Abstract
In epidemiological investigations of community legionellosis outbreaks, knowledge of the prevalence, distribution, and clinical significance (virulence) of environmental Legionella isolates is crucial for interpretation of the molecular subtyping results. To obtain such information for Legionella pneumophila serogroup 1 isolates, we used the standardized amplified fragment length polymorphism (AFLP) protocol of the European Working Group on Legionella Infection to subtype L. pneumophila SG1 isolates obtained from patients and water sources in Queensland, Australia. An AFLP genotype, termed AF1, was predominant in isolates from both patients (40.5%) and water (49.0%). The second most common AFLP genotype found in water isolates was AF16 (36.5%), but this genotype was not identified in the patient isolates. When virulence gene-based PCR assays for lvh and rtxA genes were applied to the isolates from patients and water, nearly all (65 of 66) AF1 strains had both virulence genes, lvh and rtxA. In contrast, neither the lvh nor the rtxA gene was found in the AF16 strains, except for one isolate with the rtxA gene. It appears that this may explain the failure to find this genotype in the isolates from patients even though it may be common in the environment. In view of the evidence that the AF1 genotype is the most common genotype among strains found in patients and water sources in this region, any suggested epidemiological link derived from comparing the AF1 genotype from patient isolates with the AF1 genotype from environmental isolates must be interpreted and acted on with caution. The use of virulence gene-based PCR assays applied to environmental samples may be helpful in determining the infection potential of the isolates involved.
The etiologic agents of legionellosis are members of the genus Legionella, which are ubiquitous worldwide in natural waters such as rivers and lakes (10), in potable water (29), and in man-made water systems including cooling towers and spas (9). Legionellae can invade and multiply in free-living protozoa, making it harder to eradicate legionellae from water systems and making them more virulent, as demonstrated in animal models (14). Legionellosis of humans varies from a mild respiratory illness to an acute life-threatening pneumonia, which is acquired by inhalation or aspiration of legionellae from a contaminated environmental source (34). A recent international study of the distribution of Legionella species, with Brisbane, Australia, Legionella isolates being part of the study, showed that the vast majority of cases of legionellosis are caused by L. pneumophila, and most of these are serogroup (SG) 1 strains (35), whereas L. longbeachae outbreaks have been linked to contaminated potting mix in Australia (22). In addition, certain monoclonal antibody subgroups of L. pneumophila SG 1 isolates are associated with human disease (7, 29).
The genetic basis for virulence differences in the subgroup strains have been shown to be related to the presence or absence of certain virulence genes among the strains (26, 28). The presence of these genes may affect the ability of the strains to survive in the environment and cause disease. The identified virulence genes of Legionella have been extensively reviewed (9, 14, 30). These genes include type IV secretion system genes icm/dot (32), tra1 (26), and lvh (28); type IV pilus genes pilDE (9); and other genes such as mip (macrophage infectivity potentiator) (3), rtxA, encoding the dot/icm-regulated pore-forming toxin (36), and enhC, encoding a secreted protein for interaction of L. pneumophila with host cells (5). Several other loci involved in the intracellular growth of L. pneumophila have also been identified, including mak (macrophage killing), mil (macrophage-specific infectivity loci), and pmi (protozoan and macrophage infectivity) (9). However, a recent study shows that lvh and rtxA regions are found more frequently in strains associated with human disease (26).
The source of an outbreak may be determined by linking environmental isolates to clinical isolates by various molecular subtyping methods, including monoclonal antibody typing, restriction fragment length polymorphism, pulsed-field gel electrophoresis (PFGE), ribotyping, use of random amplified polymorphic DNA, repetitive-element sequence-based PCR, and amplified fragment length polymorphism (AFLP) (11, 16, 17). It has been reported that AFLP is a rapid, discriminatory, and epidemiologically concordant molecular method in comparison studies of these methods (11, 17). Because of its simplicity and reproducibility, AFLP proved to be the most effective technique in outbreak investigation. In addition, a standardized AFLP protocol has been established by the European Working Group on Legionella Infection (EWGLI) and has been proven successful for interlaboratory comparison of AFLP results (12).
