Abstract
The occurrence of a clonal outbreak of serogroup W135 (of the electrophoretic type 37 [ET-37] clonal complex) meningococcal disease among Hajj pilgrims in 2000 has led to enhanced surveillance of the evolution of this particular serogroup, formerly considered rare, in invasive infections. Since the first case of meningococcal disease due to a serogroup W135 strain was detected in France in 1994, all isolates were characterized phenotypically. We further used phenotypic and genotypic approaches to type the 101 serogroup W135 strains isolated from patients with invasive meningococcal diseases in France in 2001 and 2002. Overall, 55% of these isolates had Hajj strain-related phenotypes (60 and 52% in 2001 and 2002, respectively), although only 45% belonged to the ET-37 clonal complex. Moreover, pulsed-field gel electrophoresis of the ET-37 clonal complex isolates showed that only 32% of the serogroup W135 isolates were indistinguishable from the 2000 Hajj-related strain. Our results suggest the continuous emergence of new genetic lineages of serogroup W135 independently of the 2000 global outbreak.
Neisseria meningitidis is a highly polymorphic bacterium and is most often a commensal organism of the human upper respiratory tract that occasionally provokes invasive infections and that may become an epidemic threat, particularly in the countries of the African meningitis belt (2, 4, 10). N. meningitidis strains have been classified into 13 serogroups on the basis of the immune specificity of the polysaccharide capsule (11, 12). Invasive infections (mainly septicemia and meningitis) are most frequently caused by five of these serogroups (serogroups A, B, C, Y, and W135). Phenotype determination by serological typing (designated as the serogroup:serotype:serosubtype) was the first useful method for the classification of meningococcal strains. However, most of the surface antigens used to determine the phenotype are subjected to phase and antigenic variations, but these variations reflect the selective pressure exerted by herd immunity rather than the clonal origins of the strains (22). Molecular typing methods have considerably improved our understanding of meningococcal epidemiology. Molecular techniques, based on the analysis of the polymorphisms at several genetic loci, are widely used to characterize meningococcal strains and show that epidemic strains can be clustered into particular clonal complexes, with each of them being composed of highly related genotypes (4). As an example, strains of serogroup A belonging to the subgroup III clonal complex were shown to be responsible for the second pandemic wave of meningococcal infections reported since the mid-1980s (2). More recently, molecular typing methods demonstrated the clonal expansion of serogroup W135 strains. Indeed, a global outbreak of serogroup W135 meningococcal disease occurred in 2000 among Hajj pilgrims returning from Saudi Arabia and their household contacts (16, 17). This clonal outbreak was caused by strains of the phenotype W135:2a:P1.5,2, which belongs to the electrophoretic type 37 (ET-37) clonal complex. However, related strains belonging to this clonal complex have been observed worldwide since 1970 (14). In 2001, meningococcal strains of serogroup W135 also belonging to the ET-37 clonal complex were involved in epidemics in Burkina Faso and Niger (20) and represented 84% of the isolates characterized during the 2002 outbreak in Burkina Faso (21). Following the outbreak in 2000, the European Commission-funded Invasive Bacterial Infections Surveillance Network established a sentinel rapid reporting system for the outbreak strain, including phenotypically similar isolates: W135:2a:P1.5,2, W135:2a:P1.2, W135:2a:P1.5, W135:2a:NST (where NST is nonserosubtypeable), W135:NT:P1.5,2 (where NT is nonserotypeable), W135:NT:P1.2, and W135:NT:P1.5. Whereas the number of cases due to Hajj-related strains decreased in 2001 and 2002 in France (6), the overall number of cases due to serogroup W135 isolates did not (18). In the work described here, we aimed to analyze the genetic evolution of serogroup W135 strains isolated in France after the outbreak in 2000 in order to evaluate their epidemiological potentials.
MATERIALS AND METHODS
Bacterial isolates and phenotypic characterization.
All isolates of N. meningitidis serogroup W135 were from patients with invasive meningococcal infections (septicemia, meningitis, arthritis, or pericarditis) and were isolated from blood and cerebrospinal, synovial, and pericardial fluid, respectively. Bacteria were grown at 37°C in a 5% CO2 atmosphere on GCG medium with Kellogg supplements (8). The phenotypic determination, based on the antigenic formula (serogroup:serotype:serosubtype) of each isolate, was performed by standard methods (1).
