Abstract
The findings of recent studies addressing the molecular characteristics of Mycobacterium tuberculosis complex isolates have initiated a discussion on the classification of M. africanum, especially of those isolates originating from East Africa (cluster F, subtype II) and displaying phenotypic and biochemical characteristics more similar to those of M. tuberculosis. To further address this question, we analyzed a representative collection of 63 M. tuberculosis complex strains comprising 30 M. africanum subtype I strains, 20 M. africanum subtype II strains, 10 randomly chosen M. tuberculosis isolates, and type strains of M. tuberculosis, M. bovis, and M. africanum for the following biochemical and molecular characteristics: single-nucleotide polymorphisms (SNPs) in gyrB and narGHJI and the presence or absence of RD1, RD9, and RD12. For all molecular markers analyzed, subtype II strains were identical to the M. tuberculosis strains tested. In contrast, the subtype I strains as well as the M. africanum type strain showed unique combinations of SNPs in gyrB and genomic deletions (the absence of RD9 and the presence of RD12), which proves their independence from M. tuberculosis and M. bovis. Accordingly, all subtype I strains displayed main biochemical characteristics included in the original species description of M. africanum. We conclude that the isolates from West Africa were proved to be M. africanum with respect to the phenotypic and genetic markers analyzed, while the isolates from East Africa must be regarded as phenotypic variants of M. tuberculosis (genotype Uganda). We propose the addition of the molecular characteristics defined here to the species description of M. africanum, which will allow clearer species differentiation in the future.
Mycobacterium africanum is a member of the M. tuberculosis complex (MTBC), which comprises the closely related species M. tuberculosis, M. bovis, M. africanum, M. microti, and M. canettii (24, 27). The close relationship of these species has been confirmed by DNA-DNA hybridization, multilocus enzyme electrophoresis, and sequencing of the 16S rRNA gene (rDNA) and the 16S to 23S rDNA internal transcribed spacer region (5, 6, 11, 14, 27). However, despite this high degree of similarity at the DNA level, the members of MTBC differ in their host ranges, geographical prevalences, and pathogenicities (27). Hence, the accurate species differentiation of clinical isolates remains necessary for epidemiological and public health purposes.
Since its first description in 1968 (3), M. africanum has been found in several regions of Africa, where it represents up to 60% of clinical strains from patients with pulmonary tuberculosis (TB) (16, 17, 20, 26). Recent surveys showed highly variable prevalences of M. africanum in different regions of Africa; e.g., approximately 5% of patients with TB from the Ivory Coast, approximately 10% of patients with TB from Cameroon (17), and at least 60% of patients with TB from Guinea-Bissau (1, 8) were found to be infected with M. africanum.
In contrast to M. tuberculosis and M. bovis, M. africanum strains show higher degrees of variability in their phenotypic attributes, which comprise characteristics of M. tuberculosis and M. bovis. This heterogeneity of M. africanum complicates its unequivocal identification and may lead to the misclassification of clinical strains. According to their biochemical characteristics, two major M. africanum subgroups have been described, and these subgroups correspond to their geographic origins in West Africa (subtype I, cluster G) or East Africa (subtype II, cluster F) (4, 16). Numerical analyses of biochemical characteristics revealed that M. africanum subtype I is more closely related to M. bovis, whereas subtype II more closely resembles M. tuberculosis (4).
More recently, the application of molecular methods has permitted new insights into the molecular characteristics and phylogeny of MTBC species to be obtained (2, 7, 8, 13, 14, 16, 26). In those studies M. africanum strains from West African countries (subtype I) were characterized by the presence of low or medium numbers of IS6110 bands and a specific spoligotype pattern with characteristics of M. bovis and M. tuberculosis. Additionally, all West African M. africanum strains showed a characteristic genomic deletion (RD9) (2) and a characteristic gyrB DNA sequence (15) that permitted their clear distinction from M. tuberculosis. A further interesting genomic region that allows the differentiation of M. africanum from M. bovis is RD12, which is present in the former organism but not the latter (2, 7, 13).
In contrast, M. africanum strains from Uganda (subtype II, East Africa) showed a high number of IS6110 copies, a spoligotype pattern similar to that of M. tuberculosis, and a gyrB sequence identical to that of M. tuberculosis (14, 16). Preliminary results indicated that M. africanum subtype II isolates might not have the RD9 deletion, which was detected in all M. africanum strains in the new evolutionary scenario developed by Brosch and coworkers (2) and Huard and coworkers (7). Taken together, these findings reiterate the close relationship between M. africanum subtype II and M. tuberculosis and question the taxonomic status of M. africanum subtype II within MTBC.