Comparative studies of Legionella isolates from patients and environmental sources in Paris, France (6, 19), and Germany (20) have shown that one subtype of L. pneumophila SG1 isolates is the predominant strain. The phenomenon has been explained by either its predominance in the environment (19) or presumably the higher infectivity of the strain (6). In addition, lack of knowledge of the prevalence, distribution, and clinical significance (virulence) of L. pneumophila SG1 isolates from water has hindered legionellosis outbreak investigations (15). Therefore, in this study we report the use of the EWGLI standardized protocol (12) applied to L. pneumophila SG1 isolates from both patients and water sources in Brisbane and neighboring regions and the detection of lvh and rtxA virulence genes in these strains. The aims of this study were to identify how many AFLP genotypes are in the patient isolates, to identify any linkage between AFLP genotypes and virulence genes lvh and rtxA in the patient isolates, and to obtain basic knowledge of the AFLP genotypes in water isolates in Queensland, Australia.
MATERIALS AND METHODS
Isolates.
A total of 37 clinical L. pneumophila SG1 isolates were referred to our laboratory in the period 1990 to 2003, during which legionellosis was a notifiable disease in all the states and territories of Australia. The number of L. pneumophila SG1 isolates from water sources was 104, consisting of 96 isolates from samples collected from cooling towers and reservoirs between July and December 2003 and 8 isolates referred to our laboratory from 1994 to 2003. The clinical isolates were from sporadic community-acquired cases of legionellosis and were not epidemiologically related to the environmental isolates, except one water isolate in 1994 which was epidemiologically related to a case. The majority of isolates from patients and water were from Brisbane and neighboring regions. Culture, identification, and serotyping were carried out in our Legionella reference laboratory for patient isolates and in two private laboratories for the majority of the water isolates, using standard methods (34). All were analyzed by AFLP subtyping.
AFLP assay.
The DNA of the strain was extracted by using the MasterPure DNA Purification kit (Epicentre, Madison, Wis.). The standard EWGLI AFLP protocol was used (12) for subtyping. Only bands within the range 300 to 3,500 bp were included in the description of the pattern. The gel analysis software BioNumerics (Applied Maths) was used to compare AFLP profiles within and between gels. The unique patterns from the samples were run on the same gel to verify the profile. The criteria for interpreting the AFLP patterns were adapted from Tenover et al. (31), who described the interpretation of PFGE patterns. However, unlike PFGE patterns, AFLP fragments in the gel represent only a part of the genome because only those fragments in the range between 300 and 3,500 bp are reliably amplified and the primer design’s having one selective nucleotide determines that only one in four of the ligated fragments can be amplifiable (12, 27). If there was no difference or a one-band difference between the samples, they were scored as indistinguishable because there is no genetic difference or one genetic difference between the samples. If two or more bands were different between the samples, they were regarded as unique types because there are two or more genetic differences between the samples.
Virulence gene detection using PCR.
Two virulence gene loci (lvh and rtxA regions) were chosen because they demonstrated a strong association with legionellosis in patients in a previous study (26). The six pairs of virulence gene-based PCR primers used in this study were lvh1/prpA-lvh2/prpA, lvh3/lvhB3- lvh4/lvhB4, lvh5/lvhB8-lvh6/lvhB9, and lvr1/lvrE-lvr2/lvrE for the amplification of the lvh region and rtx1/rtxA-rtx2/rtxA and rtx3/rtxA-rtx4/rtxA for the detection of the rtxA region. All the isolates from patients and water were used in virulence gene detection. The primer sequences and PCR conditions were the same as those described previously (26).
RESULTS
Isolates from patients.
All 37 isolates of L. pneumophila SG1 from reported cases were typeable with AFLP. With the pattern classification criteria adapted from Tenover et al. (31), 15 AFLP-distinguishable patterns were observed, and they were termed AF1 to AF15. The distribution of the genotypes of the isolates is detailed in Table 1. AF1 was the predominant genotype. This included five strains that had exactly the same profile as the other AF1 strains but without an 800-bp band (data not shown). The frequency of the AF1 type in the patient samples was 40.5% (15 of 37), and the type was identified among isolates obtained between 1990 and 2000, suggesting that AF1 is possibly more often associated with disease and/or a common source such as the water systems in Brisbane and neighboring regions.
TABLE 1.