Molecular typing of meningococcal isolates.
All isolates were typed by using three genotyping methods. We first analyzed the isolates by multilocus DNA fingerprinting (MLDF), which characterizes five genes (pilA, pilD, crgA, regF, and iga), as described previously (14). This method defines the alleles of these five genes according to their restriction patterns. Isolates belonging to the ET-37 clonal complex are characterized by the alleles 4, 1, 1a, 1, and 1 of pilA, pilD, crgA, regF, and iga, respectively (14, 18). Multilocus sequence typing (MLST) was then used to analyze the major clusters detected by MLDF analysis (all 45 isolates that were predicted to belong to the ET-37 clonal complex and 13 representative isolates of the other MLDF clusters containing more than 2 isolates). MLST was performed as described previously (13) with a new set of oligonucleotides (Table 1). Adaptors corresponding to universal forward and reverse oligonucleotides were added to the 5′ ends of the upstream and the downstream oligonucleotides, respectively. After amplification, universal forward and reverse oligonucleotides were used for sequencing. Standard pulsed-field gel electrophoresis (PFGE) was performed with the restriction enzyme SpeI (14). The restriction profiles were compared by using the Taxotron package (P. A. D. Grimont, Institut Pasteur, Paris, France).
TABLE 1.
Oligonucleotides used for MLSTa
| Oligonucleotide use and name | Sequence (5′ to 3′) |
|---|---|
| Amplification | |
| abcZ-fN1 | gttttcccagtcacgacgttgta CCGGTTTGCAAAAGCTCGACGAC |
| abcZ-rN2 | ttgtgagcggataacaatttc TTGTCAAAGAGGCGGTTGTGTTCC |
| adk-fN1 | gttttcccagtcacgacgttgtaGCACTCAGGCGCAATTCATCACC |
| adk-rN2 | ttgtgagcggataacaatttc TTTGCCCAATGCGCCCAATACTTC |
| aroE-fN1 | gttttcccagtcacgacgttgtaCTGGCGGACGAGCATTCCGAACGC |
| aroE-rN2 | ttgtgagcggataacaatttc GCGGTAATCCAGTGCGACATCGCG |
| fumC-fN1 | gttttcccagtcacgacgttgtaCGTCTGAACGATGCGCTTAAAGAC |
| fumC-rN2 | ttgtgagcggataacaatttcTCGGCAGGAACGACCAGTTCGTC |
| gdh-fN1 | gttttcccagtcacgacgttgtaGAAGGGCAAATTTACCGCATCGAC |
| gdh-rN2 | ttgtgagcggataacaatttcCCAAATCGGTTGCCAGCGGCACGG |
| pdhC-fN1 | gttttcccagtcacgacgttgtaTGCCGGCCGTACGACGCTGAACGG |
| pdhC-rN2 | ttgtgagcggataacaatttcGCGTTTCCAGCTAGGAGCTGAATC |
| pgm-fN1 | gttttcccagtcacgacgttgtaCCTGCAAGATTTGATTGCCGCGCT |
| pgm-rN2 | ttgtgagcggataacaatttcACGCATCAGACCGAAGCCGTCGGG |
| Sequencing | |
| Universal forward | gttttcccagtcacgacgttgta |
| Universal reverse | ttgtgagcggataacaatttc |
The sequences of upstream and downstream oligonucleotides (for amplification) of each gene (series N1 and N2, respectively) are given as uppercase letters. Universal forward and reverse oligonucleotides were added as adaptors in front of each oligonucleotide and are given as lowercase letters. Universal forward and reverse oligonucleotides were used for sequencing.
Statistical analysis.
Data were analyzed by the chi-square test. A P value of ≤0.05 was considered statistically significant. Due to the small number of isolates obtained before 1999, the statistical analysis was performed only for isolates recovered since 1999.
RESULTS
Phenotypic characterization of invasive meningococcal isolates of serogroup W135.