In the present study, we performed a detailed analysis of the phenotypic and genotypic properties of MTBC strains from West and East Africa that were previously classified as M. africanum according to their biochemical characteristics. On the basis of the results of those analyses, we propose an improved species description for M. africanum that includes the present results obtained with new molecular markers.
MATERIALS AND METHODS
Strains analyzed.
A total of 63 MTBC isolates comprising 30 M. africanum strains from West Africa (Ghana); 20 M. africanum strains from East Africa (Uganda); and type strains M. tuberculosis H37 (ATCC 27294), M. bovis (ATCC 19210), and M. africanum (ATCC 25420). Ten randomly chosen M. tuberculosis isolates from Germany were included as controls. The M. africanum strains from Uganda were a subset of a collection described previously (16). All clinical isolates were obtained from different patients with pulmonary TB. The patients from Ghana comprised only human immunodeficiency virus-negative individuals, whereas the patients from Uganda comprised human immunodeficiency virus-negative and -positive individuals. The main biochemical as well as genetic characteristics of the 63 MTBC isolates are summarized in Table 1.
TABLE 1.
Phenotypic and genetic characteristics of type strains M. tuberculosis H37 (ATCC 27294), M. bovis (ATCC 19210), and M. africanum (ATCC 25420) and the other strains analyzeda
| Phenotypic differentiation and genotype | Origin | Test result (% of isolates)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Colony morphologyb | Growth in presence of TCH | Type of growth on Lebek medium | RD1 | RD9 | RD12 | gyrBc | Nucleotide at narGHJI position −215 | ||
| M. tuberculosis H37 | ATCC | Eugonic | + | Aerophilic | + | + | + | MTB | T |
| M. bovis | ATCC | Dysgonic | − | Mircroaerophilic | + | − | − | MBOV | C |
| M. africanum | ATCC | Dysgonic | − | Mircroaerophilic | + | − | + | MAFRI | C |
| M. tuberculosis (n = 10) | Germany | Eugonic (100) | + (100) | Aerophilic (100) | + (100) | + (100) | + (100) | MTB | T |
| M. africanum subtype II, Uganda I and II (n = 20) | Uganda | Dysgonic (100) | + (100) | Mircroaerophilic (100) | + (100) | + (100) | + (100) | MTB | T |
| M. africanum subtype I, A1, A2, A3 (n = 30) | Ghana | Dysgonic (100) | − (100) | Mircroaerophilic (100) | + (100) | − (100) | + (100) | MAFRI | C |
Symbols and abbreviations: +, positive test result or genomic region present; −, negative test result or genomic region deleted; ATCC, American Type Culture Collection.
Growth characteristics on Löwenstein-Jensen slants.
Classification according to GenoType MTBC test results: MTB, M. tuberculosis or M. africanum subtype II; MBOV, M. bovis subsp. bovis; MAFRI, M. africanum subtype I.
Primary isolation and culturing of the mycobacterial isolates were performed as described elsewhere (10). All isolates were identified as members of MTBC by using gene probes (ACCUProbe; GenProbe, San Diego, Calif.).
Biochemical tests.
The biochemical analyses used for differentiation included colony morphology, nitrate reduction on modified Dubos broth, niacin accumulation test (isoniazid-test strips; Difco, Detroit, Mich.), and growth in the presence of thiophen-2-carboxylic acid hydrazide (TCH; 2 μg/ml). Growth characteristics on Lebek medium and bromcresol purple medium were determined as described previously (12).
IS6110 DNA fingerprinting and spoligotyping analysis.
Extraction of DNA from mycobacterial strains and DNA fingerprinting with IS6110 as a probe were performed by a standardized protocol, as described elsewhere (23, 25). Spoligotyping of strains was performed as described by Kamerbeek et al. (9). The IS6110 fingerprint patterns and the spoligotype patterns were analyzed with BioNumerics software (Windows NT, version 2.5; Applied Maths, Kortrijk, Belgium) by applying the Dice coefficient (with 1.2% position tolerance) and the unweighted pair group with mathematic averages method.
Analysis of genomic deletions, gyrB DNA polymorphisms, and the narGHJI promoter polymorphism.
