Abstract
A simple method, using commercial disks to detect Escherichia coli mutator strains, is proposed. The breakpoint for detecting strains with a mutation frequency ≥5 × 10−7 was established at ≥70 and ≥20 colonies in the inhibition zone of fosfomycin and rifampin disks, respectively, after seeding 100 μl of an overnight culture. Strains with <30 and <10 colonies in fosfomycin and rifampin inhibition zones are presumptively non-mutators.
The frequency of mutator strains in clinical isolates ranges from 1% for uropathogenic Escherichia coli isolates to 20% for Pseudomonas aeruginosa in patients with cystic fibrosis (3, 5, 9). Mutator strains may adapt more efficiently than nonmutator strains to sequential exposure to antibiotics, due to their greater facility in generating antibiotic resistance-conferring mutations (1, 2, 6, 7, 9), which in all likelihood is connected with a higher probability of therapeutic failure (4, 8). Detection of mutator strains could minimize the emergence of antibiotic-resistant mutants and the resulting risk of therapy failure, but classical methods of determining mutation frequency are cumbersome and outside the realm possibility for daily routine. Our approach is based on a slightly modified diffusion test using commercial fosfomycin and rifampin disks. The identification of hypermutable strains was made by counting the squatter colonies growing in the inhibition zone after 24 h of incubation. The basic conditions of the test were set with a group of three control strains with different mutation frequencies determined by the classic Luria-Delbrück procedure (25 replicate experiments per strain). This preliminary study served to establish the optimal bacterial inoculum in plates. This was defined as one where squatter colonies of E. coli strains with 10−6 or 10−7 mutation frequencies could be detected in the inhibition zone after 24 h of incubation at 37°C (but not E. coli with 10−8 mutation frequency). Disks with spectinomycin (100 μg), nalidixic acid (30 μg), cotrimoxazole (25 μg), and ampicillin (10 μg) (Oxoid, Basingstoke, United Kingdom) proved to be unreliable for test purposes, as the number of squatter colonies obtained in hypermutable strains was too low. The optimal bacterial load that we selected to be used in the tests with rifampin disks (30 μg) was fixed at 100 μl of overnight culture (∼2 McFarland standards). In the case of the fosfomycin disks (50 μg), a bacterial load of 100 μl of the 1:100 dilution of the overnight culture (∼0.5 McFarland standard) was chosen for further testing.
To establish the test cutoff values, that is, the number of colonies around fosfomycin and rifampin disks that could serve to detect different types of hypermutable E. coli, a control collection of 100 E. coli clinical strains with well-characterized mutation frequencies were studied. To calculate mutation frequencies in this collection, three independent colonies were inoculated in Luria-Bertani (LB) broth tubes and incubated with rotary movement at 37°C for 24 h, and different dilutions (100 μl) were seeded onto LB agar and LB-rifampin plates (100 μg/μl). Colonies on LB plates were counted after 24 h of incubation, and those on LB-rifampin plates were counted after 48 h. Mutation frequency values were estimated as the ratio of rifampin-resistant colonies to the total number of CFU. The results of the experiment indicate that the collection included 4 strong mutators (mutation frequency, ≥5 × 10−7), 28 weak mutators (mutation frequency, ≤5 × 10−7 to ≥5 × 10−8), and 68 normomutable strains.
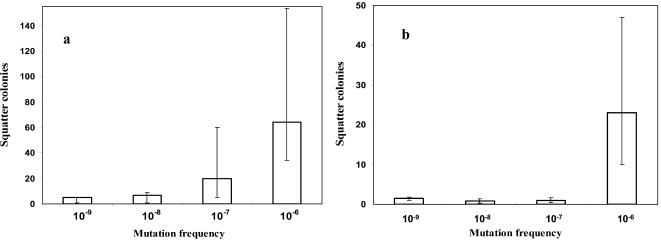
Both rifampin and fosfomycin disks clearly enabled identification of strong mutators in the conditions described above. Weak mutators were detected by the presence of squatter colonies around the fosfomycin disk but not around the rifampin disk. The number of colonies was recorded in double-blind experiments by two different observers, with an overall agreement of 90%. According to the distribution of the number of squatter colonies versus the frequency of mutation (Fig. 1), a presumptive cutoff value was established. Strains with ≥20 colonies in the fosfomycin inhibition zone using 10−2 dilution can be considered mutators. If in addition there are 10 squatter colonies directly in the rifampin inhibition zone with the overnight inoculum, then the strains can be considered strong mutators. With these criteria, the positive and negative predictive values (PPV and NPV) for mutators were 73% and 81%, respectively.
FIG. 1.
Means (bars) and ranges of the number of squatter colonies in the inhibition zones of 50-μg fosfomycin (a) and 30-μg rifampin (b) disks in strains with known mutation frequencies.