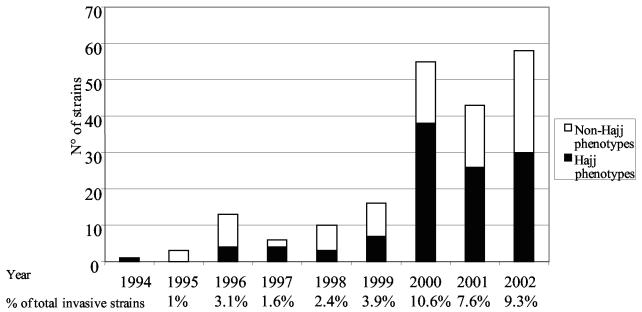
The first involvement of N. meningitidis serogroup W135 in a meningococcal disease case in France was detected in 1994. Then, from 1995 to 1999, 3 to 16 strains were isolated per year, and these represented less than 4% of the total invasive isolates. In 2000, 55 serogroup W135 isolates accounted for 10.6% of all invasive strains (Fig. 1). The increase in the year 2000 was mainly due to isolates expressing Hajj strain-related phenotypes (W135:2a:P1.5,2, W135:2a:P1.2, W135:2a:P1.5, W135:2a:NST, W135:NT:P1.5,2, W135:NT:P1.2, and W135:NT:P1.5). The proportions of isolates with Hajj strain-related phenotypes decreased from 69% (38 isolates) in 2000 to 60% (26 isolates) in 2001 and 52% (30 isolates) in 2002 (Fig. 1). The increase in the total number of isolates was associated with an increase and a diversification of the phenotypes observed, suggesting the expansion of several meningococcal lineages (Table 2). The 43 serogroup W135 strains isolated in 2001 displayed 9 different phenotypes, whereas the 58 strains isolated in 2002 displayed 14 different phenotypes. The proportion of isolates showing the W135:2a:P1.5,2 phenotype increased significantly in 2000 (P = 0.03), and the isolates corresponded to cases from the Hajj-associated outbreak (18). Only four of the cases in 2001 had a historical link to the 2001 Hajj pilgrimage (the source patient had contact with a Hajj pilgrim during the 10 days prior to his illness). The four corresponding isolates had the typical Hajj strain-related phenotype W135:2a:P1.5,2. No historical link with the Hajj pilgrimage was found among the cases detected in 2002. The proportion of strains with the phenotype W135:NT:P1.5 increased significantly in 2002 (P = 0.01) in comparison to the proportion in previous years, with only one strain detected in 1996, one strain detected in 1998, and one strain detected in 2000 (Table 2).
FIG. 1.
Distribution of meningococcal isolates with Hajj strain-related or non-Hajj strain-related phenotypes among invasive W135 strains isolated in France between 1994 and 2002. The percentage of meningococcal invasive strains of serogroup W135 is given for each year with respect to the total number of invasive strains.
TABLE 2.
Distribution of phenotypes among invasive serogroup W135 strains isolated in France in 2001 and 2002
| Phenotypea | No. of isolates
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1994 | 1995 | 1996 | 1997 | 1998 | 1999 | 2000 | 2001 | 2002 | Total | |
| 2a:P1.5,2 | 0 | 0 | 1 | 2 | 2 | 2 | 25 | 16 | 12 | 60 |
| 2a:NST | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 1 | 3 | 8 |
| 2a:P1.2 | 1 | 0 | 0 | 0 | 0 | 0 | 5 | 8 | 6 | 20 |
| 2a:P1.5 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 |
| NT:P1.2 | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 0 | 1 | 4 |
| NT:P1.5 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 7 | 10 |
| NT:P1.5,2 | 0 | 0 | 2 | 2 | 0 | 1 | 3 | 1 | 1 | 10 |
| 15:P1.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 15:P1.6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 2 |
| 2b:NST | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| 2b:P1.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| NT:P1.1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 2 |
| NT:P1.4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 3 |
| NT:P1.6 | 0 | 1 | 6 | 0 | 0 | 4 | 7 | 8 | 13 | 39 |
| NT:P1.7 | 0 | 1 | 1 | 0 | 1 | 0 | 4 | 1 | 1 | 9 |
| NT:P1.16 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 4 |
| NT:NST | 0 | 1 | 1 | 1 | 3 | 5 | 5 | 6 | 8 | 30 |
| Total | 1 (1)b | 3 (3) | 13 (7) | 6 (4) | 10 (5) | 16 (7) | 55 (10) | 43 (9) | 58 (14) | 205 |
Phenotypes are presented as the serogroup:serotype:serosubtype.
The values in parentheses are the members of different phenotypes.
These data clearly indicate that the phenotype diversity among invasive meningococcal isolates associated with serogroup W135 significantly increased after 2000.
Genotypic characterization of serogroup W135 invasive meningococcal isolates.
