the next day after the initial bDNA test. Thus, the goal in the development of our assay was to replace the combination of the bDNA and Amplicor tests with a convenient single quantitative assay having a lower limit of detection of 100 IU/ml or better.
Several groups have reported the use of quantitative real-time RT-PCR assays for HCV. Many of the early real-time RT-PCR assays were less sensitive than the second-generation Roche Amplicor assay (lower limit of detection, 100 IU/ml), but comparison is difficult for assays described before adoption of the standardized international unit (5, 8, 10, 13, 17, 18, 23, 25, 27, 28). However, more recent real-time RT-PCR assays have achieved sensitivities of approximately 100 IU/ml or better (9, 14, 19, 20). While sensitive and accurate, these assays may not be ideal for laboratories performing large test volumes, due to the necessity of manual RNA extraction of specimens. Manual extraction by less-experienced technologists may also increase inaccuracy in quantitative results, due to inter- and intraoperator variations in the extraction process.
To address the issues of efficient specimen throughput and reproducibility, we developed a fully automated real-time RT-PCR assay for HCV, in which the extraction process was performed by the MagNA Pure LC robotic extraction system. Mitsunaga et al. previously reported automated extraction for HCV RT-PCR using the QIAGEN 9604 Biorobot (15). This assay was developed for nucleic acid testing of blood products, rather than for diagnostic use on patient samples. It reportedly was able to detect 87% of an in-house-prepared positive control specimen containing 100 HCV genome equivalents/ml. In our hands, extraction via the MagNA Pure LC allowed an approximately 5.8-fold increase in sensitivity of the assay compared to the manual QIAGEN spin column extractions or the QIAGEN 9604 Biorobot (data not shown). This finding is not surprising, given the larger volume of material that can be extracted by the MagNA Pure LC (1 ml, versus 0.2 ml for the Biorobot). Similar results were reported by Germer et al., who found a threefold improvement in the COBAS Amplicor HCV test version 2.0 with the 1-ml MagNA Pure LC extraction compared to MagNA Pure LC or manual extractions using only 0.2 ml (7).
Our automated assay has proven to be highly sensitive (lower limit of detection of less than 50 IU/ml) and reproducible. In clinical use the assay showed a CV for the logarithmic results of 2.8% at the high end and 7.0% at the low end, despite the test being performed by four different individuals on two different TaqMan 7700 instruments. Implementation of the new assay has decreased the testing volume in the laboratory by about 15% due to the reduction in the amount of follow-up testing (usually a quantitative test followed by a more sensitive qualitative test, or sometimes the reverse). In addition, the new assay is more efficient. The newer test requires about 60 h of technologist time per week compared to the 90+ h per week required by our old test. Anecdotally, automation of the extraction process has had the additional benefit of reducing the risk of repetitive use injuries.
In summary, our laboratory has developed a fully automated real-time RT-PCR assay for the quantification of HCV RNA. The assay is faster and less expensive than commercial assays and is at least as sensitive. Because the extraction process is fully automated, technologist manipulation of the specimen is minimized, resulting in improved reproducibility and improved throughput. As the systems continue to improve, it is likely that automated extraction will become standard practice for clinical PCR-based assays.
Acknowledgments
We thank Elizabeth Krantz for help with the agreement plots shown in Fig. 4.
REFERENCES
- 1.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 2.Anonymous. 2002. National Institutes of Health Consensus Development Conference statement: management of hepatitis C 2002 (June 10-12, 2002). Gastroenterology 123:2082-2099. [DOI] [PubMed] [Google Scholar]
- 3.Baker, D. E. 2003. Pegylated interferon plus ribavirin for the treatment of chronic hepatitis C. Rev. Gastroenterol. Disord. 3:93-109. [PubMed] [Google Scholar]
- 4.Beld, M., R. Sentjens, S. Rebers, C. Weegink, J. Weel, C. Sol, and R. Boom. 2002. Performance of the new Bayer VERSANT HCV RNA 3.0 assay for quantitation of hepatitis C virus RNA in plasma and serum: conversion to international units and comparison with the Roche COBAS Amplicor HCV Monitor, version 2.0, assay. J. Clin. Microbiol. 40:788-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enomoto, M., S. Nishiguchi, S. Shiomi, M. Tanaka, K. Fukuda, T. Ueda, A. Tamori, D. Habu, T. Takeda, Y. Yano, and S. Otani. 2001. Comparison of real-time quantitative polymerase chain reaction with three other assays for quantitation of hepatitis C virus. J. Gastroenterol. Hepatol. 16:904-909. [DOI] [PubMed] [Google Scholar]
- 6.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 7.Germer, J. J., M. M. Lins, M. E. Jensen, W. S. Harmsen, D. M. Ilstrup, P. S. Mitchell, F. R. Cockerill III, and R. Patel. 2003. Evaluation of the MagNA Pure LC instrument for extraction of hepatitis C virus RNA for the COBAS AMPLICOR hepatitis C virus test, version 2.0. J. Clin. Microbiol. 41:3503-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawai, S., O. Yokosuka, T. Kanda, F. Imazeki, Y. Maru, and H. Saisho. 1999. Quantification of hepatitis C virus by TaqMan PCR: comparison with HCV Amplicor Monitor assay. J. Med. Virol. 58:121-126. [DOI] [PubMed] [Google Scholar]
