Abstract
Genetic and antigenic characterizations of 70 strains of adenovirus type 41 (Ad41), isolated between 1998 and 2001 from children in Japan, Vietnam, and Korea, were done by DNA restriction enzyme (RE) analysis, sequencing analysis, and monoclonal antibody (MAb)-based enzyme-linked immunosorbent assay (ELISA). Eight genome types were observed in the present study, among which D25, D26, D27, and D28 were novel genome types. These eight genome types were divided into two genome-type clusters (GTCs) based on phylogenetic analysis of the hypervariable regions (HVRs) of the hexon. GTC1 includes D1, D25, D26, D27, and D28, and the GTC2 contains D4, D12, and D22. The amino acid homologies among the members within a GTC were 97 to 100%, whereas between the members of different GTCs the homologies were 92 to 94%. The specificity of the GTC classification was confirmed by ELISA with MAb 1F, which was selected by the Ad41 prototype Tak strain. It was found that only the isolates of GTC1 but not of GTC2 reacted with MAb 1F. These results suggest that Ad41 isolates from the three countries should be classified into two subtypes. The accumulation of amino acid mutations located in HVRs of hexon are indicative for the classification of Ad41 subtype.
Adenoviruses are responsible for a wide range of disease symptoms. To date, there are 51 recognized serotypes of human adenovirus that are classified into six subgenera (A to F) based on several antigenic, morphological and molecular criteria. Among them, subgenus F, represented by the two serotypes adenovirus type 40 (Ad40) and Ad41, is associated with diarrhea in children; it is found in 1 to 20% of fecal specimens from children with acute gastroenteritis (3, 9, 12, 15, 22, 35). The two serotypes (Ad40 and Ad41) of subgenus F are termed enteric adenovirus (EAd) because of their tropism for the gastrointestinal tract (10, 32). The subgenus F adenoviruses grow poorly in most cell culture systems, in contrast to other cultivable “nonenteric” adenoviruses, and are therefore also termed “noncultivable” or “fastidious” adenovirus (7, 16).
Earlier surveys had shown that the occurrences of Ad40 and Ad41 are approximately equal (6, 11). However, several studies have recently reported a decrease in the rate of isolation of Ad40 and a concomitant increase in the rate of isolation of Ad41 (5, 8, 25, 34). After 1986, Ad41 infection became dominant over Ad40. Our previous studies confirmed that Ad41 was the prevailing serotype of adenovirus associated with acute diarrhea among children also in Japan, Vietnam, and Korea (19). The change in prevalence of Ad41 might have been caused by an antigenic drift, thus increasing the incidence of infection in susceptible individuals (3). Therefore, it is important to characterize these Ad41 strains. In the last decade, there were relatively few comprehensive epidemiological studies of subgenus F adenoviruses from children with diarrhea in Asian countries. The objectives of the present study were (i) to describe the genome types of Ad41 isolates by DNA restriction endonucleases (REs) analysis in Japan, Vietnam, and Korea and (ii) to describe the genetic and antigenic characterization of different genome types of Ad41 isolates in these three countries.
MATERIALS AND METHODS
Fecal specimens.
A total of 3,577 fecal specimens were collected from children between 1 month to 15 years of age with acute diarrhea from Japan, Vietnam, and Korea. These specimens included 1,991 specimens from Tokyo, Osaka, Saga, Maizuru, and Sapporo in Japan collected between July 1998 and June 2001; 1,355 specimens from Ho Chi Minh City in Vietnam collected between December 1999 and November 2000; and 231 specimens from Seoul in Korea collected between January 1998 and July 1999. By using polyclonal antibody-based enzyme-linked immunosorbent assay (ELISA) (24) and the combination of PCR and restriction fragment length polymorphism methods (31), adenoviruses were detected and serotyped. A total of 158 adenovirus-positive specimens were identified. Of these specimens, 101 specimens were typed as Ad41, the most common serotype, followed by Ad40 (12 isolates), Ad8 (12 isolates), Ad2 (10 isolates), Ad3 (10 isolates), Ad5 (8 isolates), and Ad31 (1 isolate). Four specimens remained untypeable. These Ad41 specimens were used for analysis in the present study.
DNA RE analysis. (i) Virus isolation.
Ad41 prototype strain Tak was obtained from the National Institute of Infectious Diseases of Japan. Ad41 specimens were isolated and propagated in Caco-2 cells (human colonic carcinoma cells). The inoculation was performed as previously described (26). The cultured fluids were harvested after infection at the time of maximal cytopathic effect (CPE).
(ii) Extraction of viral DNA.
A total of 8 ml of the cultured fluids were centrifuged at 1,353 × g for 30 min, and the cells were pelleted. In order to avoid mixing of cellular nucleic acid in the subsequent extraction of viral DNA, ultracentrifugation of the cultured supernatant was performed (40,700 × g for 3 h). After centrifugation, the supernatant was removed, and the viral pellet was obtained. Subsequent extraction was conducted by a modified Hirt method (44).
(iii) Digestion of DNA by RE.