The presence or absence of molecular regions was determined by PCR assays described previously. The strains were analyzed for the presence or absence of RD9 by a PCR protocol described by Parsons and coworkers (18), and the strains were analyzed for the presence or absence of RD12 by a PCR protocol described by Mostowy and colleagues (13). The presence of the RD1 region and of discriminatory single-nucleotide polymorphisms (SNPs) in the gyrB gene were determined by using the commercially available GenoType MTBC assay (Hain Lifescience GmbH, Nehren, Germany), as described previously (19). The T-to-C transition within the promoter of narGHJI was detected by PCR analysis with a LightCycler instrument, as described by Stermann and colleagues (22).
RESULTS AND DISCUSSION
In order to achieve an improved characterization of the species M. africanum (3, 27), a representative collection of 63 MTBC isolates was analyzed for key phenotypic and genetic characteristics. These isolates included 30 strains from Ghana (West Africa), 20 strains from Uganda (East Africa), and 10 strains from Germany, as well as type strains M. tuberculosis H37 (ATCC 27294), M. bovis ATCC 19210, M. africanum ATCC 25420. On the basis of their phenotypic characteristics (Table 1), the strains from Uganda were classified as M. africanum subtype II, the strains from Ghana were classified as M. africanum subtype I, and the strains from Germany were classified as M. tuberculosis.
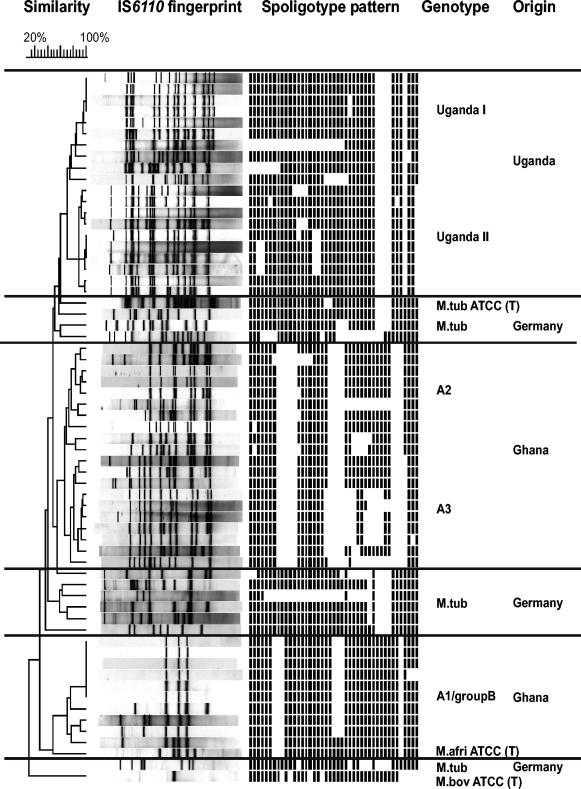
All strains were analyzed by IS6110 DNA fingerprinting and spoligotyping. In the corresponding dendrogram based on the similarity of the IS6110 fingerprinting patterns, the isolates from Uganda and Ghana formed distinct groups or genotypes that were clearly separated from the groups or genotypes containing the M. tuberculosis isolates (Fig. 1). The similarities of the IS6110 fingerprinting patterns among isolates of one group or genotype confirm the close relationship of the particular isolates. According to the IS6110 patterns and spoligotypes, the genotypes determined were assigned to the major genotypes described for M. africanum strains in previous studies, which also confirmed the representativeness of the strain collection analyzed. These genotypes are for M. africanum subtype II, genotypes Uganda I and II, from East Africa (16) and for M. africanum subtype I, genotype A1-group 4, genotype A2, and genotype A3, from West Africa (8, 17, 26). Type strain M. africanum ATCC 25420 clustered together with M. africanum subtype I strains of genotype A1-group 4.
FIG. 1.
IS6110 DNA fingerprint and spoligotype patterns of the 63 MTBC strains analyzed. The IS6110 banding patterns are ordered by similarity in a dendrogram. The position of each IS6110 band is normalized so that the banding patterns of all strains are mutually comparable. The scale depicts the similarities of the IS6110 patterns calculated with the Dice coefficient and the unweighted pair group with mathematic averages method. Genotypes were defined on the basis of the similarities of the IS6110 banding patterns compared to those of genotypes reported in previous studies (8, 16, 17, 26). M. tub, M. tuberculosis; M. afri, M. africanum; M. bov, M. bovis.
