Abstract
Gram-negative bacteria of the Burkholderia cepacia complex (Bcc) are opportunistic pathogens that can infect the lungs of cystic fibrosis (CF) patients and can be transmitted among these patients, causing epidemics in the CF community. Lipopolysaccharide (LPS) is an important virulence factor of many gram-negative bacteria, with the O antigen component of LPS being responsible for serotype specificity. The goal of this work was to develop a genetic method of determining the serotype of Bcc isolates based on the conserved gene wbiI. Homologues of wbiI are found in polysaccharide biosynthesis gene clusters in other bacteria. Primers to a conserved region of the Bcc wbiI gene were able to amplify by PCR a single product in 67 of 80 Bcc isolates tested. Sequencing and restriction enzyme digestion of this wbiI PCR product revealed sufficient DNA polymorphisms to distinguish and group various isolates. In five of nine instances, Bcc isolates of a single serotype had a single wbiI restriction fragment length polymorphism (RFLP) pattern, while isolates of the other four serotypes could have multiple wbiI RFLP types. Species determination of the Bcc isolates revealed no obvious correlation between wbiI RFLP type and species. There was also no apparent correlation between wbiI RFLP type and the ability of a single Bcc isolate to infect an individual with CF. However three of five Bcc outbreaks involved isolates with the same wbiI RFLP type, indicating that wbiI RFLP typing may be a useful tool to help track Bcc outbreaks.
The Burkholderia cepacia complex (Bcc) is a group of phenotypically similar, genetically distinct gram-negative bacteria that can infect the lungs of cystic fibrosis (CF) patients. Infection can result in asymptomatic carriage, a slow progressive decline in lung function, or cepacia syndrome, which is characterized by bacteremia, septicemia, and eventually death (16). Bcc is an increasing problem in the CF community, as certain strains can be transmitted among CF patients, causing epidemics within the community (1, 6, 27). Isolates of the ET12 and PHDC lineages, for example, were recovered from outbreaks involving CF patients from western Europe and Canada (ET12) and the mid-Atlantic region of the United States (PHDC) (3, 27).
The Bcc comprises at least nine taxonomically distinct species (previously described as genomovars) that can be distinguished based on 16S ribosomal DNA (rDNA) and recA gene sequences (8, 29). Most CF patients are infected with B. cenocepacia (genomovar III) or B. multivorans (genomovar II) (18); the ET12 and PHDC lineage isolates are B. cenocepacia (3, 30). However, all Bcc species have been isolated from infected CF patients (31). Other markers of infectivity in the Bcc, such as the genomovar, are not fully indicative of the ability of a Bcc strain to cause infection in CF patients (4, 18).
Lipopolysaccharide (LPS), which is composed of lipid A, core oligosaccharide, and O antigen, is a virulence factor in many gram-negative bacteria. The O antigen is a prominent and immunogenic component of the bacterial cell surface and thus can be the basis of bacterial serotyping. However, serological cross-reactivity among bacteria with different O antigen structures can occur. There are at least 16 different serotypes of Bcc, and most of the O antigen structures of these Bcc serotype strains are known (32). Due to lack of available sera for Bcc and the ease of genotyping, serotyping is no longer a commonly used typing method for these bacteria.
In an effort to identify a reliable typing system for Bcc strains, Rabkin et al. analyzed Bcc isolates from seven hospital outbreaks by various typing methods, including serotyping, chromosome analysis, and antimicrobial susceptibility testing (22). No correlation between serotype of a particular Bcc isolate and the ability to cause infection in humans was established in this study. However, very few, if any, Bcc isolates from the lungs of CF patients were used in this study; thus no conclusions can be made concerning the relationship between serotype and infection in CF patients (22).
Recently, it was observed that the serotypes of Bcc isolates and the species do not correlate with one another. Kenna et al. (15) determined the species of many of the Bcc isolates previously serotyped by Rabkin et al. (22). The conclusion that species and serotype do not correlate agreed with our studies using a smaller number of isolates of known serotypes (A. D. Vinion-Dubiel, T. Spilker, C. R. Dean, J. J. LiPuma, and J. B. Goldberg, Abstr. Int. Burkholderia cepacia Work. Group 7th Annu. Meet., abstr. 11, 2002).
O antigens are generally synthesized by enzymes encoded by genes in a single chromosomal location. At the 3′ end of O antigen gene loci in Pseudomonas aeruginosa, another bacterium responsible for CF lung infections, there is a conserved gene, wbpM, that is required for O antigen production (2, 11, 24). In the present study, we describe a genetic method to predict the serotype of a Bcc isolate based on restriction fragment length polymorphism (RFLP) of wbiI, the wbpM homologue in Bcc.
MATERIALS AND METHODS
Bacterial isolates.