Approximately 1 μg of viral DNA was digested with 10 U of the REs BamHI, BglI, BstEII, EcoRI, HindIII, KpnI, PstI, SacI, SmaI, and XhoI (TaKaRa Biotechnology Co., Ltd.) according to the manufacturer's instructions. After digestion, reaction products were loaded onto 1.0% agarose gel containing ethidium bromide (0.5 μg/ml) and run at 50 V for 5 h in Tris-acetate (TAE) buffer. The gel was photographed under UV light. The genome typing system proposed by Adrian et al. (1) was used as the basis for our study. By comparing the resulting patterns with the published restriction profiles of prototype strains, genome type identifications could be achieved.
PCR sequencing analysis of HVRs.
In order to understand the genetic relationships between different genome types of Ad41 strains isolated from the three countries in the present study, eight Ad41 representing strains were randomly selected from eight genome types, and their nucleotide sequences of hypervariable regions (HVRs) were determined.
Viral DNA diluted 1:100 was used as a template DNA for PCR. The primers were designed according to the method of Takeuchi et al. (37). In the first PCR, a pair of primers, HX5-3 and HX3-4, was used. The PCR product of 1,800 bp contained all seven HVRs. The second PCR was conducted by using the first PCR product as a template with a combination of the following two pairs of primers: S-29 and S-52 or S-51 and S-53. The primer pairs used are listed in Table 1. The cycling conditions of the PCR were modified in 35 cycles (94°C for 1 min, annealing at 45°C for 2 min, and primer extension at 72°C for 3 min, with a final product extension at 72°C for 7 min). The second PCR products were extracted from 1% agarose gel by using phenol and chloroform. The purified DNA was used to perform enzymatic extension reaction for DNA sequencing with a DNA sequencing kit (BigDye Terminate v3.0 cycle sequencing ready reaction; ABI Prism; Applied Biosystems, Warrington, Great Britain). After the reaction products were cleaned, samples were analyzed by autosequencer (ABI Prism 310 genetic analyzer). Evaluation of all sequence data and analysis of identity were conducted by using DNASIS software (version 1994; Hitachi Software Engineering Co., Ltd., Tokyo, Japan). The sequence alignments of nucleotide and deduced amino acid were carried out by DDBJ (http://www.ddbj.nig.ac.jp) by using the CLUSTAL W program. The phylogenetic tree was constructed with MEGA version 2.1 (18) by using the neighbor-joining method and the bootstrap test (http://www.megasoftware.net/).
TABLE 1.
Oligonucleotide primers for PCR sequencing
| Primer | Polarity | Sequence (5′-3′) | Positiona |
|---|---|---|---|
| HX5-3 | + | cac atc gcc gga cag gat gct tcg gag ta | 40-68 |
| HX3-4 | − | gtg ttg tga gcc atg ggg aag aag gtg gc | 1819-1847 |
| S-29 | + | gcc agc acr twc ttt gac at | 289-308 |
| S-52 | − | ccc atg ttg cca gtg ctg ttg tar tac a | 986-1013 |
| S-51 | + | ccc aac aga ccc aay tac | 937-956 |
| S-53 | − | aag ggg ttg acg ttg tcc at | 1555-1574 |
The sequence positions are based on the Ad3 hexon region.
Nucleotide sequence accession numbers.
Sequence data from the present study were entered in the GenBank/EMBL/DDBJ database under the following accession numbers: Ad41-D1-VN47 (AB103341), Ad41-D4-VN28 (AB103342), Ad41-D12-JP3171 (AB103343), Ad41-D22-Km079 (AB103344), Ad41-D26-JP3106 (AB103345), Ad41-D27-JP2149 (AB103346), Ad41-D25-Ks35 (AB103347), and Ad41-D28-VN1020 (AB103348).
ELISA based on MAb.
ELISA based on the use of monoclonal antibodies (MAbs) was utilized to examine the antigen specificity of Ad41 strains. ELISA was performed as described by Nishio et al. (24). Anti-Ad40 rabbit serum was used as capture antibody. The MAbs 15D (adenovirus group-specific), 12D (Ad40 type specific), and 1F (Ad41 type specific) were used as detector antibodies. In each test, cell culture supernatants of Ad41 prototype strain were included as positive controls, and phosphate-buffered saline was used as a negative control. If the optical density value was >0.2, as well as 2-fold greater than the negative control well, the specimen was considered positive.
RESULTS
Genome types of Ad41 isolates from three countries.