All spoligotypes of West African M. africanum isolates hybridized to at least one of the spacer sequences 33 to 36, whereas these spacer sequences were absent from the chromosomes of all M. africanum subtype II strains (East Africa) analyzed and 90% of the M. tuberculosis strains analyzed. Furthermore, all M. africanum subtype I strains (West Africa) failed to hybridize to spacer 39, while this spacer was present in all other strains analyzed. Thus, in accordance with the findings of previous studies (14, 26), this finding confirmed that the combination of hybridization to spacers 33 to 36 and a lack of hybridization to spacer 39 is a useful marker for the identification of M. africanum subtype I.
With respect to the presence or absence of the genomic regions analyzed (RD1, RD9, and RD12), all M. africanum subtype II and M. tuberculosis isolates as well as type strain M. tuberculosis H37 (ATCC 27294) showed identical test results. RD1, RD9, and RD12 were present in all M. africanum subtype II isolates and all M. tuberculosis isolates (Table 1). The same findings were obtained for the additional genetic markers for which tests were conducted. M. africanum subtype II and M. tuberculosis isolates had identical combinations of SNPs in the gyrB gene and carried a thymine residue instead of a cytosine residue at nucleotide −215 prior to the start codon of narG. This polymorphism was previously described to be specific for M. tuberculosis (22). Thus, isolates of genotypes Uganda I and II (which were previously named M. africanum subtype II) more likely represent phenotypic variants of M. tuberculosis than members of the M. africanum subgroup in MTBC. This conclusion is in accordance with the results and discussions presented in previous publications (15, 21) and fits well with the new phylogenetic scenario for MTBC proposed by Brosch and coworkers (2).
In contrast to genotypes Uganda I and II, all M. africanum strains from Ghana (genotype A1-group 4, genotype A2, and genotype A3) are clearly identified by a unique combination of molecular characteristics (Table 1). M. africanum isolates were separated (i) from M. tuberculosis by a lack of RD9 and by carriage of a cytosine residue at position −215 of the narGHJI promoter region and (ii) from M. bovis by the presence of RD12. Furthermore, M. africanum strains from Ghana had a unique gyrB sequence that permitted their rapid identification by gyrB PCR-restriction fragment length polymorphism analysis (15) or the commercially available GenoType MTBC test (19). All characteristics analyzed were identical to those of type strain M. africanum ATCC 25420. The results determined in this investigation concerning the presence or absence of RD9, RD12, and SNPs in the gyrB genes of M. tuberculosis, M. bovis, and M. africanum are in accordance with the findings of previous studies (2, 7, 16).
The unique combination of several independent genetic markers determined for West African M. africanum isolates is evidence of their independence from M. tuberculosis and M. bovis and confirm that these isolates represent a unique phylogenetic branch within MTBC that might have originated in West Africa. Furthermore, the West African isolates also fulfill the main phenotypic and biochemical test criteria that were given in the original species description of M. africanum (3, 27). On the contrary, although isolates of genotypes Uganda I and II from East Africa were different by some phenotypic and biochemical tests, they showed nearly identical genetic characteristics when they were compared with those of M. tuberculosis (except the typical IS6110 fingerprint and spoligotype patterns). Thus, no evidence for their independence from M. tuberculosis was found. However, these strains are the predominant cause of TB in Uganda (16) and might exhibit not only certain biochemical characteristics but also certain clinical characteristics. We therefore suggest that this interesting group of isolates be distinguished from other M. tuberculosis isolates (M. tuberculosis genotype Uganda) as genotype or variant “Uganda.” Identification of genotype “Uganda” isolates can be achieved by evaluation for the combination of phenotypic and genetic characteristics presented previously (16). Future studies are needed to more thoroughly analyze the genetic relationship of “Uganda” genotype isolates and other M. tuberculosis isolates and to address the clinical and pathological aspects of TB caused by strains of this genotype.
In conclusion, we propose that the molecular markers that we describe here, which are characteristic for the “true” M. africanum isolates, should be added to the species description of M. africanum (3, 27). M. africanum should be characterized by the absence of RD9, the presence of RD12, and the presence of a unique gyrB sequence. These markers should be used in future studies and before new isolates are designated M. africanum, which would allow clear species differentiation. These markers might also replace the use of more variable phenotypic and biochemical characteristics in the future and will contribute to the generation of more precise data on the regional prevalence and pathogenic properties of M. africanum.
Description of M. africanum.