Bcc serotype strains (for serotypes O1 to O9) were obtained from The Institute of Bacteriology in Strasbourg, France. All other Bcc isolates, including those previously used by Rabkin et al. (22), are part of the collection at the Cystic Fibrosis Foundation Burkholderia cepacia Research Laboratory and Repository at the University of Michigan. Bcc was routinely grown on Trypticase soy agar (Remel, Lenexa, Kans.) at 30 or 32°C.
PCR amplification of wbiI.
Genomic DNA templates for PCR were made as described previously (7). Briefly, 20 μl of lysis buffer (0.05 M NaOH, 0.25% sodium dodecyl sulfate [SDS]) was inoculated with a single bacterial colony and heated at 95°C for 15 min. Sterile deionized water was added to a final volume of 200 μl, and cell debris was removed by centrifugation. For a 25-μl PCR mixture, 2 μl of the supernatant was used. A portion of the wbiI gene from Bcc isolates was amplified by PCR with 0.4 μM primers Mconfor (5′CCATGCGGCCGCGTACAAGCACGTGCC3′) and Mconrev (5′TAGAGCTTCTCGCCCGGGCGCAG3′). The 25-μl PCR mixture was composed of 250 nM deoxynucleoside triphosphate, 1× QIAGEN (Valencia, Calif.) PCR buffer, 1 mM MgCl2, 2 U of Taq polymerase, and 1× Q solution (QIAGEN). PCR was performed with a GeneAmp PCR System 2400 (Perkin-Elmer, Boston, Mass.); conditions were as follows: 97°C for 5 min and then 30 cycles of 95°C for 45 s, 52°C for 30 s, and 72°C for 1 min, followed by 72°C for 5 min. PCR products were purified with QIAGEN purification kits.
RFLP analysis.
PCR products were digested with restriction enzyme BglI or MboII (2 U/reaction; New England Biolabs, Beverly, Mass.) for 2 h at 37°C in appropriate buffers. DNA fragments were separated on a 3% agarose gel and stained with ethidium bromide.
Sequence analysis.
Sequences of wbiI PCR fragments were determined by the Biomolecular Research Facility at the University of Virginia using primers Mconfor and Mconrev. Genes with similarity to wbiI from B. cenocepacia J2315 were obtained by searching GenBank with BLASTX (www.ncbi.nlm.nih.gov/BLAST). Nucleotide sequences were analyzed with SeqWeb software programs BESTFIT, PILEUP, and PRETTY (Accelrys, Inc.).
Determination of Bcc species.
The species of Bcc isolates were determined by PCR analysis of the recA and 16S rDNA genes (17, 19) or by determining the sequence of the recA gene, as described previously (19).
Determination of serotype.
Anti-Bcc sera for serotype O1 to O9 strains were described previously (13, 33). Crude LPS preparations from Bcc isolates were obtained by lysing bacteria in SDS-LPS buffer (2% β-mercaptoethanol, 2% SDS, 5% glycerol, 0.05 M Tris [pH 6.8], 0.002% bromophenol blue) and heating at 95°C for 15 min. The lysed bacteria were then treated with proteinase K for 1 h at 60°C. Twelve to 20 μl of this crude LPS preparation was separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes. The membranes were immunoblotted with a 1:1,000 dilution of anti-Bcc sera, followed with an anti-rabbit monoclonal antibody conjugated to alkaline phosphatase (Sigma, St. Louis, Mo.) and detected with BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium (Sigma). LPS was purified from Bcc isolates by the method described by Dean et al. (10), separated by SDS-PAGE, and silver stained as described by Tsai and Frasch (28).
Nucleotide sequence accession numbers.
The wbiI PCR product sequences for the Bcc serotype O1 to O9 strains are deposited in the GenBank database at the National Center for Biotechnology Information under accession numbers AY523830 to AY523838, respectively.
RESULTS
wbiI gene of Bcc.
Genes similar to P. aeruginosa wbpM can be found among polysaccharide biosynthesis gene clusters from a variety of bacteria, including Vibrio cholerae and Staphylococcus aureus (2). A search of the B. cenocepacia strain J2315 genome, currently being sequenced by the Wellcome Trust SangerInstitute (http://www.sanger.ac.uk/Projects/B_cenocepacia/),revealed a single gene of 1,881 nucleotides (nt) on chromosome 1 with similarity to wbpM from P. aeruginosa PAO1 (67.8% nucleotide identity). The GenBank protein database was used to identify homologues to the amino acid translation of the J2315 gene. WbiI proteins from B. mallei and B. pseudomallei were most similar, with the corresponding genes having 83.9% nucleotide identity with the J2315 gene. Due to its similarity to other Burkholderia species genes, the B. cenocepacia J2315 gene was designated wbiI.