Distribution of AFLP genotypes of L. pneumophila SG1 isolates from patients and water
| Isolate type and yr | Total no. of isolates | No. of type:
|
Other AF type(s) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| AF1 | AF2 | AF3 | AF4 | AF5 | AF11 | AF13 | AF16 | |||
| Patient | ||||||||||
| 1990 | 1 | 1 | ||||||||
| 1992 | 5 | 2 | AF7, AF8, AF14 | |||||||
| 1993 | 4 | 1 | 1 | 1 | AF9 | |||||
| 1994 | 3 | 1 | 1 | AF10 | ||||||
| 1996 | 7 | 5 | 1 | AF12 | ||||||
| 1997 | 1 | 1 | ||||||||
| 1998 | 3 | 1 | 2 | |||||||
| 1999 | 5 | 1 | 1 | 1 | AF6, AF15 | |||||
| 2000 | 4 | 4 | ||||||||
| 2001 | 1 | 1 | ||||||||
| 2002 | 2 | 1 | 1 | |||||||
| 2003 | 1 | 1 | ||||||||
| Total | 37 | 15 | 3 | 4 | 2 | 3 | 1 | 1 | 0 | 8a |
| Water | 104 | 51 | 0 | 1 | 1 | 0 | 1 | 1 | 38 | AF17-AF26 (one for each, except two for AF23) |
Value is number of isolates.
Isolates from water.
To test if the high frequency of the AF1 strain in the patient samples was a result of a high frequency of the AF1 strain in water, as suggested in other reports (19, 21), L. pneumophila SG1 isolates from Brisbane and neighboring regions were subtyped by AFLP. The results showed that 16 AFLP types were identified in the water isolates, of which 11 were new genotypes and 5 were the same as in those in isolates from patients. This suggests that there is a difference in the distributions of L. pneumophila SG1 strains between patient and water isolates. However, the AFLP typing results also show that the AF1 type was the predominant type in water isolates, with a frequency of 49.0% (51 of 104). Therefore, the high frequency of the AF1 type in patients may be due to the high frequency of the AF1 type in water, as there is more chance of humans being exposed to this strain. Interestingly, a second common strain (AF16) was identified in water isolates, with a 36.5% (38 of 104) frequency. However, type AF16 was not identified in the L. pneumophila SG1 strains from patients.
Detection of virulence genes lvh and rtxA in isolates from patients and water.
The virulence genes lvh and rtxA have a strong association with legionellosis (26). Virulence PCR assays for lvh and rtxA were applied to isolates to determine if they played a role in the significantly different distributions of the AF1 and AF16 types in isolates from both patient and water samples. The same primers as those described previously were used (26). Figure 1 shows the positive PCR products in strains ATCC 33152 (Philadelphia-1) and 03M1684 (an isolate from water) for the six pairs of primers. The detection of lvh and rtxA in the isolates from patients and water is summarized in Table 2. The results show that AF1 strains from patients and water had a significantly high frequency of lvh and rtxA (14 of 15 and 51 of 51 for lvh and 15 of 15 and 51 of 51 for rtxA for patient and water isolates, respectively). In contrast, AF16 strains from water had a frequency of 0 of 38 for lvh and 1 of 38 for rtxA. This suggests that the AF16 strains were less virulent and might not be able to cause disease in humans. For 22 isolates of the AF2 to AF15 types from patients, 14 isolates had both lvh and rtxA genes, 7 isolates had rtxA but not lvh, and 1 isolate had lvh but not rtxA. The 15 isolates from water with the remaining AF types had virulence gene frequencies similar to those of the other isolates from patients: nine isolates had both lvh and rtxA, and six isolates had rtxA but not lvh.
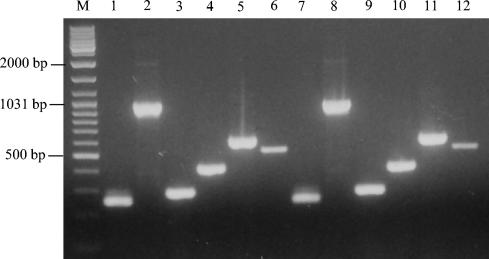
FIG. 1.