The description of M. africanum is the same as those given for M. africanum by Castets et al. (3) and Wayne and Kubica (27) and for isolates of cluster G in the publication of David et al. (4). In addition to these characteristics, isolates of M. africanum lack RD9, are positive for RD12, and present a specific combination of gyrB gene polymorphisms: nucleotide 675 is C, nucleotide 756 is G, nucleotide 1311 is T, nucleotide 1410 is C, and nucleotide 1450 is T. The type strain is Rist 3419 (ATCC 25420), which has the characteristics described for the taxon.
Acknowledgments
We thank K. Ott, B. Schlüter, I. Radzio, T. Ubben, and P. Vock (Borstel, Germany) and S. Maass (Hannover, Germany) for excellent technical assistance. We are grateful to the members of the working group on a consensus definition of M. africanum of EU Concerted Action Project QLK2-CT-2000-00630, who contributed to the initiation and design of this study by their fruitful suggestions and discussions.
Parts of this work were supported by the Robert Koch Institute, Berlin, Germany, and the EU Concerted Action Project “New generation genetic markers and techniques for the epidemiology and control of tuberculosis” (QLK2-CT-2000-00630).
REFERENCES
- 1.Bonard, D., P. Msellati, L. Rigouts, P. Combe, D. Coulibaly, I., M. Coulibaly, and F. Portaels. 2000. What is the meaning of repeated isolation of Mycobacterium africanum? Int. J. Tuberc. Lung Dis. 4:1176-1180. [PubMed] [Google Scholar]
- 2.Brosch, R., S. V. Gordon, M. Marmiesse, P. Brodin, C. Buchrieser, K. Eiglmeier, T. Garnier, C. Gutierrez, G. Hewinson, K. Kremer, L. M. Parsons, A. S. Pym, S. Samper, D. van Soolingen, and S. T. Cole. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc. Natl. Acad. Sci. USA 99:3684-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castets, M., H. Boisvert, F. Grumbach, M. Brunel, and N. Rist. 1968. Tuberculosis bacilli of the African type: preliminary note. Rev. Tuberc. Pneumol. 32:179-184. [PubMed] [Google Scholar]
- 4.David, H. L., M. T. Jahan, A. Jumin, J. Grandry, and E. H. Lehman. 1978. Numerical taxonomy analysis of Mycobacterium africanum. Int. J. Syst. Bacteriol. 28:464-472. [Google Scholar]
- 5.Feizabadi, M. M., I. D. Robertson, D. V. Cousins, and D. J. Hampson. 1996. Genomic analysis of Mycobacterium bovis and other members of the Mycobacterium tuberculosis complex by isoenzyme analysis and pulsed-field gel electrophoresis. J. Clin. Microbiol. 34:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frothingham, R., H. G. Hills, and K. H. Wilson. 1994. Extensive DNA sequence conservation throughout the Mycobacterium tuberculosis complex. J. Clin. Microbiol. 32:1639-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huard, R. C., L. C. de Oliveira Lazzarini, W. R. Butler, D. van Soolingen, and J. L. Ho. 2003. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. J. Clin. Microbiol. 41:1637-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Källenius, G., T. Koivula, S. Ghebremichael, S. E. Hoffner, R. Norberg, E. Svensson, F. Dias, B. L. Marklund, and S. B. Svenson. 1999. Evolution and clonal traits of Mycobacterium tuberculosis complex in Guinea-Bissau. J. Clin. Microbiol. 37:3872-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamerbeek, J., L. Schouls, A. Kolk, M. van Agterveld, D. van Soolingen, S. Kuijper, A. Bunschoten, H. Molhuizen, R. Shaw, M. Goyal, and J. D. A. van Embden. 1997. Simultaneous detection and strains differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J. Clin. Microbiol. 35:907-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent, P. T., and G. P. Kubica. 1985. Public health mycobacteriology. A guide for the level III laboratory. U.S. Department of Health and Human Services, Centers for Disease Control, Atlanta, Ga.