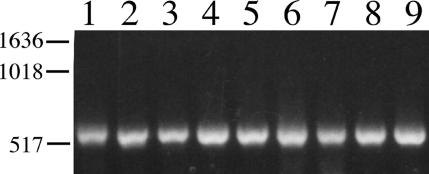
As wbpM is conserved among P. aeruginosa strains of different serotypes (24), it was of interest to determine if wbiI was likewise conserved among Bcc isolates of various serotypes. Primers Mconfor and Mconrev, which would ideally amplify by PCR a 3′ conserved region in wbiI and its homologues, were designed (Fig. 1). Indeed, these primers were able to amplify by PCR a single product in P. aeruginosa strains (data not shown). An approximately 550-nt fragment was likewise amplified from the nine Bcc serotype strains (Fig. 2), indicating that wbiI is conserved among Bcc strains of various serotypes.
FIG. 1.
Alignment of J2315 wbiI homologues. Numbers at the ends of each sequence represent nucleotide designations in the whole gene. The consensus sequence is shown on the next-to-last line. Underlined sequences were used to design primers Mconfor and Mconrev, shown on the bottom line. Homologues used are as follows: wbiI from J2315 (www.sanger.ac.uk), wbiI from Burkholderia pseudomallei (AF064070), wbpM from P. aeruginosa PAO1 (www.pseudomonas.com), cap5D from S. aureus (U81973), wbfY from V. cholerae (AB012957), flaA1 HP0804 from Helicobacter pylori 26695 (AE000595), cap8E from S. aureus (U73374).
FIG. 2.
PCR products from Bcc serotype O1 to O9 strains obtained with primers Mconfor and Mconrev to a conserved region of the wbiI gene. DNA size markers in base pairs are on the left. Lane 1, CIP 8235 (serotype O1); lane 2, CIP 8236 (O2); lane 3, CIP 8237 (O3); lane 4, CIP 8238 (O4); lane 5, CIP 8239 (O5); lane 6, CIP 8240 (O6); lane 7, ATCC 17759 (O7); lane 8, CDC99 (O8); lane 9, CDC86 (O9).
RFLP of Bcc wbiI.
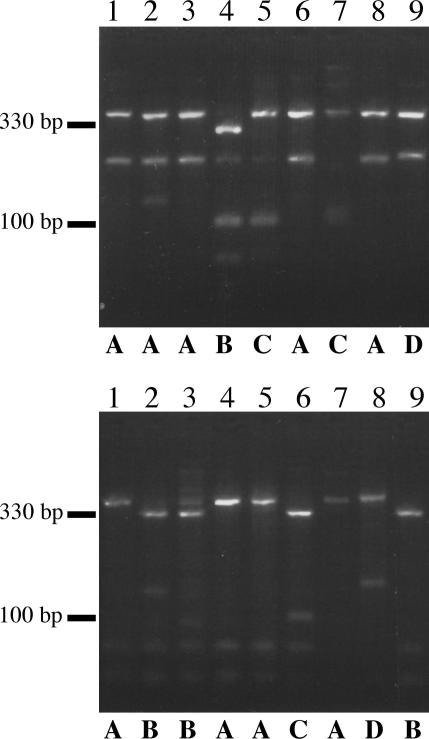
Sequencing of the wbiI PCR products from the Bcc O1 to O9 serotype strains revealed that the 542-nt products were, on average, 91% identical, with no insertions or deletions. There were, however, some single-nucleotide polymorphisms that affected restriction enzyme recognition sites for BglI (GCCN5GGC) and MboII (GAAGA). These polymorphisms were observed experimentally when the wbiI PCR products were digested with BglI or MboII (Fig. 3). There were four different RFLP patterns observed for each restriction enzyme (A to D), with a total of seven different combinations (Tables 1 and 2).
FIG. 3.
RFLP of wbiI PCR products from the Bcc O1 to O9 serotype strains. wbiI PCR products from Bcc O1 to O9 serotype strains were digested with BglI (top) or MboII (bottom). Four different patterns observed for each restriction enzyme digestion, labeled A to D, are shown below. Lanes 1 to 9 are as in Fig. 2, DNA size markers are shown to the left. The extra band in lane 2 is also observed in the undigested PCR product and is thus not included in pattern designation.
TABLE 1.