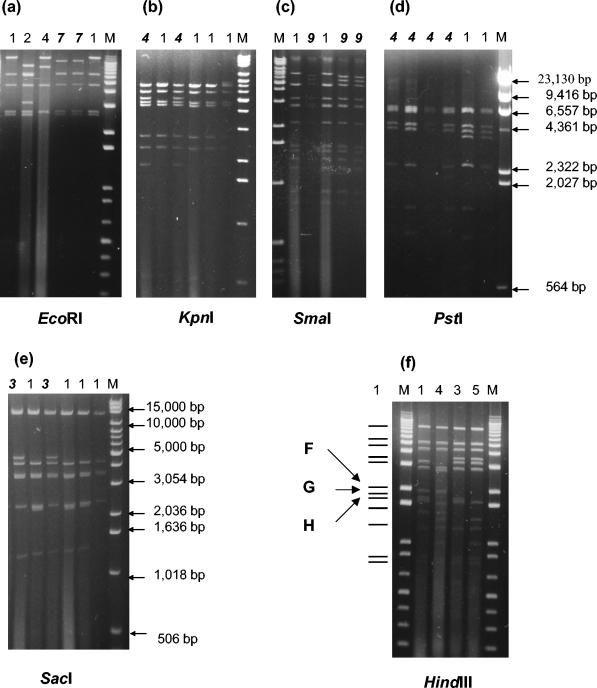
Seventy Ad41 fecal specimens were isolated successfully, and RE analyses were performed with 10 different REs: BamHI, BglI, BstEII, EcoRI, HindIII, KpnI, PstI, SacI, SmaI, and XhoI. To confirm the genome type, restriction patterns were compared to that of the Ad41 prototype Tak strain. The results of enzyme code and genome type determinations obtained in the present study are shown in Table 2. New restriction patterns were observed in cleavage with enzyme EcoRI, KpnI, PstI, SacI, and SmaI. According to the nomenclature system, the new patterns were named as follows: code 7 of EcoRI, code 4 of KpnI, code 4 of PstI, code 3 of SacI, and code 9 of SmaI (Fig. 1a to e).
TABLE 2.
Genome types of Ad41 isolates in Japan, Vietnam, and Korea
| Country | Genome typeb | Enzyme codea
|
Total no. of isolates | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BamHI | BglI | BstEII | EcoRI | HindIII | KpnI | PstI | SacI | SmaI | XhoI | |||
| Japan | D26 | 2 | 2 | 1 | 2 | 1 | 4 | 1 | 3 | 4 | 2 | 26 |
| D12 | 2 | 1 | 2 | 1 | 5 | 1 | 1 | 1 | 2 | 1 | 17 | |
| D27 | 2 | 2 | 1 | 7 | 1 | 1 | 1 | 1 | 4 | 2 | 4 | |
| Vietnam | D28 | 2 | 1 | 4 | 4 | 4 | 2 | 4 | 1 | 9 | 1 | 10 |
| D4 | 3 | 1 | 1 | 1 | 3 | 1 | 1 | 1 | 4 | 1 | 4 | |
| D1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | |
| Korea | D22 | 2 | 1 | 2 | 1 | 5 | 1 | 3 | 1 | 2 | 1 | 4 |
| D25 | 1 | 2 | 1 | 2 | 1 | 4 | 1 | 3 | 4 | 2 | 1 | |
| D26 | 2 | 2 | 1 | 2 | 1 | 4 | 1 | 3 | 4 | 2 | 2 | |
The boldface numbers indicate a novel pattern.
Novel genome types are indicated in boldface.
FIG. 1.
Novel restriction patterns with EcoRI (a), KpnI (b), PstI (d), SacI (e), and SmaI (c). The numbers above the lanes are the enzyme code; lane M contains molecular weight standards (1-kb DNA extension ladder; Life Technologies), except for lane M of PstI. Lane M of PstI contains molecular weight standards (Marker II, λDNA HindIII digested; TaKaRa). Enzyme code “1” refers to the pattern of the prototype. The boldface italic numbers mark the novel patterns. (f) Restriction patterns with HindIII observed in the present study. The numbers above the lanes are the enzyme code; lane M contains molecular weight standards. The marker lines on the left give the model pattern of enzyme code 1. The arrowed letters F, G, and H refer to the names of the fragments according to the physical map given by Takiff et al. (40). The patterns of codes 3 and 5 lack fragment F.
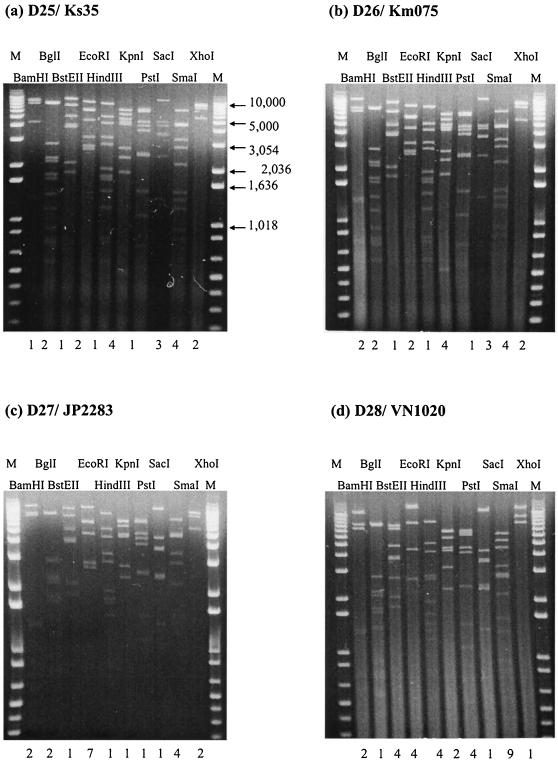
Analysis of 70 Ad41 isolates from three countries revealed the existence of eight genome types in the present study. Of these genome types, four (D1, D4, D12, and D22) were previously reported by van der Avoort et al. (43). In addition, four novel genome types D25, D26, D27, and D28 are reported here for the first time. The photographs of representative strains of these novel genome types cleaved by the panel endonucleases are shown in Fig. 2. They accounted for 61% (43 of 70) of all Ad41 isolates. The novel genome type D26 had enzyme code 2212141341 and was closely related to another novel genome type D25, differing only in cleavage with BamHI.