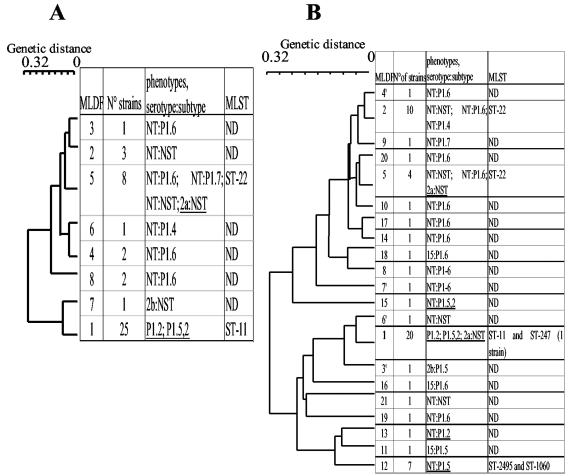
We next focused our analysis on the molecular typing of the serogroup W135 invasive strains isolated in 2001 and 2002. We typed all 101 isolates genetically using a sequential approach. All isolates were first typed by the MLDF method, which was recently used as a rapid approach to the identification of strains belonging to the ET-37 clonal complex (14, 18). Forty-five isolates were hence classified as ET-37 on the basis of their pilA4, pilD1, crgA1a, regF1, and iga1 alleles, which defined MLDF type 1 (Table 3; Fig. 2). MLST of all 45 of these isolates showed that 44 isolates belonged to sequence type 11 (ST-11) and 1 isolate belonged to ST-247. ST-11 is the major sequence type of the ET-37 clonal complex, and ST-247 is a minor member of this clonal complex that differs from ST-11 by the allele of the fumC gene. These data confirm that isolates of MLDF type 1 belonged to the ET-37 clonal complex. Isolates belonging to the ET-37 clonal complex represented 58 and 34% of W135 invasive isolates in 2001 and 2002, respectively. This decrease was not significant (P = 0.08). The decrease in the percentage of W135 (ET-37) isolates in 2002 was due to a decrease in the number of these isolates between 2001 and 2002 (25 and 20, respectively) and to the increase in the number of W135 non ET-37 isolates (18 and 38, respectively). The W135 non ET-37 isolates displayed several different genotypes, which increased in incidence from 7 in 2001 to 20 in 2002 (Fig. 2). MLDF type 5 was the second most frequent MLDF type in 2001, while MLDF types 2, 5, and 12 were observed more frequently in 2002 (Fig. 2). MLST analysis of six representative isolates revealed that strains of MLDF types 2 and 5 belonged to ST-22. The seven isolates with the W135:NT:P1.5 phenotype were MLDF type 12. All seven of these isolates were recovered in 2002 and were identical by MLDF. By MLST analysis they were found to belong to sequence types ST-2495 (6 isolates) and ST-1060 (1 isolate). ST-2495 and ST-1060 are highly related, and the sequences of their fumC genes differ by only one nucleotide. It is noteworthy that three isolates belonging to MLDF type 5 (ST-22) showed a Hajj strain-related phenotype (W135:2a:NST) (Fig. 2). However, none of the strains showing non-Hajj strain-related phenotypes belonged to the MLDF type 1, ET-37 clonal complex, according to the results of MLDF and MLST analyses (Fig. 2 and Table 3). Other genotypes were also distributed on the MLDF-based dendrogram but were represented by no more than two isolates, which may reflect horizontal genetic exchanges between strains (Fig. 2). These data demonstrate an extensive genetic diversity among the W135 invasive strains isolated in France in 2001 and 2002.
TABLE 3.
Characterization of serogroup W135 N. meningitidis strains isolated from patients with invasive infections in France in 2001 and 2002
| Yr | No. of isolates with:
|
||||
|---|---|---|---|---|---|
| Hajj strain-related phenotypes
|
Non-Hajj strain-related phenotypes
|
Total | |||
| ET-37 | Non-ET-37 | ET-37 | Non-ET-37 | ||
| 2001 | 25 | 1 | 0 | 17 | 43 |
| 2002 | 20 | 10 | 0 | 28 | 58 |
| Total | 45 | 11 | 0 | 45 | 101 |
FIG. 2.