Characteristics of select Bcc isolatesa
| Group and isolate | Species (genomovar) | Serotypeb | wbil RFLP typec | Source/lineage | Reference |
|---|---|---|---|---|---|
| Serotype representatives | |||||
| CIP 8235 | B. stabilis (IV) | O1 | A/A | Environmental | 13 |
| CIP 8236 | B. cenocepacia (III) | O2 | A/B | Nosocomial | 13 |
| CIP 8237 | B. cenocepacia (III) | O3 | A/B | Nosocomial | 13 |
| CIP 8238 | B. cenocepacia (III) | O4 | B/A | Nosocomial | 13 |
| CIP 8239 | Indeterminate | O5 | C/A | Nosocomial | 13 |
| CIP 8240 | Indeterminate | O6 | A/C | Nosocomial | 13 |
| ATCC 17759 | B. cepacia (I) | O7 | C/A | Environmental | 13 |
| CDC 99 | Indeterminate | O8 | A/D | Unknown | 33 |
| CDC 86 | B. cenocepacia (III) | O9 | D/B | Nosocomial | 33 |
| Species representatives | |||||
| ATCC 25416 | B. cepacia (I) | O7 | C/A | Environmental | 20 |
| ATCC 17616 | B. multivorans (II) | NT-R | Unique | Environmental | 20 |
| J2315 | B. cenocepacia (III) | NT-R | A/B | CF/ET12 | 20 |
| LMG 14294 | B. stabilis (IV) | MT | Unique | CF | 20 |
| PC259 | B. vietnamiensis (V) | O3 | Multiple PCR | CF | 20 |
| AU0645 | B. dolosa (VI) | NT-R | A/B | CF | 9 |
| AMMD | B. ambifaria (VII) | O8 | Unique | Environmental | 9 |
| AU1293 | B. anthina (VIII) | NT-S | Multiple PCR | CF | 9 |
| BC011 | B. pyrrocinia (IX) | NT-S | A/A | Environmental | 9 |
| CF outbreak representatives | |||||
| K56-2 | B. cenocepacia (III) | MT | A/B | CF/ET12 | 14 |
| Bc7 | B. cenocepacia (III) | NT-R | A/B | CF/ET12 | 14 |
| PC184 | B. cenocepacia (III) | NT-R | A/D | CF/Midwest | 5 |
| AU1054 | B. cenocepacia (III) | NT-S | A/B | CF/PHDC | 3 |
| AU2130 | B. dolosa (VI) | NT-R | A/B | CF/SLC6 | 1 |
| AU0066 | B. multivorans (II) | Not tested | Multiple PCR | CF/OHBM | 1 |
Bcc isolates listed are the nine serotype strains, nine species (genomovar) isolates, and isolates from five outbreaks among CF patients.
NT-R, nontypeable, rough; NT-S, nontypeable, smooth; MT, reactive with multiple antisera.
BgiI RFLP type/MboII RFLP type; unique, unique RFLP type; multiple PCR, multiple PCR products.
TABLE 2.
wbiI RFLP types of 80 Bcc isolates tested
| RFLP type (BgiI/MboII) | No. of Bcc isolates | % of total |
|---|---|---|
| A/A | 6 | 8 |
| A/B | 20 | 25 |
| A/C | 3 | 4 |
| A/D | 13 | 16 |
| B/A | 1 | 1 |
| C/A | 9 | 11 |
| D/B | 8 | 10 |
| Unique RFLP type | 7 | 9 |
| Multiple PCR products | 13 | 16 |
PCR amplification and RFLP analysis of the conserved region of wbiI were performed on a total of 80 Bcc isolates, including the O1 to O9 serotype strains. More than one PCR product was obtained from 13 of 80 isolates (16.25%), likely as a result of nonspecific annealing by the primers, so RFLP analysis was not performed. Of the remaining 67 isolates, 7 had unique BglI or MboII wbiI RFLP patterns not observed among the nine serotype strains (Table 2). For the other 60 isolates, which produced RFLP patterns similar to those of the nine serotype strains, most (20 of 80, 25%) had BglI RFLP pattern A and MboII RFLP pattern B (shorthand notation, A/B), followed by those of type D/B (17.5%) (Table 2). Three isolates from each Bcc outbreak described by Rabkin et al. (22) were shown to have the same wbiI RFLP type, indicating the expected clonality of these outbreak strains (data not shown).
Correlation between wbiI RFLP type and serotype.
To analyze the relationship between wbiI RFLP and serotype of Bcc, the serotypes for 57 of 80 Bcc isolates were identified. The serotypes of 23 isolates had been reported previously (22), while 34 isolates were typed by immunoblotting of crude LPS preparations with the anti-Bcc O1 to O9 sera. Thirteen of the isolates tested did not react with any of the nine anti-Bcc sera, including isolates from four different outbreaks among CF patients (Tables 1 and 3). Ten of the 13 nonserotypeable isolates did not express O antigen, while 3 expressed O antigen, as visualized by silver staining. The demonstration that three isolates expressing O antigen did not react with the nine anti-Bcc sera confirmed previous observations that there are more than nine O antigen-specific serotypes among Bcc isolates (26, 32) (Table 3 and data not shown).