FIG. 2.
(a to d) Restriction profiles of novel Ad41 genome type strains isolated from Japan, Vietnam, and Korea, obtained in the present study. (a) D25/Ks35; (b) D26/Km075; (c) D27/JP2283; (d) D28/VN1020. The numbers under the lanes are the enzyme code; lane M contains molecular weight standards.
In Japan, the novel genome type D26 predominates with 26 isolates, followed by D12 and D27 with 17 and 4 isolates, respectively. In 3 years, D26 and D12 were observed every year. From July 1999 to June 2000, the new genome type D27 appeared, but it was not found the next year. In Vietnam, D28, represented by 10 isolates, was the most common. D4 and D1 were observed in 4 and 2 isolates, respectively. In Korea, D22 predominated with four isolates. Interestingly, the novel genome type D26 was observed in Korea as well. It was present in two isolates, whereas only one isolate of novel genome type D25 was detected.
Genetic analyses of HVRs for eight genome types of Ad41.
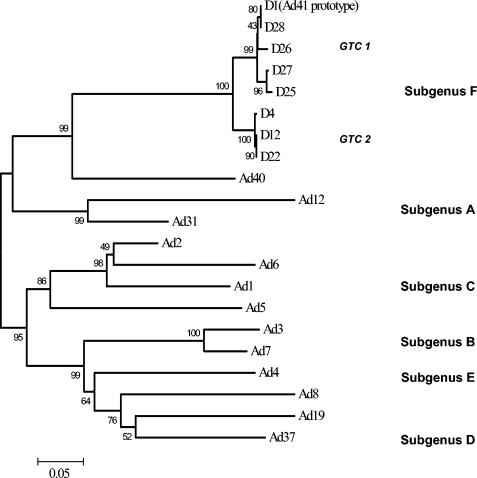
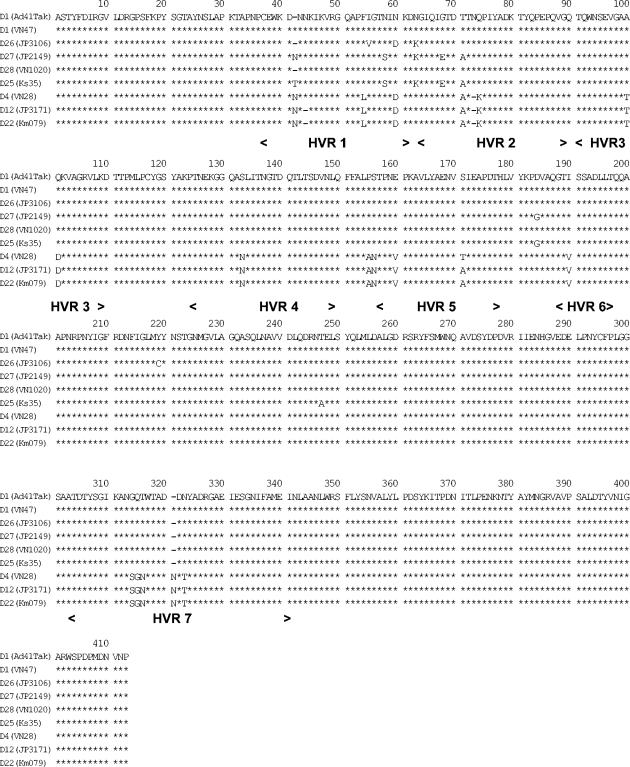
The HVRs of eight representative strains from eight genome types obtained in the present study were sequenced. The DNA sequences, containing seven hexon HVRs, with a total of 1,232 to 1,235 nucleotides, from the eight strains were determined. Alignments of the predicted amino acid sequences were performed. A phylogenetic tree based on the deduced amino acid sequences of eight representing genome types was subsequently constructed (Fig. 3). It revealed that in the eight genome types representing strains, two genome type clusters (GTCs) are present. GTC1 includes D1 (Ad41 prototype Tak strain), D28, D26, D27, and D25, and GTC2 consists of D4, D12, and D22. The amino acid homology rates in all seven HVRs among the eight representative strains are shown in Table 3. The highest amino acid homologies were observed within a GTC (97 to 100%). Between the members of different GTCs, the homologies were 92 to 94%. The alignments of HVRs predicted amino acid sequences are shown in Fig. 4. The same substitutions in amino acid (aa) 54 (HVR1), aa 74 (HVR2), aa 100 to 101 (HVR3), aa 133 (HVR4), aa 155 to 156, aa 160 (HVR5), aa 190 (HVR6), aa 314 to 316, and aa 323 (HVR7) were presented between GTC1 and GTC2 strains. The members of GTC2 have a common deletion in aa 73 and an insertion in aa 320.
FIG. 3.
Phylogenetic tree based on the deduced amino acid sequences of HVRs of Ad reference strains and Ad41 strains isolated from Japan, Vietnam, and Korea. Two GTCs, GTC1 and GTC2, are shown. The sequences of reference strains previously reported are listed in GenBank under the indicated accession numbers: Ad12 (X73487) (36), Ad31 (74661) (29), Ad1 (X67709) (28), Ad2 (J01917) (2), Ad5 (M73260) (17), Ad6 (X67710) (28), Ad3 (X76549) (30), Ad7 (Z48571) (21), Ad4 (X84646) (30), Ad8 (X74663) (29), Ad19 (X98359) (43), Ad37 (X98360) (43), Ad40 (X51782) (42), and Ad41 (X51783) (41).