MLDF-based dendrogram obtained by the unweighted pair-group method of averages algorithm showing the genetic distances among invasive W135 strains isolated in 2001 (A) and 2002 (B) in France. Genetic distances were calculated on the basis of the amplified fragment length restriction polymorphisms of the pilA, pilD, crgA, regF, and iga genes. Each MLDF group was assigned an arbitrary number. The number of strains in each group as well as the phenotypes observed in each group is indicated. The corresponding sequence type was determined for the major MLDF group. Hajj strain-related phenotypes are underlined. ND, not determined.
Further discrimination among serogroup W135 isolates within the ET-37 clonal complex.
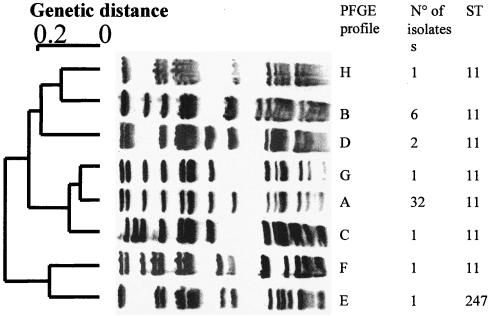
We further analyzed all isolates that belonged to the ET-37 clonal complex by PFGE, as this method has previously been shown to discriminate between different members within the ET-37 clonal complex (14, 15). A total of eight distinct profiles were distinguished by PFGE analysis with the SpeI restriction enzyme (Fig. 3). The Hajj strain-related profile (profile A) was found for 71% of the ET-37 strains (32 isolates, including 15 recovered in 2001 and 17 recovered in 2002). The four strains isolated in 2001 that had an epidemiological link to the Hajj pilgrimage all had this profile. Thirteen isolates belonging to the ET-37 clonal complex (10 isolates in 2001 and 3 isolates in 2002) were distinct from the 2000 outbreak strain and represented 29% of the ET-37 strains. Seven different PFGE profiles were observed among these 13 isolates. Six isolates had one non-Hajj strain-related profile (profile B), whereas the seven remaining isolates had six other profiles (Fig. 3). Profile B was also observed for invasive W135 isolates that have been circulating in France since 1994 (data not shown). These data indicate that the ET-37 strains isolated in France during 2001 and 2002 represent several strains of this clonal complex. Strains identical to Hajj-related strains accounted for 32% of all serogroup W135 isolates.
FIG. 3.
PFGE profiles observed among strains belonging to the ET-37 clonal complex. The dendrogram on the left was obtained by using the Taxotron package and the unweighted pair-group method of averages algorithm. Each PFGE group was assigned an arbitrary letter. All 45 strains were typed by MLST. The number of strains in each group and their sequence types are indicated. Profile A corresponds to the strain from the Hajj 2000 outbreak.
DISCUSSION
The incidence of invasive serogroup W135 strains has increased gradually in France since 1994, and the proportion of invasive meningococcal strains has reached 10% (3). Since the global outbreak among Hajj pilgrims and their contacts in 2000 due to serogroup W135 (ET-37) strains, the expansion of these strains has been monitored particularly closely in Europe. Our data show that strains with Hajj strain-related phenotypes have been the most frequent W135 invasive strains in France since 2000, accounting for 56 of 101 isolates in 2001 and 2002. This was also the case in England and Wales (7). However, MLDF and MLST analyses showed that only 45 of these isolates belonged to the ET-37 clonal complex, whereas 11 other isolates did not. Other genotypes were observed for several isolates. Seven isolates with the W135:NT:P1.5 phenotype, all isolated in 2002, clustered together (MLDF cluster 12) and belonged to two highly related genotypes, ST-2495 and ST-1060. Isolates belonging to the ST-22 clonal complex were also observed.
By using PFGE, only 32 of the 45 serogroup W135 (ET-37) isolates were indistinguishable from the Hajj-related strain. The 13 other isolates differed from the 2000 Hajj-related strain by at least three bands by PFGE analysis (except for isolates with PFGE profile G). Similar results were also recently obtained in Sweden when Hajj-related strains were compared to local W135 strains (15). Other non-Hajj-related ET-37 strains were observed prior to 2000 in several countries and may correspond to local strains (14). Several of the unique profiles shown in Fig. 3 may reflect the evolution of the Hajj-related strains over a 2-year period as a result of genetic drift. However, W135 (ET-37) strains of profile B (Fig. 3) have been detected in France since 1994 (18). Profile B was observed in six strains in 2001 and 2002 (Fig. 3) and may hence correspond to local strains that were circulating in France before the Hajj strain-related outbreak. Similar results have also been obtained in other countries, particularly in Africa (5, 9; our unpublished data). Our work underlines the need to use several molecular approaches to provide more discriminatory data for reliable epidemiological analysis.