Positive PCR products in strains ATCC 33152 (Philadelphia-1) and 03M1684 (an isolate from water) for six pairs of primers. Lane M, GeneRuler DNA ladder mix (Fermentas); the representative DNA sizes are indicated. Lanes 1 to 6, results for ATCC 33152 with the primer pairs of lvh1/prpA-lvh2/prpA, lvh3/lvhB3-lvh4/lvhB4, lvh5/lvhB8-lvh6/lvhB9, and lvr1/lvrE-lvr2/lvrE for the lvh region and rtx1/rtxA-rtx2/rtxA and rtx3/rtxA-rtx4/rtxA for the rtxA region. Lanes 7 to12, results for 03M1684 with the same primer pairs.
TABLE 2.
Detection of virulence genes lvh and rtxA in isolates from patients and water
| Source | AFLP type(s) | No. of isolates | No. with gene/total
|
|
|---|---|---|---|---|
| lvh | rtxA | |||
| Patients | AF1 | 15 | 14/15 | 15/15 |
| AF2-AF15 | 14 | 14/14 | 14/14 | |
| 7 | 0/7 | 7/7 | ||
| 1 | 1/1 | 0/1 | ||
| Total | 37 | 29/37 | 36/37 | |
| Water | AF1 | 51 | 51/51 | 51/51 |
| AF16 | 38 | 0/38 | 1/38 | |
| Other AF types | 9 | 9/9 | 9/9 | |
| 6 | 0/6 | 6/6 | ||
| Total | 104 | 60/104 | 67/104 | |
DISCUSSION
In this study, AF1 was identified as the predominant AFLP genotype in L. pneumophila SG1 isolates from clinical samples (40.5%) and water (49.0%). Similar observations have been reported in Paris, France: a high proportion of L. pneumophila SG1 isolates from patients and the environment originate from a single strain (1, 19). These authors first suggested that it was abundance rather than greater pathogenicity that explained one predominant type in both patient and water isolates. In the current study, the high frequency of the AF1 strain from patients is clearly demonstrated to be associated with two factors. (i) AF1 strains are widely distributed in the water systems of Brisbane and neighboring regions (Table 1). Therefore, humans have a higher chance of exposure to the AF1 strain of L. pneumophila SG1 from the environment. (ii) AF1 strains are virulent and disease associated, as 14 of 15 and 51 of 51 strains from patients and water, respectively, had both lvh and rtxA virulence genes. The AF1 strain is more likely to have higher infectivity. In addition, the patient isolates were collected from Brisbane and surrounding regions in the period 1990 to 2003, suggesting that the AF1 strain persisted widely in southeast Queensland for over 10 years. Our data and other published reports indicate that certain strains of L. pneumophila are persistent in the environment (1, 19, 24).
The surprising finding in this study was that the second common AFLP type, AF16, was identified with a significantly high frequency of 36.5% (38 of 104) in the water isolates but not in patient isolates. The difference in distributions appears to be related to the fact that AF16 strains do not have the lvh and rtxA virulence genes, except one isolate with rtxA, making them less virulent strains (Table 2). The results also suggest that the endemic strains in Brisbane and neighboring regions are AF1 and AF16. This differs from the French studies, in which only one endemic strain was predominant (1, 19).
The virulence differences among subgroup strains of legionellae have been shown to be related to the presence or absence of certain virulence genes within the genome of the strains (26, 28). The results of the PCR assays for virulence genes may help us in understanding the molecular epidemiology and pathogenesis of Legionella. Two of these genes, lvh and rtxA, have shown a strong association with legionellosis (26). lvh encodes a second type IV secretion system of Legionella and interacts with the dot/icm-encoded type IV secretion system for transfer of plasmids by conjugation and enhancement of infectivity (25, 30). The lvh-encoded type IV secretion system is used by Bordetella pertussis for secretion of proteins involved in pathogenesis (33). rtxA encodes a dot/icm-regulated pore-forming toxin, which is involved in adherence, cytotoxicity, and pore formation in addition to its role in cellular entry (4, 36). Almost all AF1 strains from patients have both lvh and rtxA genes, suggesting a strong association between these genes and virulence (Table 2). Other AF2 to AF15 strains showed a 68.2% frequency (15 of 22) of lvh and a 95.5% frequency (21 of 22) of rtxA. It is evident that strains which have both lvh and rtxA genes would be more virulent than strains that have either lvh or rtxA alone. However, infection and clinical manifestation are also related to the immunity status and other risk factors of the patient at the time of infection. Possibly, that is the reason that seven strains without lvh virulence genes still caused infections. Strains without the lvh and rtxA genes, such as AF16, may not be capable of infection, as demonstrated by the absence of the AF16 type among the patient isolates (Table 1).