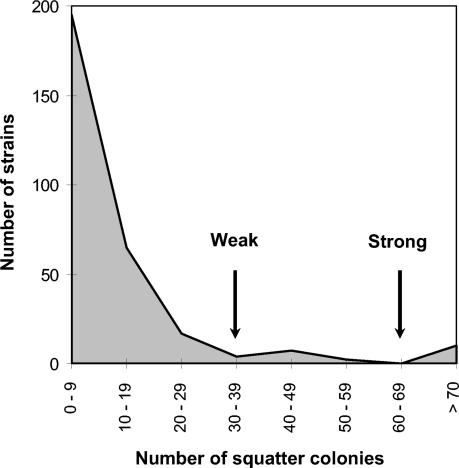
Once the feasibility of finding an acceptable correlation between disk detection test and mutation frequencies in the control collection of strains was established, a prospective blind study was made using a collection of 300 consecutively isolated uropathogenic E. coli clinical strains. In order to reduce the workload, the test was simplified by seeding 100 μl of overnight culture for both fosfomycin and rifampin. The distribution of strains according to the number of squatter colonies in the fosfomycin inhibition zone is shown in Fig. 2. This distribution indicates that >30 squatter colonies around the fosfomycin disk are highly unusual in the collection. This limit could therefore presumptively be considered the limit of the normomutable population. Strains with more than 70 colonies around the fosfomycin disk might be presumptively considered strong mutators, while a number of colonies between 30 and 70 could correspond to weak mutators. Rifampin disks were used to refine this classification. Strains with <30 colonies around the fosfomycin inhibition zone and also having less than 10 colonies around the rifampin inhibition zone should be considered normomutable. Some strains with >70 colonies in fosfomycin and <10 in rifampin or <30 colonies in fosfomycin and >10 colonies in rifampin were presumptively classified as weak mutators. By using these population-based criteria, we identified 3 strong mutators and 20 putative weak mutators in this collection of 300 E. coli strains.
FIG. 2.
Distribution of squatter colonies around 50-μg fosfomycin disks of 300 consecutively isolated E. coli urinary tract pathogenic strains. The arrows indicate the breakpoints established to identify weak and strong mutator strains.
To confirm or reject this classification, we determined mutation frequencies of 50 selected strains by conventional methods. These strains included all 23 putative mutators identified in the disk test. All three strains presumptively classified as hypermutators and 15 of 20 of the presumptive weak mutators were confirmed by conventional mutation frequency testing. The PPV for the identification of weak mutators was 60%, and the NPV was 88%. With lower inocula (10−2 dilution of overnight cultures), the PPV for weak mutators was higher (72%), but at the expense of a reduction in the NPV (65%). If strong mutators were included in the analysis, the overall PPV and NPV of the disk detection method for any type of mutators rose to 75 and 84%, respectively.
Based on these results, we have proposed the following simple method for detecting hypermutable strains in the clinical microbiological laboratory. Blood agar plates were seeded with 100 μl of overnight LB (or brain heart infusion [data not shown]) broth cultures, and rifampin (50 μg) and fosfomycin (30 μg) disks were placed on the surface. Inhibition zones were examined after 24 h of incubation at 37°C. Strains with >70 colonies around the fosfomycin disk and >10 colonies around the rifampin disk have been considered to correspond to strong mutators. Strains with >30 colonies but <70 colonies around the fosfomycin disk or >70 around the fosfomycin but <10 around the rifampin disk can be presumptively considered weak mutators. Strains with <30 colonies in fosfomycin and <10 in rifampin are presumptively nonmutators. Normomutator strains should not yield ≥20 colonies around the fosfomycin disk in a plate seeded with 100 μl of the 10−2 dilution of the overnight culture.
As the expected prevalence of strong mutators in bacterial populations is in the range of 1% (2, 5; Baquero et al., submitted for publication), we suggest implementation of this method only in the case of samples from patients with long-term antibiotic treatment. The introduction of this procedure is expected to favor the use of antibiotic combinations in patients infected by mutator strains (4) and to establish barrier measures for preventing the spread of these dangerous organisms in the hospital environment.
Acknowledgments
This work was supported by QLK2-CT-2001-00873 Project of the European Union.
REFERENCES
- 1.Blázquez, J. 2003. Hypermutation as a factor contributing to the acquisition of antimicrobial resistance. Clin. Infect. Dis. 37:1201-1209. [DOI] [PubMed] [Google Scholar]
- 2.Chopra, I., A. J. O'Neill, and K. Miller. 2003. The role of mutators in the emergence of antibiotic-resistance bacteria. Drug Resist. Updat. 6:137-145. [DOI] [PubMed] [Google Scholar]
- 3.Denamur, E., S. Bonacorsi, A. Giraud, P. Duriez, F. Hilali, C. Amorin, E. Bingen, A. Andremont, B. Picard, F. Taddei, and I. Matic. 2002. High frequency of mutator strains among human uropathogenic Escherichia coli isolates. J. Bacteriol. 184:605-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giraud, P., I. Matic, M. Radman, M. Fons, and F. Taddei. 2002. Mutator bacteria as a risk in treatment of infectious diseases. Antimicrob. Agents Chemother. 46:863-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 6.Lindgren, P. K., Å. Karlsson, and D. Hughes. 2003. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob. Agents Chemother. 47:3222-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martínez, J. L., and F. Baquero. 2000. Mutation frequencies and antibiotic resistance. Antimicrob. Agents Chemother. 44:1771-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller, K., A. J. O'Neill, and I. Chopra. 2002. Response of Escherichia coli hypermutators to selection pressure with antimicrobial agents from different classes. J. Antimicrob. Chemother. 49:925-934. [DOI] [PubMed] [Google Scholar]
- 9.Oliver, A., R. Cantón, P. Campo, F. Baquero, and J. Blázquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]