TABLE 3.
Homology comparison of HVRs between Ad41 isolates and prototype strains Ad40 and Ad41
| HVR | % Homologya with HVR:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ad40 | D1 | D26 | D27 | D28 | D25 | D4 | D12 | D22 | |
| Ad40 (prototype) | 72 | 72 | 72 | 72 | 72 | 72 | 72 | 72 | |
| D1 (Ad41 prototype) | 68 | 98 | 98 | 100 | 98 | 94 | 95 | 95 | |
| D26 (JP3106) | 68 | 98 | 98 | 98 | 98 | 94 | 94 | 94 | |
| D27 (JP2149) | 67 | 98 | 99 | 98 | 98 | 94 | 94 | 94 | |
| D28 (VN1020) | 68 | 100 | 98 | 98 | 98 | 94 | 95 | 95 | |
| D25 (Ks35) | 67 | 98 | 97 | 99 | 98 | 94 | 94 | 94 | |
| D4 (VN28) | 66 | 94 | 93 | 93 | 94 | 92 | 99 | 99 | |
| D12 (JP3171) | 66 | 94 | 93 | 93 | 94 | 93 | 99 | 99 | |
| D22 (Km079) | 66 | 94 | 93 | 93 | 94 | 93 | 99 | 100 | |
The upper right data are the percent homologies of the nucleotide sequences; the lower left data are the percent homologies of the amino acid sequences.
FIG. 4.
Alignments of the deduced amino acid sequences of hexon, including seven HVRs from eight Ad41 genome type strains. The regions of seven HVRs are given. The borders of the seven HVRs are in accordance with the literature (37). Asterisks indicate the identical residues to the sequence of D1.
Antigen specificity of Ad41 isolates.
MAbs were used to examine the antigen specificity in all Ad41 isolates belonging to eight different genome types. All of these isolates reacted with MAb 15D; none of them reacted with MAb 12D. On the other hand, two antigenic subtypes among eight genome types were distinguished on the basis of the specificity of the MAb 1F selected by Ad41 prototype Tak strain. The isolates that belonged to genome types D1, D28, D26, D27, and D25, were recognized by MAb 1F, but the members of D4, D12, and D22 could not be distinguished with the same MAb 1F.
DISCUSSION
In this study, the genome types of Ad41 strains isolated from fecal specimens in Japan, Vietnam, and Korea were determined. In addition, the genetic and antigenic characterizations are given.
EAd are known as fastidious adenoviruses since they do not grow well in conventional cell lines. There are varied reports in the literature concerning the ability of EAd to replicate in different cell lines, such as Chang conjunctival cells, Hep-2 cells, 293 cells, and PLC/PRF/5 cells. The 293 cells, an Ad5-transformed HEK cell line that contains and expresses the Ad5 early region E1A and E1B, were considered useful for the growth of many Ad41 strains (40), but infection yields are still low. Recently, Pinto et al. (27) have reported the use of Caco-2 cell line in isolating laboratory strains of human enteric viruses, including the group A rotavirus type 3, astrovirus serotype 1, and Ad5, Ad40, and Ad41. In their study, for EAd, apparent CPE was observed with laboratory strains, whereas wild-type Ad40 and Ad41 strains failed to induce CPE in Caco-2 cells, though the amplification of viral nucleic acid was confirmed by dot blot hybridization. In our study, Caco-2 cells were used to propagate EAd from fecal specimens. Seventy Ad41 and ten Ad40 (unpublished data) strains from clinical specimens were successfully isolated. The present study confirmed that Caco-2 cells are a useful cell line for research on diarrheal viruses.
Among these genome types observed in Japan, Vietnam, and Korea, four novel genome types (D25, D26, D27, and D28) were observed for the first time, and the other four genome types (D1, D4, D12 and D22) were reported in earlier studies (13, 14, 43). Genome type D12 was present in 17 isolates in Japan over a period of 3 years. This genome type was first isolated in 1982 from The Netherlands and was highly prevalent in the 1980s (43). In Sweden, an identical D12 was also reported, specimens of which were mainly obtained from an outbreak of gastroenteritis in a long-term pediatric ward at a hospital of Stockholm in 1988; it was then found sporadically for several years afterward (13). Another genome type D22 observed in Korea was first isolated during 1984 in The Netherlands, too (43). In the present study, genome type D4 identified in Ho Chi Minh City, Vietnam, was also first found in Malaysia in 1979 (14). These phenomena may indicate that genome types of Ad41 have a scattered global distribution. It is quite possible that the novel genome types observed in the present study are also present in Europe and other continents. Another possibility is that, because of limited research work these novel genome types were not detected.
