Abstract
Members of the Bacteroides fragilis group are among the most common anaerobic bacterial isolates in clinical specimens. Metronidazole, a 5-nitroimidazole, is often used as empirical therapy for anaerobic infections. Susceptibility testing is not routinely performed because of nearly universal susceptibility of Bacteroides spp. to this agent. We report a case of metronidazole-resistant Bacteroides fragilis in the United States and demonstrate the presence of the nimA gene, encoding a nitroreductase previously shown to mediate resistance to 5-nitroimidazole antimicrobial agents in B. fragilis strains from Europe and Africa. Because clinical failures in Bacteroides infections have been associated with the use of inactive antimicrobial agents, clinicians need to be aware of the possibility of metronidazole-resistant B. fragilis strains in the United States and the importance of susceptibility testing in selected situations.
Emerging problems of resistance to antimicrobial agents make the treatment of bacterial infections an increasing challenge. An inability to rely on stereotypic resistance patterns can lead to alterations in empirical regimens for common infections or require additional susceptibility testing, thereby increasing the costs of medical care. For clinical isolates of anaerobic bacteria, susceptibility testing is frequently not performed. Rather, treatment is guided by the periodic assessment of susceptibility patterns among stocked isolates or from national surveillance studies (1, 20-22). Members of the Bacteroides fragilis group, the most common isolates from clinically significant anaerobic infections, have been associated with increased mortality rates, increased length of hospital stay (15), and clinical failures when inappropriately treated (14). They can be resistant to some agents used in the treatment of anaerobic infections, such as penicillin and clindamycin (11). However, as seen in large surveillance studies, they have remained almost universally susceptible to metronidazole, a 5-nitroimidazole, despite widespread use of this agent over the past 30 years (1, 20-22). Metronidazole resistance has been reported in Europe (6) and Africa (11), most commonly attributable to the presence of one of five known nim nitroreductase genes (7), but metronidazole resistance has not previously been reported in B. fragilis isolates from the Western Hemisphere (22). We now report a serious infection involving a metronidazole-resistant B. fragilis isolate recovered from a patient in Seattle, Washington, and characterize the likely mechanism of resistance.
CASE REPORT
A 60-year-old female sustained an open fracture of her left ankle after falling into a storm drain while backpacking through Ghana in December 2002. Treatment in Ghana consisted of temporary pinning of her lateral malleolus and subsequent open reduction and internal fixation. No course of antibiotics was prescribed. Purulent drainage from the wound ensued, and the patient traveled to London for further treatment, in which all hardware was removed and necrotic bone and infected tissue were debrided on 12/25/02. An external fixation of the ankle was performed, treatment with clindamycin (450 mg orally, four times daily) and amoxicillin (500 mg orally, thrice daily) was initiated, and the patient returned home to the United States. In March 2003 the patient was admitted to Harborview Medical Center, Seattle, Wash., with increasing pain and swelling of the left ankle. A 4- by 4-cm abscess with extension to necrotic areas of the talus and fibula was found, and therapy with imipenem (500 mg intravenously every 6 h) was initiated on 3/11/03. Intraoperative cultures yielded B. fragilis susceptible to clindamycin but resistant to metronidazole and Morganella morganii. Outpatient therapy was started with clindamycin (900 mg intravenously every 8 h) and cefepime (2 g intravenously every 8 h). Three cultures over a 2-week period remained positive for B. fragilis, and so further debridement of necrotic bone was done. An ankle fusion was performed 2 months into treatment, with intraoperative cultures negative. Antibiotics were continued for another month. There was little evidence of bone regeneration, and so an amputation was considered. The patient declined this course of treatment, and the antibiotics were continued for an additional 3 months along with hyperbaric oxygen therapy. She remains without evidence of recurrent infection or ongoing inflammation but requires assistance with walking.
MATERIALS AND METHODS
Isolation and antimicrobial susceptibility testing.