In this study, the data appear to suggest that isolates with the AF1 genotype have linkage to the two disease-associated virulence genes lvh and rtxA (Table 2). In view of this, environmental isolates of L. pneumophila SG1 with the AF1 genotype pose a greater infection potential. Therefore, immediate measures should be taken such as keeping people away from the infection source and implementing a control strategy for the presence of L. pneumophila SG1. Current Australia/New Zealand Standards (2) for microbial control of air handling and water systems (AS3666 series) operate within the premise that “increased risk is associated with increased concentration of micro-organisms.” There is no recognition within the risk framework that specific genotypes or indeed specific Legionella species pose a greater infection potential. It may be pertinent that upon isolation of L. pneumophila SG1 with the AF1 genotype, the higher-risk control strategy (online decontamination) be employed regardless of the concentration detected (the current decontamination trigger is set at 1,000 CFU/ml).
Epidemiological investigations rely on the discriminatory power of subtyping methods to assist in the identification of the possible sources of Legionella infections by relating clinical and environmental strains. In this study, the AF1 type was predominant among strains isolated from patients (Table 1). Therefore, when two or more cases involving the AF1 type appear within a time frame indicative of an outbreak, interpreting this as a single outbreak should be done with caution and be supported by sound epidemiological data and two or more molecular subtyping results (see below). It has also been found in several studies (1, 8, 18, 23) that endemic clones of L. pneumophila causing apparently unrelated cases of legionellosis have the same molecular genotype. In addition, the results of AF1 type matching of patient and environmental isolates must be interpreted with caution, as the AF1 type was predominant in our study (49.0% of water isolates [Table 1]). To increase the discriminatory power, the best approach would be to apply a combination of two or more molecular subtyping methods in the investigation. Gaia et al. (13) reported that the combination of a sequence-based typing (SBT) and monoclonal antibody subtyping increased the discriminatory power from 0.92 with SBT alone to 0.97. In an epidemiological investigation, the use of a combination of methods such as AFLP combined with other molecular typing methods, such as PFGE and SBT, will increase the discriminatory power so that the most stringent criteria are met for identifying the environmental source of infections.
Heath et al. (15) performed an epidemiological investigation of a legionellosis outbreak in western Sydney using restriction fragment length polymorphism and random amplified polymorphic DNA methods. Due to the lack of knowledge of the prevalence of molecular subtypes and of the distribution and virulence characteristics of L. pneumophila SG1 strains in water samples, the interpretation of the results in the investigation was hindered. The results of our study provide data and some insight into the possible associations between virulent clinical isolates and strains detected in water sources in Queensland, which would be of value in investigating any further outbreak of legionellosis in the geographical region studied.
Acknowledgments
We are grateful to Helen Smith, Lester Hiley, and Leonie Barnett of Queensland Health Scientific Services for helpful discussion. We acknowledge Glen Pinna of Biotech Laboratories and Margaret Wimberley of EML Consulting Services Queensland Pty. Ltd. for providing some of the water isolates and the clinicians and laboratory and hospital staff of Queensland Health who submitted isolates to this laboratory over the period of this study.
REFERENCES
- 1.Aurell, H., J. Etienne, F. Forey, M. Reyrolle, P. Girardo, P. Farge, B. Decludt, C. Campese, F. Vandenesch, and S. Jarraud. 2003. Legionella pneumophila serogroup 1 strain Paris: endemic distribution throughout France. J. Clin. Microbiol. 41:3320-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Australian/New-Zealand-Standard. 2000. Air-handling and water systems of buildings—microbial control: part 3. Performance-based maintenance of cooling water systems. Standards Australia, Sydney, Australia.