In studying the profiles of Ad41 isolates obtained with 10 restriction enzymes in three countries, it was found that almost all restriction patterns that appeared in Korean strains were also present in Japanese strains and, in contrast, Vietnamese isolates always exhibited different patterns. For example, Japanese and Korean Ad41 isolates have the patterns 1 and 5 upon cleavage with enzyme HindIII, but Vietnamese strains showed patterns 3 and 4, along with pattern 1. In addition, novel genome type D26 was observed in both Japan and Korea. These data might represent a gradient of antigenic drift reflecting increased travel activities between the two countries.
The occurrences of novel genome types reflect the proceeding mutation of viral DNA. Hexons are important capsid proteins of adenovirus. It was reported that seven HVRs located in hexons take part in the neutralization reaction and contain the epitopes of serotype determination (4). It is interesting to estimate what role these regions might have in identical adenovirus serotypes. Previous studies revealed that hexon contains HVRs that are conserved in different genome types of Ad3 and Ad7 belonging to the subgenus B (20, 38). In the present study, the sequence analyses of HVRs of different genome types of Ad41 isolates from three countries also demonstrated a high level of conservation. The homologies ranged from 92 to 100% and the eight genome types of Ad41 were divided into two clusters according to the predicted amino acid sequences of HVRs. The classification of GTCs had a curious coincidence with the result of ELISA with MAb 1F. It has been confirmed that the MAb 1F may react with hexon protein of Ad41 (24). Therefore, the results presented here suggest that HVRs of hexons may contain epitopes that are associated with the Ad41 subtype.
In comparing the ten enzyme codes, one difference can be found in cleavage with HindIII between GTC1 and GTC2. The members of GTC1 have the HindIII code 1 and 4; the members of GTC2 have codes 3 and 5. Physical map of Ad41 was published by Takiff et al. (39). Tak strain HindIII fragments F and H were determined to lie between map units 51 and 60, which is within the confines of the hexon gene. van der Avoort et al. (43) revealed that D4, D12, and D22 could not react with MAbs 1-23, 3-10, 3-18, and 7-14 and lacked fragment F in common (Fig. 1f). However, in the present study, the nucleotide sequence analysis of hexon-containing HVRs confirmed that the guanine-for-adenine substitution in nucleotide 696, which leads to failure of HindIII-cut viral DNA of GTC2 isolates, was a conservative substitution.
Recently, ELISA with MAbs specifically reacting with Ad40 and Ad41 is widely used for the diagnosis of EAd in fecal specimens. It is a rapid, readily available means for identification of Ad40 and Ad41. MAb 1F was used successfully for screening Ad41 in clinical specimens (24). In the present study, the Ad41 isolates belonging to genome types D4, D12, and D22 did not react with MAb 1F. The application limitation of MAbs was reported in earlier studies (5, 23, 33, 43). Due to virus strain mutation, the MAb may be safely used for screening purposes only temporarily. The genetic variation within one type of adenovirus can evidently affect the diagnosis capability of highly specific MAbs. It has been reported that a commercial MAb-based ELISA could not demonstrate the presence of a highly prevalent genome type of Ad41 in Canada (33). In South Africa, the locally circulating adenoviruses were not detected with a commercial kit utilizing MAbs directed against type-specific epitopes on the adenovirus hexon (23). For Japanese specimens, MAb 1F distinguished all of the Ad41 strains isolated from 1982 to 1988 (24) but failed to react with some Ad41 isolates in 1998 to 2001. These results also confirmed that antigenic drift on the surface of adenovirus occurred frequently. Hexon and fiber compose most of the external capsid of adenovirus. It may be difficult to find other, better targets for reagent antibodies except the hexon and fiber. The use of pooled MAbs to hexon may provide a practicable way to avoid omissions in detection such as the ones described above when a single MAb was used.
In conclusion, eight genome types of Ad41 were found in Japan, Vietnam, and Korea. Four novel genome types were investigated for the first time in the present study. The existence of two GTCs and the antigenic difference between them may reflect the accumulation of amino acid mutations located in HVRs of the hexon. These amino acid mutations may affect classification of the Ad41 subtype. These data contribute to our understanding of the molecular epidemiological characterization of adenoviruses in these countries. Further studies are needed to obtain additional established strains of the Ad41 genome types in order to clarify the genetic and antigenic relationships between them.
Acknowledgments
The grant sponsor of this study was the Asian Development Bank-Japan Scholarship Program. This study was supported by grants-in-aid from the Ministry of Education and the Ministry of Health, Labor, and Welfare of Japan. This study was also supported by Japan Food Hygiene Association and the Sumitomo Foundation.