The metronidazole-resistant B. fragilis clinical strain (JC303) was initially isolated on brucella agar plates that had been incubated for 48 h in an anaerobic BBL GasPak (Becton Dickinson, Sparks, Md.). Initial identification was made by using a Rapid Ana II panel (Remel, Lenexa, Kans.) with confirmation by Vitek ANI (bioMerieux, Durham, N.C.) following incubation in a Shel Lab Bactron anaerobic chamber (Shel Lab, Cornelius, Oreg.). Antibiotic susceptibilities were determined by Etest (AB Biodisk, Solna, Sweden), with metronidazole results confirmed by an agar dilution method (13). All sources of metronidazole were stored at 4°C in the dark. The B. fragilis strain ATCC 25285 was used as a metronidazole-susceptible control. Interpretive criteria were obtained from NCCLS guidelines (13).
PCR and DNA sequencing.
Cultures of JC303 and ATCC 25285 B. fragilis strains were grown overnight in Trypticase soy yeast extract broth, and total cellular DNA was extracted by use of a QIAGEN DNeasy kit (QIAGEN, Valencia, Calif.). PCR was performed for the nim resistance genes using the primers NIM-3 and NIM-5 (23), which were designed to produce a 480-bp fragment. As a control, the katB gene for catalase was amplified using the primers KatB1 (5′-GTAGCAGGAGAACGCGGAGCTGCT-3′) and KatB2 (5′-GTTCATCCGCAGGCATCAGTCGGA-3′) to produce a 1,021-bp fragment. The high-fidelity, proofreading DNA polymerase enzyme PfuUltra (Stratagene) was used in the PCR with the protocol of 94°C for 4 min followed by 30 cycles at 94°C for 30 s, an annealing temperature of 62°C for 30 s, and an extension temperature of 72°C for 2 min. A final extension step of 4 min at 72°C completed the reaction. DNA sequencing of the purified PCR product was performed utilizing a BigDye Terminator protocol (18).
RESULTS
Identification and antimicrobial susceptibility testing.
JC303 was found to be catalase positive, spot indole negative, and resistant to colistin, kanamycin, vancomycin, and bile. The Vitek ANI system identified the organism as B. fragilis at a 99% confidence level, thus confirming the Rapid Ana II panel result of B. fragilis at the same confidence level. Sequencing of 16S ribosomal DNA further confirmed the identity of the organism as B. fragilis.
Strain JC303 was found to be beta-lactamase positive, and the Etest (Table 1) demonstrated susceptibility to clindamycin, amoxicillin-clavulanate, and moxifloxacin (MICs, 0.25 μg/ml) but resistance to metronidazole, with an MIC of >256 μg/ml. Close scrutiny of the Etest zone revealed no inner zone of clearance, but there was a discernible lighter growth pattern at >32 μg/ml. Due to this atypical phenotype, agar dilution testing was performed with metronidazole, demonstrating an MIC of 32 μg/ml. An expanded antibiotic susceptibility profile obtained with the Etest is shown in Table 1. The patient's isolate was resistant to ampicillin, cefotaxime, ceftriaxone, metronidazole, penicillin, piperacillin, and tetracycline. The MIC results for the quality control strain ATCC 25285 were all within acceptable limits.
TABLE 1.
MICs for B. fragilis strain JC303
| Drug | MIC (μg/ml) | Interpretationa |
|---|---|---|
| Amoxicillin-clavulanic acid | 4.0 | S |
| Ampicillin | >256 | R |
| Ampicillin-sulbactam | 4.0 | S |
| Cefotaxime | >256 | R |
| Cefotetan | 8.0 | S |
| Ceftriaxone | >32 | R |
| Chloramphenicol | 2.0 | S |
| Clindamycin | 2.0 | S |
| Imipenem | 0.25 | S |
| Metronidazole | >32 | R |
| Penicillin | >32 | R |
| Piperacillin | >256 | R |
| Piperacillin-tazobactam | 1.0 | S |
| Tetracycline | 32 | R |
| Ticarcillin-clavulanic acid | 8.0 | S |
| Trovafloxacin | 0.125 | S |
Susceptibilities were determined by Etest except for metronidazole, for which agar dilution was performed. Interpretive criteria are per NCCLS guidelines (10). S, sensitive; R, resistant; I, intermediate.