- 9.Kleiber, J., T. Walter, G. Haberhausen, S. Tsang, R. Babiel, and M. Rosenstraus. 2000. Performance characteristics of a quantitative, homogeneous TaqMan RT-PCR test for HCV RNA. J. Mol. Diagn. 2:158-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komurian-Pradel, F., G. Paranhos-Baccala, M. Sodoyer, P. Chevallier, B. Mandrand, V. Lotteau, and P. Andre. 2001. Quantitation of HCV RNA using real-time PCR and fluorimetry. J. Virol. Methods 95:111-119. [DOI] [PubMed] [Google Scholar]
- 11.Lau, D. T., D. E. Kleiner, M. G. Ghany, Y. Park, P. Schmid, and J. H. Hoofnagle. 1998. 10-year follow-up after interferon-alpha therapy for chronic hepatitis C. Hepatology 28:1121-1127. [DOI] [PubMed] [Google Scholar]
- 12.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 13.Martell, M., J. Gomez, J. I. Esteban, S. Sauleda, J. Quer, B. Cabot, R. Esteban, and J. Guardia. 1999. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J. Clin. Microbiol. 37:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng, Q., C. Wong, A. Rangachari, S. Tamatsukuri, M. Sasaki, E. Fiss, L. Cheng, T. Ramankutty, D. Clarke, H. Yawata, Y. Sakakura, T. Hirose, and C. Impraim. 2001. Automated multiplex assay system for simultaneous detection of hepatitis B virus DNA, hepatitis C virus RNA, and human immunodeficiency virus type 1 RNA. J. Clin. Microbiol. 39:2937-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitsunaga, S., K. Fujimura, C. Matsumoto, R. Shiozawa, S. Hirakawa, K. Nakajima, K. Tadokoro, and T. Juji. 2002. High-throughput HBV DNA and HCV RNA detection system using a nucleic acid purification robot and real-time detection PCR: its application to analysis of posttransfusion hepatitis. Transfusion 42:100-106. [DOI] [PubMed] [Google Scholar]
- 16.Morishima, C., M. Chung, K. W. Ng, D. J. Brambilla, and D. R. Gretch. 2004. Strengths and limitations of commercial tests for hepatitis C virus RNA quantification. J. Clin. Microbiol. 42:421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris, T., B. Robertson, and M. Gallagher. 1996. Rapid reverse transcription-PCR detection of hepatitis C virus RNA in serum by using the TaqMan fluorogenic detection system. J. Clin. Microbiol. 34:2933-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nozaki, A., and N. Kato. 2002. Quantitative method of intracellular hepatitis C virus RNA using LightCycler PCR. Acta Med. Okayama 56:107-110. [DOI] [PubMed] [Google Scholar]
- 19.Puig, M., K. Mihalik, M. Y. Yu, S. M. Feinstone, and M. E. Major. 2002. Sensitivity and reproducibility of HCV quantitation in chimpanzee sera using TaqMan real-time PCR assay. J. Virol. Methods 105:253-263. [DOI] [PubMed] [Google Scholar]
- 20.Ratge, D., B. Scheiblhuber, O. Landt, J. Berg, and C. Knabbe. 2002. Two-round rapid-cycle RT-PCR in single closed capillaries increases the sensitivity of HCV RNA detection and avoids amplicon carry-over. J. Clin. Virol. 24:161-172. [DOI] [PubMed] [Google Scholar]
- 21.Reichard, O., G. Norkrans, A. Fryden, J. H. Braconier, A. Sonnerborg, and O. Weiland. 1998. Comparison of 3 quantitative HCV RNA assays—accuracy of baseline viral load to predict treatment outcome in chronic hepatitis C. Scand. J. Infect. Dis. 30:441-446. [DOI] [PubMed] [Google Scholar]
- 22.Saldanha, J., A. Heath, N. Lelie, G. Pisani, M. Nubling, M. Yu, et al. 2000. Calibration of HCV working reagents for NAT assays against the HCV international standard. Vox Sang. 78:217-224. [DOI] [PubMed] [Google Scholar]
- 23.Schroter, M., B. Zollner, P. Schafer, R. Laufs, and H. H. Feucht. 2001. Quantitative detection of hepatitis C virus RNA by LightCycler PCR and comparison with two different PCR assays. J. Clin. Microbiol. 39:765-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shiratori, Y., N. Kato, H. Yoshida, R. Nakata, M. Ihori, F. Imazeki, O. Yokosuka, T. Kawase, T. Katamoto, T. Unuma, A. Nakamura, F. Ikegami, K. Hirota, and M. Omata. 2000. Sustained viral response is rarely achieved in patients with high viral load of HCV RNA by excessive interferon therapy. Dig. Dis. Sci. 45:565-574. [DOI] [PubMed] [Google Scholar]
- 25.Takeuchi, T., A. Katsume, T. Tanaka, A. Abe, K. Inoue, K. Tsukiyama-Kohara, R. Kawaguchi, S. Tanaka, and M. Kohara. 1999. Real-time detection system for quantification of hepatitis C virus genome. Gastroenterology 116:636-642. [DOI] [PubMed] [Google Scholar]
- 26.Trimoulet, P., P. Halfon, E. Pohier, H. Khiri, G. Chene, and H. Fleury. 2002. Evaluation of the VERSANT HCV RNA 3.0 assay for quantification of hepatitis C virus RNA in serum. J. Clin. Microbiol. 40:2031-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White, P. A., Y. Pan, A. J. Freeman, G. Marinos, R. A. Ffrench, A. R. Lloyd, and W. D. Rawlinson. 2002. Quantification of hepatitis C virus in human liver and serum samples by using LightCycler reverse transcriptase PCR. J. Clin. Microbiol. 40:4346-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang, J. H., J. P. Lai, S. D. Douglas, D. Metzger, X. H. Zhu, and W. Z. Ho. 2002. Real-time RT-PCR for quantitation of hepatitis C virus RNA. J. Virol. Methods 102:119-128. [DOI] [PubMed] [Google Scholar]