TABLE 3.
wbiI RFLP type versus serotype for 57 Bcc isolates
| wbiI RFLP type | No. of Bcc isolates of serotypea:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| O1 | O2 | O3 | O4 | O5 | O6 | O7 | O8 | O9 | MT | NT-R | NT-S | |
| A/A | 2 | 1 | ||||||||||
| A/B | 7 | 3 | 1 | 6 | 1 | |||||||
| A/C | 1 | |||||||||||
| A/D | 4 | 4 | ||||||||||
| B/A | 1 | |||||||||||
| C/A | 2 | 4 | 2 | |||||||||
| D/B | 3 | 4 | 1 | |||||||||
| Unique RFLP type | 1 | 1 | 1 | |||||||||
| Multiple PCR products | 4 | 1 | 2 | |||||||||
MT, reactive with multiple antisera; NT-R, nontypeable, rough; NT-S, nontypable, smooth.
Bcc isolates of serotypes O2, O5, O6, O7, and O9 each had a single wbiI RFLP type, while serotype O1 and O4 isolates had one of two wbiI RFLP types. When a single PCR product could be obtained from O3 isolates, it was wbiI RFLP type A/B (Table 3). These results indicate that the wbiI RFLP type can be inferred from the serotype of a Bcc isolate. However, some Bcc isolates of different serotypes had the same wbiI RFLP pattern. For example, wbiI RFLP type C/A strains could correspond to serotype O4, O5, or O7 and both serotype O1 and O8 isolates were wbiI RFLP type A/D (Table 3). Thus, by using the wbiI RFLP type, it is possible to restrict the potential serotype of an isolate. However, the wbiI RFLP typing scheme cannot be used to predict the specific serotype of a Bcc isolate.
Correlations with species.
The species was determined for all 80 Bcc isolates used in the present study. At least two Bcc isolates from each species were represented, including 12 Bcc isolates that did not group with any of the current nine species and thus were deemed indeterminate (Table 4). Bcc isolates with a single wbiI RFLP type were often found in more than one species. Likewise, isolates of a single species had different wbiI RFLP types (Table 4), thereby indicating that there is no correlation between species and wbiI RFLP type.
TABLE 4.
wbiI RFLP type versus species for 80 Bcc isolates
| wbiI RFLP type (BglI/MboII) | No. of Bcc isolates of species:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| B. cepacia | B. multivorans | B. cenocepacia | B. stabilis | B. vietnamiensis | B. dolosa | B. ambifaria | B. anthina | B. pyrrocinia | Indeterminant | |
| A/A | 3 | 3 | ||||||||
| A/B | 17 | 2 | 1 | |||||||
| A/C | 1 | 1 | 1 | |||||||
| A/D | 1 | 6 | 1 | 4 | ||||||
| B/A | 1 | |||||||||
| C/A | 2 | 2 | 5 | |||||||
| D/B | 8 | |||||||||
| Unique RFLP type | 1 | 1 | 1 | 2 | 1 | 1 | ||||
| Multiple PCR products | 3 | 1 | 5 | 3 | 1 | |||||
Previously, it was determined that species and serotypes of Bcc isolates do not correlate (15). The present study, which used many of the Bcc isolates tested in the study by Kenna et al. (15) with an additional 25 Bcc isolates, including the serotype O1 to O9 strains, confirmed this conclusion (Table 1 and data not shown).
Correlation with source of Bcc isolates.
As Bcc infection of the lungs of CF patients is an increasing problem in the CF community, it is important to identify factors that may indicate the propensity of a Bcc isolate to infect individuals with CF. The origins of 72 of the Bcc isolates tested in the present study were known. Twenty-three Bcc isolates were obtained from the lungs of CF patients, while 49 were non-CF, isolated either from nosocomial infections or environmental sources. Bcc isolates with a single wbiI RFLP type could be found among both CF and non-CF isolates, indicating that there was no correlation between wbiI RFLP type and the ability of a Bcc isolate to infect CF patients (Table 5).
TABLE 5.
wbiI RFLP type and prevalence in CF and non-CF sources
| wbiI RFLP type (BglI/MboII) | No. of:
|
|
|---|---|---|
| CF isolates | Non-CF isolates | |
| A/A | 1 | 5 |
| A/B | 9 | 9 |
| A/C | 2 | 1 |
| A/D | 2 | 9 |
| B/A | 0 | 1 |
| C/A | 1 | 7 |
| D/B | 1 | 7 |
| Unique RFLP type | 1 | 6 |
| Multiple PCR products | 6 | 4 |
CF patients are more likely to be infected with B. cenocepacia (genomovar III) or B. multivorans (genomovar II), although all species have been isolated from the lungs of CF patients (30). To determine whether wbiI RFLP type, in addition to species, could predict the ability of a Bcc isolate to infect CF patients, the wbiI RFLP type and species were cross-referenced with the sources of the Bcc isolates. B. cenocepacia isolates had more than one wbiI RFLP type (Table 4), and B. cenocepacia isolates of the same wbiI RFLP type were found among both CF and non-CF sources (Table 1 and data not shown). The same was true for isolates of B. multivorans (Table 4 and data not shown). Thus, the ability of a Bcc isolate to infect the lungs of CF patients cannot be predicted by the wbiI RFLP type coupled with species.