PCR and DNA sequencing.
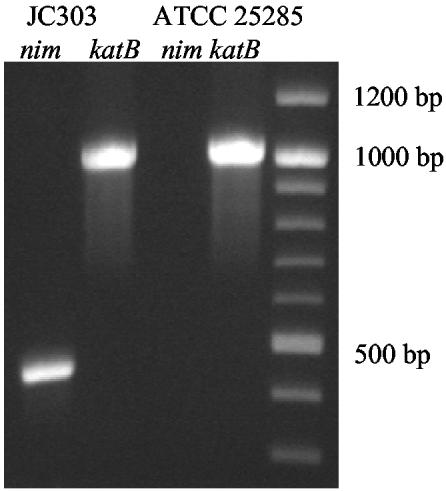
PCR with nim gene-specific primers (18) revealed the presence of a 480-bp nim gene fragment in JC303 not found in ATCC 25285 (Fig. 1). Direct sequencing of the PCR product demonstrated 100% identity to the published nimA gene sequence (11).
FIG. 1.
The predicted 480-bp fragment generated from the nimA-specific primers NIM-3 and NIM-5 (11) (lane 1). Sequencing showed the amplified product to have 100% identity to the published nimA nitroreductase gene sequence (11). Lanes 2 and 4 show the 1,021-bp fragment from KatB, which was used as a control.
DISCUSSION
As the appreciation for anaerobic bacteriology has grown, it has become apparent that the isolation of Bacteroides species from clinical isolates is a significant finding. Studies by Redondo et al. (15) and Nguyen et al. (14) have linked Bacteroides bacteremia to higher mortality rates, which can be further affected by the use of inactive antimicrobials in the treatment regimen.
The class of 5-nitroimidazole drugs (metronidazole, tinidizole, and ornidizole) exerts antimicrobial actions via inhibition of DNA synthesis (19). This action requires intracellular reduction of the nitro group of these prodrugs to produce reactive radical species. Although the 5-nitroimidazole drugs were introduced into clinical practice in 1960, resistance of B. fragilis to these drugs was not described until 1978 (10) and remains rare (1, 20-22). The first metronidazole-resistant Bacteroides isolates were found to have altered end products of glucose metabolism that conferred distinctive biochemical and growth characteristics along with metronidazole MICs of ≥64 (3, 12). The characterization of the nim gene (8) revealed a more prevalent mechanism for metronidazole resistance. Five nim genes (nimA to -E) (7) have now been discovered, either chromosomal or on mobilizable plasmids (2, 16), encoding a nitroreductase that catalyzes drug uptake and reduction without the formation of damage-inducing nitroradicals (4). Moderate- to high-level 5-nitroimidazole resistance is conferred by these genes, with metronidazole MICs ranging from 4 to 32 μg/ml (16). Although multiple antimicrobial resistance determinants may be found clustered together on plasmids, insertion elements, or transposons, this does not seem to be the case for the nim loci (20). Therefore, the finding of metronidazole resistance is not a surrogate for resistance to other antimicrobials, and additional susceptibility testing should be performed.
Although B. fragilis strains with nim-associated metronidazole resistance have been isolated from diverse geographic locations in the Eastern Hemisphere, they are not yet highly prevalent. This may reflect that even though some nim loci are carried on transferable genetic elements (i.e., plasmids), they may not be highly mobilizable in vivo. Nevertheless, the present case demonstrates how resistant bacteria can spread globally even in the absence of selective pressure, as our patient most likely acquired her resistant organism while in Ghana, where she had not been treated with metronidazole. Whether the infection was acquired during the injury in the storm drain in Ghana or subsequently from endogenous intestinal colonization remains speculative. Resistant strains of pathogenic bacteria can spread when patients are transferred between health care facilities, as described for some outbreaks of methicillin-resistant Staphylococcus aureus (5, 17). This underscores the need for health care facilities to be aware of their own susceptibility patterns, because utilizing national surveillance studies of anaerobic susceptibility to guide therapy may not necessarily be a reliable indicator of local trends (9). This further emphasizes the need for increased awareness of possible resistant species and for more local surveillance and susceptibility testing when anaerobes may be clinically significant, even for organisms with heretofore-stereotypic resistance profiles. Clinicians in the United States should be aware of the possibility of encountering imported B. fragilis strains resistant to metronidazole, and they should consider susceptibility testing in selected clinical situations.