REFERENCES
- 1.Adrian, T., B. Best, and R. Wigand. 1985. A proposal for naming adenovirus genome types, exemplified by adenovirus type 6. J. Gen. Virol. 66:2685-2691. [DOI] [PubMed] [Google Scholar]
- 2.Akusjarvi, G., P. Alestrom, M. Pettersson, M. Lager, H. Jornvall, and U. Pettersson. 1984. The gene for the adenovirus 2 hexon polypeptide. J. Biol. Chem. 259:13976-13979. [PubMed] [Google Scholar]
- 3.Brown, M. 1990. Laboratory identification of adenoviruses associated with gastroenteritis in Canada from 1983 to 1986. J. Clin. Microbiol. 28:1525-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford-Miksza, L., and D. P. Schnurr. 1996. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J. Virol. 70:1836-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Jong, J. C., K. Bijlsma, A. G. Wermenbol, M. W. Verweij-Uijterwaal, H. G. van der Avoort, D. J. Wood, A. S. Bailey, and A. D. Osterhaus. 1993. Detection, typing, and subtyping of enteric adenoviruses 40 and 41 from fecal samples and observation of changing incidences of infections with these types and subtypes. J. Clin. Microbiol. 31:1562-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Jong, J. C., R. Wigand, A. H. Kidd, G. Wadell, J. G. Kapsenberg, C. J. Muzerie, A. G. Wermenbol, and R. G. Firtzlaff. 1983. Candidate adenoviruses 40 and 41: fastidious adenoviruses from human infant stool. J. Med. Virol. 11:215-231. [DOI] [PubMed] [Google Scholar]
- 7.Gary, G. W., Jr., J. C. Hierholzer, and R. E. Black. 1979. Characteristics of noncultivable adenoviruses associated with diarrhea in infants: a new subgroup of human adenoviruses. J. Clin. Microbiol. 10:96-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimwood, K., R. Carzino, G. L. Barnes, and R. F. Bishop. 1995. Patients with enteric adenovirus gastroenteritis admitted to an Australian pediatric teaching hospital from 1981 to 1992. J. Clin. Microbiol. 33:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrmann, J. E., N. R. Blacklow, D. M. Perron-Henry, E. Clements, D. N. Taylor, and P. Echeverria. 1988. Incidence of enteric adenoviruses among children in Thailand and the significance of these viruses in gastroenteritis. J. Clin. Microbiol. 26:1783-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobsson, P. A., M. E. Johansson, and G. Wadell. 1979. Identification of an enteric adenovirus by immunoelectroosmophoresis (IEOP) technique. J. Med. Virol. 3:307-312. [DOI] [PubMed] [Google Scholar]
- 11.Jarecki-Khan, K., S. R. Tzipori, and L. E. Unicomb. 1993. Enteric adenovirus infection among infants with diarrhea in rural Bangladesh. J. Clin. Microbiol. 31:484-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarecki-Khan, K., and L. E. Unicomb. 1992. Seroprevalence of enteric and nonenteric adenoviruses in Bangladesh. J. Clin. Microbiol. 30:2733-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson, M. E., M. A. Andersson, and P. A. Thorner. 1994. Adenoviruses isolated in the Stockholm area during 1987-1992: restriction endonuclease analysis and molecular epidemiology. Arch. Virol. 137:101-115. [DOI] [PubMed] [Google Scholar]
- 14.Kidd, A. H. 1984. Genome variants of adenovirus 41 (subgroup G) from children with diarrhoea in South Africa. J. Med. Virol. 14:49-59. [DOI] [PubMed] [Google Scholar]
- 15.Kidd, A. H., B. P. Cosgrove, R. A. Brown, and C. R. Madeley. 1982. Faecal adenoviruses from Glasgow babies: studies on culture and identity. J. Hyg. 88:463-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidd, A. H., and C. R. Madeley. 1981. In vitro growth of some fastidious adenoviruses from stool specimens. J. Clin. Pathol. 34:213-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kinloch, R., N. Mackay, and V. Mautner. 1984. Adenovirus hexon: sequence comparison of subgroup C serotypes 2 and 5. J. Biol. Chem. 259:6431-6436. [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Arizona State University, Tempe. [DOI] [PubMed]
- 19.Li, L. 2003. Molecular epidemiological study of adenovirus among children with acute gastroenteritis in Japan, Vietnam, and Korea. M.S. thesis. University of Tokyo, Tokyo, Japan.
- 20.Li, Q., and G. Wadell. 1999. Genetic variability of hexon loops 1 and 2 between seven genome types of adenovirus serotype 7. Arch. Virol. 144:1739-1749. [DOI] [PubMed] [Google Scholar]
- 21.Li, Q. G., K. Lindman, and G. Wadell. 1997. Hydropathic characteristics of adenovirus hexons. Arch. Virol. 142:1307-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore, P., A. D. Steele, G. Lecatsas, and J. J. Alexander. 1998. Characterisation of gastroenteritis-associated adenoviruses in South Africa. S Afr. Med. J. 88:1587-1592. [PubMed] [Google Scholar]