The wbiI RFLP types for five isolates of the ET12 outbreak strain (14) and one isolate from each of the PHDC (3), Midwest (5), SLC6, and OHBM (1) outbreak strains were determined. All five ET12 lineage isolates had the same wbiI RFLP type of A/B, indicating that, as anticipated, these isolates are clonal. The B. cenocepacia PHDC outbreak isolate and the B. dolosa SLC6 outbreak isolate were also wbiI RFLP type A/B, while the B. cenocepacia Midwest outbreak isolate was wbiI RFLP type D/B (Table 1). A single PCR product could not be obtained from the B. multivorans OHBM outbreak isolate (Table 1). While three of five Bcc outbreak strains had wbiI RFLP type A/B, this trait alone is not indicative of the epidemic potential of a Bcc strain; however, monitoring this genetic trait may help track these outbreak strains.
Serotyping and subsequent testing for the presence of O antigen in ET12 isolates demonstrated that four of the five isolates did not express O antigen while K56-2 expressed O antigen but reacted with multiple anti-Bcc O1 to O9 sera (Table 1). The SLC6 and Midwest isolates, like most ET12 isolates, also did not express O antigen, while the nontypeable PHDC isolate expressed a low-molecular-weight O antigen (Table 1 and data not shown). This indicates that the presence of O antigen may not be necessary for Bcc to infect the CF lung or for transmission of a Bcc strain among CF patients.
DISCUSSION
Recent studies of Bcc have focused on identifying factors, whether phenotypic or genotypic, that could be used to indicate the potential of a Bcc strain to cause infections in the lungs of individuals with CF. A virulence factor common to all gram-negative bacteria is LPS. The O antigen portion of LPS is the basis for phenotypically classifying bacterial strains by using specific antisera. While serotyping is still used to classify some bacterial strains, specific antisera are not always readily available, and some bacterial strains lack O antigen, making them nontypeable.
To avoid the above-stated limitations of serotyping, a genetic method to delineate the serotype of a Bcc isolate was developed in the present study. The wbiI gene of Bcc was the target of this genetic serotyping method due to its conservation among most Bcc isolates (Fig. 2 and Table 2) and the known role of homologues to wbiI in O antigen biosynthesis (2). wbiI RFLP typing of Bcc isolates for which the serotype was known revealed that Bcc isolates of a single serotype usually have the same wbiI RFLP type, indicating that wbiI correlates with serotype and thus with O antigen (Table 3). However, this genetic method of serotyping has its limitations. For example, Bcc isolates of a single wbiI RFLP type may have different serotypes. Also, only Bcc serotype O1 to O9 strains and anti-Bcc O1 to O9 sera were used to establish this genetic typing method (Table 3). The latter limitation is the likely reason that three nonserotypeable but O antigen-producing Bcc isolates were identified in this study.
The wbiI RFLP type is not indicative of the ability of a Bcc isolate to infect CF patients or to be transmitted from patient to patient (Tables 1 and 5). Likewise, serotype, or even the ability to express O antigen, does not correlate with the ability of a Bcc strain to infect CF patients (Table 1 and data not shown). Several of the CF outbreak isolates, including four of five ET12 lineage isolates, did not express O antigen (Table 1). P. aeruginosa, another CF pathogen, expresses O antigen when it first infects the lungs of CF patients but loses the ability to express O antigen when chronically infecting the lung (12, 21). It is unknown if Bcc also loses expression of O antigen once it has established a chronic CF lung infection. As the outbreak isolates analyzed in the present study were isolated from the lungs of CF patients, it is possible that the strains originally expressed O antigen and then lost the ability to express O antigen during chronic lung infection, similar to P. aeruginosa. However, as these outbreak strains have been demonstrated to be transmitted from patient to patient and not acquired from an environmental source (27), it is more likely that O antigen is not necessary for transmission of Bcc strains.
Bacteria can be classified by phenotypic or genetic classification schemes. With the ease and specificity of genetic typing methods, bacteria are now commonly classified genotypically rather than phenotypically. Previously, it was demonstrated that Bcc serotype, a phenotypic characteristic, and species, a genetic characteristic, do not correlate (15). This is similar to what has been found in other bacteria (23), where serotype alone cannot be used to discriminate among different species. The present study has extended the above statement to conclude that species and wbiI RFLP, another genetic typing scheme, do not correlate (Table 4).