- 11.Kirschner, P., B. Springer, U. Vogel, A. Meier, A. Wrede, M. Kiekenbeck, F. C. Bange, and E. C. Böttger. 1993. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J. Clin. Microbiol. 31:2882-2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meissner, G., and K. Schröder. 1969. The so-called “African Tuberculosis strains” from the tropical West Africa. Zentbl. Bakteriol. Parasitenkd. Infektkrankh. Hyg. Abt. 1 Orig. 211:69-81. [PubMed] [Google Scholar]
- 13.Mostowy, S., D. Cousins, J. Brinkman, A. Aranaz, and M. A. Behr. 2002. Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J. Infect. Dis. 186:74-80. [DOI] [PubMed] [Google Scholar]
- 14.Niemann, S., E. Richter, and S. Rüsch-Gerdes. 2000. Differentiation among members of the Mycobacterium tuberculosis complex by molecular and biochemical features: evidence for two pyrazinamide-susceptible subtypes of M. bovis J. Clin. Microbiol. 38:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niemann, S., D. Harmsen, S. Rüsch-Gerdes, and E. Richter. 2000. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphisms. J. Clin. Microbiol. 38:3231-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niemann, S., S. Rüsch-Gerdes, M. L. Joloba, C. C. Whalen, D. Guwatudde, J. J. Ellner, K. Eisenach, N. Fumokong, J. L. Johnson, T. Aisu, R. D. Mugerwa, A. Okwera, and S. K. Schwander. 2002. Mycobacterium africanum subtype II is associated with two distinct genotypes and is a major cause of human tuberculosis in Kampala, Uganda. J. Clin. Microbiol. 40:3398-3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niobe-Eyangoh, S. N., C. Kuaban, P. Sorlin, P. Cunin, J. Thonnon, C. Sola, N. Rastogi, V. Vincent, and M. C. Gutierrez. 2003. Genetic biodiversity of Mycobacterium tuberculosis complex strains from patients with pulmonary tuberculosis in Cameroon. J. Clin. Microbiol. 41:2547-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parsons, L. M., R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Bretzel, D. Van Soolingen, Y. M. Hale, and M. Salfinger. 2002. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol. 40:2339-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richter, E., M. Weizenegger, S. Rüsch-Gerdes, and S. Niemann. 2003. Differentiation of clinical Mycobacterium tuberculosis isolates by Genotype MTBC assay. J. Clin. Microbiol. 41:2672-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schwander, S., S. Rüsch-Gerdes, A. Mateega, T. Lutalo, S. Tugume, C. Kityo, R. Rubaramira, P. Mugyenyi, A. Okwera, R. Mugerwa T. Aisu, R. Moser, K. Ochen, B. M'Bonye, and M. Dietrich. 1995. A pilot study of antituberculosis combinations comparing rifabutin with rifampicin in the treatment of HIV-1 associated tuberculosis. A single-blind randomized evaluation in Ugandan patients with HIV-1 infection and pulmonary tuberculosis. Tuberc. Lung. Dis. 76:210-218. [DOI] [PubMed] [Google Scholar]
- 21.Sola, C., N. Rastogi, M. C. Gutierrez, V. Vincent, R. Brosch, and L. Parsons. 2002. Is Mycobacterium africanum subtype II (Uganda I and Uganda II) a genetically well-defined subspecies of the Mycobacterium tuberculosis complex? J. Clin. Microbiol. 41:1345-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stermann, M., A. Bohrssen, C. Diephaus, S. Maass, and Bange. 2003. Polymorphic nucleotide within the promoter of nitrate reductase (NarGHJI) is specific for Mycobacterium tuberculosis. J. Clin. Microbiol. 41:3252-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Embden, J. D. A., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, T. M. Shinnick, and P. M. Small. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Soolingen, D., T. Hoogenboezem, P. E. W. de Haas, P. W. M. Hermans, M. A. Koedam, K. S. Teppema, P. J. Brennan, G. S. Besra, F. Portaels, J. Top, L. M. Schouls, and J. D. A. van Embden. 1997. A novel pathogenic taxon of the Mycobacterium tuberculosis complex canetti: characterization of an exceptional isolate from Africa. Int. J. Syst. Bacteriol. 47:1236-1245. [DOI] [PubMed] [Google Scholar]
- 25.van Soolingen, D. 2001. Molecular epidemiology of tuberculosis and other mycobacterial infections: main methodologies and achievements. J. Intern. Med. 249:1-26. [DOI] [PubMed] [Google Scholar]
- 26.Viana-Niero, C., C. Gutierrez, C. Sola, I. Filliol, F. Boulahbal, V. Vincent, and N. Rastogi. 2001. Genetic diversity of Mycobacterium africanum clinical strains based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J. Clin. Microbiol. 39:57-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wayne, L. G., and G. P. Kubica. 1986. The mycobacteria, p. 1435-1457. In P. H. A. Sneath and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams & Wilkins Co., Baltimore, Md. [Google Scholar]