- 23.Moore, P. L., A. D. Steele, and J. J. Alexander. 2000. Relevance of commercial diagnostic tests to detection of enteric adenovirus infections in South Africa. J. Clin. Microbiol. 38:1661-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nishio, O., M. Ooseto, K. Takagi, Y. Yamasita, Y. Ishihara, and S. Isomura. 1990. Enzyme-linked immunosorbent assay employing monoclonal antibodies for direct identification of enteric adenoviruses (Ad40,41) in feces. Microbiol. Immunol. 34:871-877. [DOI] [PubMed] [Google Scholar]
- 25.Noel, J., A. Mansoor, U. Thaker, J. Herrmann, D. Perron-Henry, and W. D. Cubitt. 1994. Identification of adenoviruses in faeces from patients with diarrhoea at the Hospitals for Sick Children, London, 1989-1992. J. Med. Virol. 43:84-90. [DOI] [PubMed] [Google Scholar]
- 26.Pieniazek, D., N. J. Pieniazek, D. Macejak, J. Coward, M. Rayfield, and R. B. Luftig. 1990. Differential growth of human enteric adenovirus 41 (TAK) in continuous cell lines. Virology 174:239-249. [DOI] [PubMed] [Google Scholar]
- 27.Pinto, R. M., J. M. Diez, and A. Bosch. 1994. Use of the colonic carcinoma cell line CaCo-2 for in vivo amplification and detection of enteric viruses. J. Med. Virol. 44:310-315. [DOI] [PubMed] [Google Scholar]
- 28.Pring-Akerblom, P., and T. Adrian. 1993. The hexon genes of adenoviruses of subgenus C: comparison of the variable regions. Res. Virol. 144:117-127. [DOI] [PubMed] [Google Scholar]
- 29.Pring-Akerblom, P., and T. Adrian. 1994. Type- and group-specific polymerase chain reaction for adenovirus detection. Res. Virol. 145:25-35. [DOI] [PubMed] [Google Scholar]
- 30.Pring-Akerblom, P., F. E. Trijssenaar, and T. Adrian. 1995. Sequence characterization and comparison of human adenovirus subgenus B and E hexons. Virology 212:232-236. [DOI] [PubMed] [Google Scholar]
- 31.Saitoh-Inagawa, W., A. Oshima, K. Aoki, N. Itoh, K. Isobe, E. Uchio, S. Ohno, H. Nakajima, K. Hata, and H. Ishiko. 1996. Rapid diagnosis of adenoviral conjunctivitis by PCR and restriction fragment length polymorphism analysis. J. Clin. Microbiol. 34:2113-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schofield, K. P., D. J. Morris, A. S. Bailey, J. C. de Jong, and G. Corbitt. 1994. Gastroenteritis due to adenovirus type 41 in an adult with chronic lymphocytic leukemia. Clin. Infect. Dis. 19:311-312. [DOI] [PubMed] [Google Scholar]
- 33.Scott-Taylor, T., G. Ahluwalia, B. Klisko, and G. W. Hammond. 1990. Prevalent enteric adenovirus variant not detected by commercial monoclonal antibody enzyme immunoassay. J. Clin. Microbiol. 28:2797-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott-Taylor, T. H., and G. W. Hammond. 1995. Local succession of adenovirus strains in pediatric gastroenteritis. J. Med. Virol. 45:331-338. [DOI] [PubMed] [Google Scholar]
- 35.Soares, C. C., E. M. Volotao, M. C. Albuquerque, F. M. da Silva, T. R. de Carvalho, C. M. Nozawa, R. E. Linhares, and N. Santos. 2002. Prevalence of enteric adenoviruses among children with diarrhea in four Brazilian cities. J. Clin. Virol. 23:171-177. [DOI] [PubMed] [Google Scholar]
- 36.Sprengel, J., B. Schmitz, D. Heuss-Neitzel, C. Zock, and W. Doerfler. 1994. Nucleotide sequence of human adenovirus type 12 DNA: comparative functional analysis. J. Virol. 68:379-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeuchi, S., N. Itoh, E. Uchio, K. Aoki, and S. Ohno. 1999. Serotyping of adenoviruses on conjunctival scrapings by PCR and sequence analysis. J. Clin. Microbiol. 37:1839-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuchi, S., A. Oshima, N. Itoh, N. Kitamura, E. Uchio, K. Aoki, and S. Ohno. 1998. Analysis of adenovirus type 7 hexon hypervariable region. Nippon Ganka Gakkai Zasshi. 102:570-575. (In Japanese.) [PubMed] [Google Scholar]
- 39.Takiff, H. E., W. Reinhold, C. F. Garon, and S. E. Straus. 1984. Cloning and physical mapping of enteric adenoviruses (candidate types 40 and 41). J. Virol. 51:131-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiemessen, C. T., and A. H. Kidd. 1995. The subgroup F adenoviruses. J. Gen. Virol. 76(Pt. 3):481-497. [DOI] [PubMed] [Google Scholar]
- 41.Toogood, C. I., and R. T. Hay. 1988. DNA sequence of the adenovirus type 41 hexon gene and predicted structure of the protein. J. Gen. Virol. 69(Pt. 9):2291-2301. [DOI] [PubMed] [Google Scholar]
- 42.Toogood, C. I., R. Murali, R. M. Burnett, and R. T. Hay. 1989. The adenovirus type 40 hexon: sequence, predicted structure and relationship to other adenovirus hexons. J. Gen. Virol. 70(Pt. 12):3203-3214. [DOI] [PubMed] [Google Scholar]
- 43.van der Avoort, H. G., A. G. Wermenbol, T. P. Zomerdijk, J. A. Kleijne, J. A. van Asten, P. Jensma, A. D. Osterhaus, A. H. Kidd, and J. C. de Jong. 1989. Characterization of fastidious adenovirus types 40 and 41 by DNA restriction enzyme analysis and by neutralizing monoclonal antibodies. Virus Res. 12:139-157. [DOI] [PubMed] [Google Scholar]
- 44.Wadell, G., and J. C. de Jong. 1980. Restriction endonucleases in identification of a genome type of adenovirus 19 associated with keratoconjunctivitis. Infect. Immun. 27:292-296. [DOI] [PMC free article] [PubMed] [Google Scholar]