- 3.Cianciotto, N. P., B. I. Eisenstein, C. H. Mody, G. B. Toews, and N. C. Engleberg. 1989. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect Immun. 57:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cirillo, S. L., L. E. Bermudez, S. H. El-Etr, G. E. Duhamel, and J. D. Cirillo. 2001. Legionella pneumophila entry gene rtxA is involved in virulence. Infect. Immun. 69:508-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cirillo, S. L., J. Lum, and J. D. Cirillo. 2000. Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146:1345-1359. [DOI] [PubMed] [Google Scholar]
- 6.Doleans, A., H. Aurell, M. Reyrolle, G. Lina, J. Freney, F. Vandenesch, J. Etienne, and S. Jarraud. 2004. Clinical and environmental distributions of Legionella strains in France are different. J. Clin. Microbiol. 42:458-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dournon, E., W. F. Bibb, P. Rajagopalan, N. Desplaces, and R. M. McKinney. 1988. Monoclonal antibody reactivity as a virulence marker for Legionella pneumophila serogroup 1 strains. J. Infect. Dis. 157:496-501. [DOI] [PubMed] [Google Scholar]
- 8.Drenning, S. D., J. E. Stout, J. R. Joly, and V. L. Yu. 2001. Unexpected similarity of pulsed-field gel electrophoresis patterns of unrelated clinical isolates of Legionella pneumophila, serogroup 1. J. Infect. Dis. 183:628-632. [DOI] [PubMed] [Google Scholar]
- 9.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fliermans, C. B., W. B. Cherry, L. H. Orrison, S. J. Smith, D. L. Tison, and D. H. Pope. 1981. Ecological distribution of Legionella pneumophila. Appl. Environ. Microbiol. 41:9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fry, N. K., S. Alexiou-Daniel, J. M. Bangsborg, S. Bernander, M. C. Pastoris, J. Etienne, B. Forsblom, V. Gaia, J. H. Helbig, D. Lindsay, P. C. Luck, C. Pelaz, S. A. Uldum, and T. G. Harrison. 1999. A multicenter evaluation of genotypic methods for the epidemiologic typing of Legionella pneumophila serogroup 1: results of a pan-European study. Clin. Microbiol. Infect. 5:462-477. [DOI] [PubMed] [Google Scholar]
- 12.Fry, N. K., J. M. Bangsborg, A. Bergmans, S. Bernander, J. Etienne, L. Franzin, V. Gaia, P. Hasenberger, B. Baladron Jimenez, D. Jonas, D. Lindsay, S. Mentula, A. Papoutsi, M. Struelens, S. A. Uldum, P. Visca, W. Wannet, and T. G. Harrison. 2002. Designation of the European Working Group on Legionella Infection (EWGLI) amplified fragment length polymorphism types of Legionella pneumophila serogroup 1 and results of intercentre proficiency testing using a standard protocol. Eur. J. Clin. Microbiol. Infect. Dis. 21:722-728. [DOI] [PubMed] [Google Scholar]
- 13.Gaia, V., N. K. Fry, T. G. Harrison, and R. Peduzzi. 2003. Sequence-based typing of Legionella pneumophila serogroup 1 offers the potential for true portability in legionellosis outbreak investigation. J. Clin. Microbiol. 41:2932-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harb, O. S., L. Y. Gao, and Y. Abu Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 15.Heath, T. C., C. Roberts, B. Jalaludin, I. Goldthrope, and A. G. Capon. 1998. Environmental investigation of a legionellosis outbreak in western Sydney: the role of molecular profiling. Aust. N.Z. J. Public Health 22:428-431. [DOI] [PubMed] [Google Scholar]
- 16.Joly, J. R., R. M. McKinney, J. O. Tobin, W. F. Bibb, I. D. Watkins, and D. Ramsay. 1986. Development of a standardized subgrouping scheme for Legionella pneumophila serogroup 1 using monoclonal antibodies. J. Clin. Microbiol. 23:768-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonas, D., H. G. Meyer, P. Matthes, D. Hartung, B. Jahn, F. D. Daschner, and B. Jansen. 2000. Comparative evaluation of three different genotyping methods for investigation of nosocomial outbreaks of Legionnaires' disease in hospitals. J. Clin. Microbiol. 38:2284-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanser, J. 1996. Out of the air vent. Today's Life Sci. February:34-37. [Google Scholar]
- 19.Lawrence, C., M. Reyrolle, S. Dubrou, F. Forey, B. Decludt, C. Goulvestre, P. Matsiota-Bernard, J. Etienne, and C. Nauciel. 1999. Single clonal origin of a high proportion of Legionella pneumophila serogroup 1 isolates from patients and the environment in the area of Paris, France, over a 10-year period. J. Clin. Microbiol. 37:2652-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luck, P. C., and J. H. Helbig. 2002. Typing of Legionella strains isolated from patients and environmental sources in Germany, 1990-2000, p. 267-270. In R. Marre, Y. A. Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Luck (ed.), Legionella. ASM Press, Washington, D.C.