In France, the number of invasive meningococcal strains of serogroup W135 has not decreased since 2000, but after 2000 only four cases were historically linked to the Hajj. Hajj strain-related phenotypes represented 69% of invasive W135 strains isolated in 2000 in France, but other genetic lineages were present and the incidence of those lineages has increased since the mid-1990s (Fig. 1) (18). In France, strains of serogroup W135 have now become the strains that are the most frequently isolated from patients with meningococcal diseases, after serogroups B and C (3).
The Hajj 2000 outbreak probably led to the expansion of a particular clone within the ET-37 clonal complex, but other ET-37 strains corresponding to local strains, such as those with PFGE profile B, seem to be isolated frequently. Moreover, other genetic lineages, such as the strains belonging to ST-2495 and ST-22, are now expanding and seem to make more important contributions than the ET-37 clonal complex to the genetic diversification of the meningococci associated with serogroup W135 (Fig. 2). The reason for the expansion of W135 strains is not yet clear. The strategies used to control meningococcal infection due to serogroup W135 should not apply only to Hajj pilgrims. Since N. meningitidis is naturally competent for transformation, genetic variants are expected to be continuously generated through horizontal DNA transfer and the reassortment of genes between different strains. By these mechanisms and from the selective pressure of herd immunity, changes in virulence and/or transmissibility may be responsible for the continuing emergence of new strains with polymorphic genotypes until the potential expansion of a new epidemic clone (19). Therefore, surveillance and characterization of invasive meningococcal strains by several fine molecular typing approaches are essential to monitor patients with meningococcal disease for the emergence of new strains.
Acknowledgments
This work was supported by the Institut Pasteur and the European Commission (QLK2-CT-2001-01436).
We thank Annie Guiyoule and René Pirès for excellent technical assistance.
REFERENCES
- 1.Abdillahi, H., and J. T. Poolman. 1988. Typing of group-B Neisseria meningitidis with monoclonal antibodies in the whole-cell ELISA. J. Med. Microbiol. 26:177-180. [PubMed] [Google Scholar]
- 2.Achtman, M. 1994. Clonal spread of serogroup A meningococci: a paradigm for the analysis of microevolution in bacteria. Mol. Microbiol. 11:15-22. [DOI] [PubMed] [Google Scholar]
- 3.Antignac, A., M. Ducos-Galand, A. Guiyoule, R. Pires, J. M. Alonso, and M. K. Taha. 2003. Neisseria meningitidis strains isolated from invasive infections in France (1999-2002): phenotypes and antibiotic susceptibility patterns. Clin. Infect. Dis. 37:912-920. [DOI] [PubMed] [Google Scholar]
- 4.Caugant, D. A. 1998. Population genetics and molecular epidemiology of Neisseria meningitidis. APMIS 106:505-525. [PubMed] [Google Scholar]
- 5.Fonkoua, M. C., M. K. Taha, P. Nicolas, P. Cunin, J. M. Alonso, R. Bercion, J. Musi, and P. M. Martin. 2002. Recent increase in meningitis caused by Neisseria meningitidis serogroups A and W135, Yaounde, Cameroon. Emerg. Infect. Dis. 8:327-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hahne, S., S. Handford, and M. Ramsay. 2002. W135 meningococcal carriage in Hajj pilgrims. Lancet 360:2089-2090. [DOI] [PubMed] [Google Scholar]
- 7.Hahne, S. J., S. J. Gray, J. F. Aguilera, N. S. Crowcroft, T. Nichols, E. B. Kaczmarski, and M. E. Ramsay. 2002. W135 meningococcal disease in England and Wales associated with Hajj 2000 and 2001. Lancet 359:582-583. [DOI] [PubMed] [Google Scholar]