Studies of O antigen gene clusters from other bacterial species indicate that O antigen loci have undergone horizontal gene transfer (25). One piece of evidence is that O antigen loci are often located in low-G+C regions compared with the remainder of the bacterial genome (25). The O antigen gene cluster, including wbiI, in B. cenocepacia J2315 is part of a low-G+C island in the genome (A. Baldwin, M. Holden, I. Blaby, M. Perucchini, A. Rossbach, P. Vandamme, J. Parkill, and E. Mahenthiralingam, Abstr. 17th Annu. N. Am. Cystic Fibrosis Conf., abstr. 297, 2003). Species within the Bcc can be differentiated based on analyses of 16S rDNA and the recA gene (17, 19). The recA gene is located near nt 1041000 on chromosome 1 of B. cenocepacia J2315, while the wbiI gene is located around nt 3409000 of the same chromosome (http://www.sanger.ac.uk/Projects/B_cenocepacia/). Since wbiI and recA gene sequences do not correlate, this suggests that the evolution of O antigen loci is distinct from that of recA, a conserved housekeeping gene, and that Bcc O antigen loci may have undergone horizontal gene transfer.
As has been observed for other Bcc strain markers (16), wbiI RFLP type or serotype is not predictive of the ability of a Bcc strain to cause disease in CF patients. However, wbiI RFLP typing can be added to other available tools that rapidly discriminate Bcc isolates and assist in tracking potential outbreak strains.
Acknowledgments
This work was supported by grants from the National Institutes of Health (J.J.L., AI054411-01; J.B.G., AI050230) and the Cystic Fibrosis Foundation (J.J.L., LIPUMA97P0; J.B.G., GOLDBE03P0). A.D.V.-D. was supported by the National Institutes of Health through a University of Virginia Infectious Diseases training grant (AI07046).
We thank Beatrice Muller of the Institute of Bacteriology, Strasbourg, France, for technical assistance. The J2315 wbiI sequence data were produced by the Burkholderia cenocepacia Sequencing Group at the Sanger Institute and can be obtained from http://www.sanger.ac.uk/Projects/B_cenocepacia/.
REFERENCES
- 1.Biddick, R., T. Spilker, A. Martin, and J. J. LiPuma. 2003. Evidence of transmission of Burkholderia cepacia, Burkholderia multivorans and Burkholderia dolosa among persons with cystic fibrosis. FEMS Microbiol. Lett. 228:57-62. [DOI] [PubMed] [Google Scholar]
- 2.Burrows, L. L., R. V. Urbanic, and J. S. Lam. 2000. Functional conservation of the polysaccharide biosynthetic protein WbpM and its homologues in Pseudomonas aeruginosa and other medically significant bacteria. Infect. Immun. 68:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, J. S., K. A. Witzmann, T. Spilker, R. J. Fink, and J. J. LiPuma. 2001. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J. Pediatr. 139:643-649. [DOI] [PubMed] [Google Scholar]
- 4.Clode, F. E., M. E. Kaufmann, H. Malnick, and T. L. Pitt. 2000. Distribution of genes encoding putative transmissibility factors among epidemic and nonepidemic strains of Burkholderia cepacia from cystic fibrosis patients in the United Kingdom. J. Clin. Microbiol. 38:1763-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coenye, T., and J. J. LiPuma. 2002. Multilocus restriction typing: a novel tool for studying global epidemiology of Burkholderia cepacia complex infection in cystic fibrosis. J. Infect. Dis. 185:1454-1462. [DOI] [PubMed] [Google Scholar]
- 6.Coenye, T., and J. J. LiPuma. 2003. Population structure analysis of Burkholderia cepacia genomovar III: varying degrees of genetic recombination characterize major clonal complexes. Microbiology 149:77-88. [DOI] [PubMed] [Google Scholar]
- 7.Coenye, T., T. Spilker, A. Martin, and J. J. LiPuma. 2002. Comparative assessment of genotyping methods for epidemiologic study of Burkholderia cepacia genomovar III. J. Clin. Microbiol. 40:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coenye, T., P. Vandamme, J. R. Govan, and J. J. LiPuma. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coenye, T., P. Vandamme, J. J. LiPuma, J. R. Govan, and E. Mahenthiralingam. 2003. Updated version of the Burkholderia cepacia complex experimental strain panel. J. Clin. Microbiol. 41:2797-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean, C. R., C. V. Franklund, J. D. Retief, M. J. Coyne, Jr., K. Hatano, D. J. Evans, G. B. Pier, and J. B. Goldberg. 1999. Characterization of the serogroup O11 O-antigen locus of Pseudomonas aeruginosa PA103. J. Bacteriol. 181:4275-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean, C. R., and J. B. Goldberg. 2002. Pseudomonas aeruginosa galU is required for a complete lipopolysaccharide core and repairs a secondary mutation in a PA103 (serogroup O11) wbpM mutant. FEMS Microbiol. Lett. 210:277-283. [DOI] [PubMed] [Google Scholar]