REFERENCES
- 1.Aldridge, K. E., D. Ashcraft, K. Cambre, C. L. Pierson, S. G. Jenkins, and J. E. Rosenblatt. 2001. Multicenter survey of the changing in vitro antimicrobial susceptibilities of clinical isolates of Bacteroides fragilis group, Prevotella, Fusobacterium, Porphyromonas, and Peptostreptococcus species. Antimicrob. Agents Chemother. 45:1238-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Breuil, J., O. Patey, A. Dublanchet, and C. Burnat. 1990. Plasmid and non-plasmid mediated reduced sensitivity to metronidazole in the Bacteroides fragilis group. Scand. J. Infect. Dis. 22:247-248. [DOI] [PubMed] [Google Scholar]
- 3.Britz, M. L., and R. G. Wilkinson. 1979. Isolation and properties of metronidazole-resistant mutants of Bacteroides fragilis. Antimicrob. Agents Chemother. 16:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlier, J. P., N. Sellier, M. N. Rager, and G. Reysset. 1997. Metabolism of a 5-nitroimidazole in susceptible and resistant isogenic strains of Bacteroides fragilis. Antimicrob. Agents Chemother. 41:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deplano, A., W. Witte, W. J. V. Leeuwen, Y. Brun, and M. J. Struelens. 2000. Clonal dissemination of epidemic methicillin-resistant Staphylococcus aureus in Belgium and neighboring countries. Clin. Microbiol. Infect. 6:239-245. [DOI] [PubMed] [Google Scholar]
- 6.Dublanchet, A., J. Caillon, J. P. Emond, H. Chardon, and H. B. Drugeon. 1986. Isolation of Bacteroides strains with reduced sensitivity to 5-nitroimidazoles. Eur. J. Clin. Microbiol. 5:346-347. [DOI] [PubMed] [Google Scholar]
- 7.Fang, H., C. Edlund, M. Hedberg, and C. E. Nord. 2002. New findings in beta-lactam and metronidazole resistant Bacteroides fragilis group. Int. J. Antimicrob. Agents 19:361-370. [DOI] [PubMed] [Google Scholar]
- 8.Haggoud, A., G. Reysset, H. Azeddoug, and M. Sebald. 1994. Nucleotide sequence analysis of two 5-nitroimidazole resistance determinants from Bacteroides strains and of a new insertion sequence upstream of the two genes. Antimicrob. Agents Chemother. 38:1047-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hecht, D. W., J. R. Osmolski, and J. P. O'Keefe. 1993. Variation in the susceptibility of Bacteroides fragilis group isolates from six Chicago hospitals. Clin. Infect. Dis. 16(Suppl. 4):S357-S360. [DOI] [PubMed] [Google Scholar]
- 10.Ingham, H. R., S. Eaton, C. W. Venables, and P. C. Adams. 1978. Bacteroides fragilis resistant to metronidazole after long-term therapy. Lancet i:214. [DOI] [PubMed] [Google Scholar]
- 11.Lubbe, M. M., K. Stanley, and L. J. Chalkley. 1999. Prevalence of nim genes in anaerobic/facultative anaerobic bacteria isolated in South Africa. FEMS Microbiol. Lett. 172:79-83. [DOI] [PubMed] [Google Scholar]
- 12.Narikawa, S., T. Suzuki, M. Yamamoto, and M. Nakamura. 1991. Lactate dehydrogenase activity as a cause of metronidazole resistance in Bacteroides fragilis NCTC 11295. J. Antimicrob. Chemother. 28:47-53. [DOI] [PubMed] [Google Scholar]