- 21.Luck, P. C., H. M. Wenchel, and J. H. Helbig. 1998. Nosocomial pneumonia caused by three genetically different strains of Legionella pneumophila and detection of these strains in the hospital water supply. J. Clin. Microbiol. 36:1160-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montanaro-Punzengruber, J. C., L. Hicks, W. Meyer, and G. L. Gilbert. 1999. Australian isolates of Legionella longbeachae are not a clonal population. J. Clin. Microbiol. 37:3249-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruckler, J. M., L. A. Mermel, R. F. Benson, C. Giorgio, P. K. Cassiday, R. F. Breiman, C. G. Whitney, and B. S. Fields. 1995. Comparison of Legionella pneumophila isolates by arbitrarily primed PCR and pulsed-field gel electrophoresis: analysis from seven epidemic investigations. J. Clin. Microbiol. 33:2872-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rangel-Frausto, M. S., P. Rhomberg, R. J. Hollis, M. A. Pfaller, R. P. Wenzel, C. M. Helms, and L. A. Herwaldt. 1999. Persistence of Legionella pneumophila in a hospital's water system: a 13-year survey. Infect. Control Hosp. Epidemiol. 20:793-797. [DOI] [PubMed] [Google Scholar]
- 25.Ridenour, D. A., S. L. Cirillo, S. Feng, M. M. Samrakandi, and J. D. Cirillo. 2003. Identification of a gene that affects the efficiency of host cell infection by Legionella pneumophila in a temperature-dependent fashion. Infect. Immun. 71:6256-6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samrakandi, M. M., S. L. Cirillo, D. A. Ridenour, L. E. Bermudez, and J. D. Cirillo. 2002. Genetic and phenotypic differences between Legionella pneumophila strains. J. Clin. Microbiol. 40:1352-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savelkoul, P. H., H. J. Aarts, J. de Haas, L. Dijkshoorn, B. Duim, M. Otsen, J. L. Rademaker, L. Schouls, and J. A. Lenstra. 1999. Amplified-fragment length polymorphism analysis: the state of an art. J. Clin. Microbiol. 37:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segal, G., J. J. Russo, and H. A. Shuman. 1999. Relationships between a new type IV secretion system and the icm/dot virulence system of Legionella pneumophila. Mol. Microbiol. 34:799-809. [DOI] [PubMed] [Google Scholar]
- 29.Stout, J. E., J. Joly, M. Para, J. Plouffe, C. Ciesielski, M. J. Blaser, and V. L. Yu. 1988. Comparison of molecular methods for subtyping patients and epidemiologically linked environmental isolates of Legionella pneumophila. J. Infect. Dis. 157:486-495. [DOI] [PubMed] [Google Scholar]
- 30.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogesesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 31.Tenover, F., R. Arbeit, R. Goering, P. Mickelsen, B. Murray, D. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed- field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel, J. P., and R. R. Isberg. 1999. Cell biology of Legionella pneumophila. Curr. Opin. Microbiol. 2:30-34. [DOI] [PubMed] [Google Scholar]
- 33.Winans, S. C., D. L. Burns, and P. J. Christie. 1996. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 4:64-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winn, W. C. J. 1999. Legionella, p. 572-585. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 35.Yu, V. L., J. F. Plouffe, M. C. Pastoris, J. E. Stout, M. Schousboe, A. Widmer, J. Summersgill, T. File, C. M. Heath, D. L. Paterson, and A. Chereshsky. 2002. Distribution of Legionella species and serogroups isolated by culture in patients with sporadic community-acquired legionellosis: an international collaborative survey. J. Infect. Dis. 186:127-128. [DOI] [PubMed] [Google Scholar]
- 36.Zink, S. D., L. Pedersen, N. P. Cianciotto, and Y. Abu-Kwaik. 2002. The Dot/Icm type IV secretion system of Legionella pneumophila is essential for the induction of apoptosis in human macrophages. Infect. Immun. 70:1657-1663. [DOI] [PMC free article] [PubMed] [Google Scholar]