- 8.Kellogg, D. S., Jr., W. L. Peacock, Jr., W. E. Deacon, L. Brown, and D. I. Pirkle. 1963. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J. Bacteriol. 85:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kwara, A., R. A. Adegbola, P. T. Corrah, M. Weber, M. Achtman, G. Morelli, D. A. Caugant, and B. M. Greenwood. 1998. Meningitis caused by a serogroup W135 clone of the ET-37 complex of Neisseria meningitidis in West Africa. Trop. Med. Int. Health 3:742-746. [DOI] [PubMed] [Google Scholar]
- 10.Lapeyssonnie, L. 1968. Comparative epidemiologic study of meningococcic cerebrospinal meningitis in temperate regions and in the meningitis belt in Africa. Attempt at synthesis. Med. Trop. (Marseille) 28:709-720. [PubMed] [Google Scholar]
- 11.Liu, T. Y., E. C. Gotschlich, F. T. Dunne, and E. K. Jonssen. 1971. Studies on the meningococcal polysaccharides. II. Composition and chemical properties of the group B and group C polysaccharide. J. Biol. Chem. 246:4703-4712. [PubMed] [Google Scholar]
- 12.Liu, T. Y., E. C. Gotschlich, E. K. Jonssen, and J. R. Wysocki. 1971. Studies on the meningococcal polysaccharides. I. Composition and chemical properties of the group A polysaccharide. J. Biol. Chem. 246:2849-2858. [PubMed] [Google Scholar]
- 13.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayer, L. W., M. W. Reeves, N. Al-Hamdan, C. T. Sacchi, M. K. Taha, G. W. Ajello, S. E. Schmink, C. A. Noble, M. L. Tondella, A. M. Whitney, Y. Al-Mazrou, M. Al-Jefri, A. Mishkhis, S. Sabban, D. A. Caugant, J. Lingappa, N. E. Rosenstein, and T. Popovic. 2002. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain but clonal expansion within the electrophoretic type-37 complex. J. Infect. Dis. 185:1596-1605. [DOI] [PubMed] [Google Scholar]
- 15.Molling, P., A. Backman, P. Olcen, and H. Fredlund. 2001. Comparison of serogroup W-135 meningococci isolated in Sweden during a 23-year period and those associated with a recent Hajj pilgrimage. J. Clin. Microbiol. 39:2695-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popovic, T., C. T. Sacchi, M. W. Reeves, A. M. Whitney, L. W. Mayer, C. A. Noble, G. W. Ajello, F. Mostashari, N. Bendana, J. Lingappa, R. Hajjeh, and N. E. Rosenstein. 2000. Neisseria meningitidis serogroup W135 isolates associated with the ET-37 complex. Emerg. Infect. Dis. 6:428-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taha, M. K., M. Achtman, J. M. Alonso, B. Greenwood, M. Ramsay, A. Fox, S. Gray, and E. Kaczmarski. 2000. Serogroup W135 meningococcal disease in Hajj pilgrims. Lancet 356:2159. [DOI] [PubMed] [Google Scholar]
- 18.Taha, M. K., A. Antignac, P. Renault, A. Perrocheau, D. Levy-bruhl, P. Nicolas, and J. M. Alonso. 2001. Clonal spread of Neisseria meningitidis W135. Presse Med. 30:1535-1538. [PubMed] [Google Scholar]
- 19.Taha, M. K., A. E. Deghmane, A. Antignac, M. L. Zarantonelli, M. Larribe, and J. M. Alonso. 2002. The duality of virulence and transmissibility in Neisseria meningitidis. Trends Microbiol. 10:376-382. [DOI] [PubMed] [Google Scholar]
- 20.Taha, M. K., I. Parent Du Chatelet, M. Schlumberger, I. Sanou, S. Djibo, F. de Chabalier, and J. M. Alonso. 2002. Neisseria meningitidis serogroups W135 and A were equally prevalent among meningitis cases occurring at the end of the 2001 epidemics in Burkina Faso and Niger. J. Clin. Microbiol. 40:1083-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.World Health Organization. 2002. Meningococcal disease, serogroup W135, Burkina Faso: preliminary report 2002. Wkly. Epidemiol. Rec. 77:152-155. [PubMed] [Google Scholar]
- 22.Zhu, P., A. van der Ende, D. Falush, N. Brieske, G. Morelli, B. Linz, T. Popovic, I. G. Schuurman, R. A. Adegbola, K. Zurth, S. Gagneux, A. E. Platonov, J. Y. Riou, D. A. Caugant, P. Nicolas, and M. Achtman. 2001. Fit genotypes and escape variants of subgroup III Neisseria meningitidis during three pandemics of epidemic meningitis. Proc. Natl. Acad. Sci. USA 98:5234-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]