- 13.National Committee for Clinical Laboratory Standards. 1997. Methods for antimicrobial susceptibility testing of anaerobic bacteria, 4th ed. Approved standard M11-A4. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 14.Nguyen, M. H., V. L. Yu, A. J. Morris, L. McDermott, M. W. Wagener, L. Harrell, and D. R. Snydman. 2000. Antimicrobial resistance and clinical outcome of Bacteroides bacteremia: findings of a multicenter prospective observational trial. Clin. Infect. Dis. 30:870-876. [DOI] [PubMed] [Google Scholar]
- 15.Redondo, M. C., M. D. Arbo, J. Grindlinger, and D. R. Snydman. 1995. Attributable mortality of bacteremia associated with the Bacteroides fragilis group. Clin. Infect. Dis. 20:1492-1496. [DOI] [PubMed] [Google Scholar]
- 16.Reysset, G. 1996. Genetics of 5-nitroimidazole resistance in Bacteroides species. Anaerobe 2:59-69. [DOI] [PubMed] [Google Scholar]
- 17.Roman, R. S., J. Smith, M. Walker, S. Byrne, K. Ramotar, B. Dyck, A. Kabani, and L. E. Nicolle. 1997. Rapid geographic spread of a methicillin-resistant Staphylococcus aureus strain. Clin. Infect. Dis. 25:698-705. [DOI] [PubMed] [Google Scholar]
- 18.Rosenblum, B. B., L. G. Lee, S. L. Spurgeon, S. H. Khan, S. M. Menchen, C. R. Heiner, and S. M. Chen. 1997. New dye-labeled terminators for improved DNA sequencing patterns. Nucleic Acids Res. 25:4500-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sigeti, J. S., D. G. Guiney, Jr., and C. E. Davis. 1983. Mechanism of action of metronidazole on Bacteroides fragilis. J. Infect. Dis. 148:1083-1089. [DOI] [PubMed] [Google Scholar]
- 20.Snydman, D. R., L. McDermott, G. J. Cuchural, Jr., D. W. Hecht, P. B. Iannini, L. J. Harrell, S. G. Jenkins, J. P. O'Keefe, C. L. Pierson, J. D. Rihs, V. L. Yu, S. M. Finegold, and L. Gorbach. 1996. Analysis of trends in antimicrobial resistance patterns among clinical isolates of Bacteroides fragilis group species from 1990 to 1994. Clin. Infect. Dis. 23(Suppl. 1):S54-S65. [DOI] [PubMed] [Google Scholar]
- 21.Snydman, D. R., N. V. Jacobus, L. A. McDermott, S. Supran, G. J. Cuchural, Jr., S. Finegold, L. Harrell, D. W. Hecht, P. Iannini, S. Jenkins, C. Pierson, J. Rihs, and S. L. Gorbach. 1999. Multicenter study of in vitro susceptibility of the Bacteroides fragilis group, 1995 to 1996, with comparison of resistance trends from 1990 to 1996. Antimicrob. Agents Chemother. 43:2417-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snydman, D. R., N. V. Jacobus, L. A. McDermott, R. Ruthazer, E. J. Goldstein, S. M. Finegold, L. J. Harrell, D. W. Hecht, S. G. Jenkins, C. Pierson, R. Venezia, J. Rihs, and S. L. Gorbach. 2002. National survey on the susceptibility of Bacteroides fragilis group: report and analysis of trends for 1997-2000. Clin. Infect. Dis. 35:S126-S134. [DOI] [PubMed] [Google Scholar]
- 23.Trinh, S., and G. Reysset. 1996. Detection by PCR of the nim genes encoding 5-nitroimidazole resistance in Bacteroides spp. J. Clin. Microbiol. 34:2078-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]