- 12.Hancock, R. E., L. M. Mutharia, L. Chan, R. P. Darveau, D. P. Speert, and G. B. Pier. 1983. Pseudomonas aeruginosa isolates from patients with cystic fibrosis: a class of serum-sensitive, nontypeable strains deficient in lipopolysaccharide O side chains. Infect. Immun. 42:170-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidt, A., H. Monteil, and C. Richard. 1983. O and H serotyping of Pseudomonas cepacia. J. Clin. Microbiol. 18:738-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, W. M., S. D. Tyler, and K. R. Rozee. 1994. Linkage analysis of geographic and clinical clusters in Pseudomonas cepacia infections by multilocus enzyme electrophoresis and ribotyping. J. Clin. Microbiol. 32:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kenna, D. T., V. A. Barcus, R. J. Langley, P. Vandamme, and J. R. Govan. 2003. Lack of correlation between O-serotype, bacteriophage susceptibility and genomovar status in the Burkholderia cepacia complex. FEMS Immunol. Med. Microbiol. 35:87-92. [DOI] [PubMed] [Google Scholar]
- 16.LiPuma, J. J. 2003. Burkholderia and emerging pathogens in cystic fibrosis. Semin. Respir. Crit. Care Med. 24:681-692. [DOI] [PubMed] [Google Scholar]
- 17.LiPuma, J. J., B. J. Dulaney, J. D. McMenamin, P. W. Whitby, T. L. Stull, T. Coenye, and P. Vandamme. 1999. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol. 37:3167-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LiPuma, J. J., T. Spilker, L. H. Gill, P. W. Campbell III, L. Liu, and E. Mahenthiralingam. 2001. Disproportionate distribution of Burkholderia cepacia complex species and transmissibility markers in cystic fibrosis. Am. J. Respir. Crit. Care Med. 164:92-96. [DOI] [PubMed] [Google Scholar]
- 19.Mahenthiralingam, E., J. Bischof, S. K. Byrne, C. Radomski, J. E. Davies, Y. Av-Gay, and P. Vandamme. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Penketh, A., T. Pitt, D. Roberts, M. E. Hodson, and J. C. Batten. 1983. The relationship of phenotype changes in Pseudomonas aeruginosa to the clinical condition of patients with cystic fibrosis. Am. Rev. Respir. Dis. 127:605-608. [DOI] [PubMed] [Google Scholar]
- 22.Rabkin, C. S., W. R. Jarvis, R. L. Anderson, J. Govan, J. Klinger, J. LiPuma, W. J. Martone, H. Monteil, C. Richard, S. Shigeta, et al. 1989. Pseudomonas cepacia typing systems: collaborative study to assess their potential in epidemiologic investigations. Rev. Infect. Dis. 11:600-607. [DOI] [PubMed] [Google Scholar]
- 23.Raetz, C. R. H., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raymond, C. K., E. H. Sims, A. Kas, D. H. Spencer, T. V. Kutyavin, R. G. Ivey, Y. Zhou, R. Kaul, J. B. Clendenning, and M. V. Olson. 2002. Genetic variation at the O-antigen biosynthetic locus in Pseudomonas aeruginosa. J. Bacteriol. 184:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reeves, P. P., and L. Wang. 2002. Genomic organization of LPS-specific loci. Curr. Top. Microbiol. Immunol. 264:109-135. [PubMed] [Google Scholar]
- 26.Soldatkina, A., A. Knirel'Iu, N. V. Tanatar, and I. Zakharova. 1989. Immunologic and structural studies of lipopolysaccharides from Pseudomonas cepacia. Mikrobiol. Zh. 51:32-38. [PubMed] [Google Scholar]
- 27.Sun, L., R. Z. Jiang, S. Steinbach, A. Holmes, C. Campanelli, J. Forstner, U. Sajjan, Y. Tan, M. Riley, and R. Goldstein. 1995. The emergence of a highly transmissible lineage of cbl+ Pseudomonas (Burkholderia) cepacia causing CF centre epidemics in North America and Britain. Nat. Med. 1:661-666. [DOI] [PubMed] [Google Scholar]
- 28.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 29.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 30.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahenthiralingam, J. J. LiPuma, and J. R. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 31.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J. R. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 32.Vinion-Dubiel, A. D., and J. B. Goldberg. 2003. Lipopolysaccharide of Burkholderia cepacia complex. J. Endotoxin Res. 9:201-213. [DOI] [PubMed] [Google Scholar]
- 33.Werneburg, B., and H. Monteil. 1989. New serotypes of Pseudomonas cepacia. Res. Microbiol. 140:17-20. [DOI] [PubMed] [Google Scholar]