Abstract
We examined microevolution in a series of Candida albicans strains isolated from patients with catheter-related candidemia. Sixty-one isolates (29 from blood, 18 from catheters, 10 from urine, and 4 from other sites) were obtained from 15 patients who were admitted to the same hospital over a 3-year period. Isolates were analyzed by using Southern hybridization with the C1 fragment of Ca3 as a probe (C1 fingerprinting) and pulsed-field gel electrophoresis (PFGE). PFGE typing consisted of electrophoretic karyotyping (EK) and restriction endonuclease analysis of genomic DNA (REAG) by using SfiI (REAG-S) and BssHII (REAG-B). When catheter isolates were compared with blood isolates from the same patient, catheter isolates from 5 of 14 patients (36%) exhibited minor band differences (microevolution) relative to blood isolates in either C1 fingerprinting (n = 4), REAG-S (n = 3), or REAG-B (n = 5) profiles, although they had identical EK patterns. However, the other sequential isolates from each patient, which had identical EK patterns, showed the same REAG and C1 fingerprinting patterns. Both fingerprinting methods revealed that two distinct genotypes were shared by isolates from seven patients in a neonatal intensive care unit, suggesting two nosocomial clusters. Except for two catheter isolates from the index patients of each cluster, no consecutive isolates collected from each of the two clusters showed any microevolution during the 2- or 7-month cluster periods. The findings suggest that in catheter-related candidemia, some C. albicans strains undergo microevolution during catheter colonization.
Within the genus Candida, the species Candida albicans is the most common fungus isolated from humans (15) and accounts for 50 to 70% of all nosocomial bloodstream infections (BSI) resulting from Candida species (1, 3, 14). In recent years, it has become clear that C. albicans produces genetically altered variants at a high rate. The majority of commensal and infecting populations of C. albicans from the same individuals are clonal in origin but undergo microevolution at the site of colonization and through recurrent episodes of infection (11, 21, 24). These microevolutionary changes are due to genomic reorganization involving the repetitive RPS element and can be identified in clonal populations by using the Ca3 probe or the C1 fragment of the Ca3 probe (11, 16).
Marco et al. (12) reported that microevolution of the colonizing strain occurs in one-third of patients with candidemia caused by C. albicans. However, the relationship between microevolution of C. albicans and subsequent BSI remains uncertain, because C. albicans commonly produces long-term colonization at various body sites, even in healthy individuals (15, 24), and most candidemia arises from the patient's own endogenous flora (18, 26). In the case of catheter-related BSI, the catheter may serve as the source of fungemia, as organisms proliferate on the catheter surface, from which the organism may enter the bloodstream and then spread to other sites (6). Therefore, examination of microevolution in serial isolates of C. albicans from patients with catheter-related candidemia may reveal genetic evolution of C. albicans strains during the progression of BSI.
This study used DNA fingerprinting by Southern blot hybridization with the 0.98-kb C1 fragment of the Ca3 probe (C1 fingerprinting) and pulsed-field gel electrophoresis (PFGE) to examine serial C. albicans isolates from 15 patients with catheter-related candidemia who were admitted to the same hospital over a 3-year period. The purpose of this study was to determine whether clonal strains of C. albicans from catheter-related candidemia exhibit microevolution during the progression of infection or during nosocomial spread among patients over time.
MATERIALS AND METHODS
Microorganisms.
Sixty-one C. albicans isolates were obtained from 15 patients with catheter-related candidemia. The patients were admitted to Chonnam National University Hospital over a three-year period (between 2000 and 2002), and all were diagnosed as having catheter-related candidemia. Cases of candidemia were defined as catheter related if no other source of infection was found and if the semiquantitative catheter tip culture yielded more than 15 colonies of the same C. albicans strain (19). The C. albicans isolates were collected from cultures of various clinical specimens before, during, or after collection of the isolates that caused candidemia. The isolates were cultured from blood (29 isolates from 15 patients), catheter tip (18 isolates from 15 patients), urine (10 isolates from 7 patients), cerebrospinal fluid (CSF) (2 isolates from 1 patient), and respiratory (2 isolates from 2 patients) specimens, as part of routine diagnostic procedures. C. albicans was identified by assessing germ tube and chlamydospore formation and by examining API 20C (BioMérieux, Marcy L'Etoile, France) or ATB 32C (BioMérieux) sugar assimilation patterns.
Southern hybridization with the C1 fragment of the Ca3 probe.
Southern blot hybridization was performed by using previously described methods (10). Briefly, cells from single clonal colonies were transferred to YPD broth (1% yeast extract, 2% Bacto Peptone, 2% glucose) (Difco, Detroit, Mich.) and grown to the late log phase. The DNA from each isolate was prepared by the method of Scherer and Stevens (20), digested with EcoRI, and separated on 0.8% agarose gels. The gels were stained with ethidium bromide to assess loading and transferred to nitrocellulose membranes. The DNA fragments on the membranes were then hybridized with the radiolabeled C1 probe as previously described (11, 21). The blots were exposed to XAR-S film (Eastman Kodak Co., Rochester, N.Y.). The autoradiograms were examined visually to assess both band position and band intensity, in order to determine whether C. albicans isolates from the same patient or the same clone showed genetic variation (i.e., minor genotypes) suggestive of microevolution (11). The C1 fingerprints were assessed for evidence of microevolution by examining differences in the number of bands for isolates from clinical specimens compared with the original blood isolate from each patient and whether any addition (1+) or loss (1−) of bands occurred. This procedure was repeated for all isolates analyzed at least twice to ascertain pattern relatedness and to ensure reproducibility.
PFGE analysis.
PFGE analysis was performed by using a procedure described previously (26). It involved electrophoretic karyotyping (EK) and restriction endonuclease analysis of genomic DNA (REAG) with BssHII (REAG-B) or SfiI (REAG-S). Briefly, for each Candida isolate, one colony from 48-h Sabouraud dextrose agar cultures was incubated overnight at 37°C in 10 ml of YPD broth. A 150-μl aliquot of cell suspension was mixed with 30 U of lyticase (Sigma, St. Louis, Mo.) and 150 μl of 1.6% agarose (FMC BioProducts, Hercules, Calif.) that was melted and held at 50 to 55°C. Aliquots of the mixture of cells and agarose were placed in individual molds to form agarose plugs and allowed to harden for 20 min at room temperature. The agarose plugs were removed from the molds, placed in 500 μl of a lyticase buffer containing 50 mM EDTA and 100 U of lyticase per ml for 2 h, and then washed once in 2 ml of distilled water. The plugs were incubated in proteinase K solution (50 mM EDTA and 100 μg of proteinase K [Invitrogen, Carlsbad, Calif.]) for 16 to 18 h at 50°C and finally washed five times in 50 mM sodium EDTA (pH 8.0).
For EK, Candida chromosomal DNA was separated by PFGE with the GenePath system (Bio-Rad, Hercules, Calif.). Electrophoresis was performed for 48 h in a 0.7% agarose gel (SeaKem GTG agarose; FMC BioProducts) in 0.5× TBE buffer (0.1 M Tris, 0.09 M boric acid, 0.01 M EDTA [pH 8.0]) at 4 V/cm, with initial and final switch times of 90 and 325 s, respectively. After electrophoresis, the gels were stained with a 0.5-μg/ml ethidium bromide solution and photographed under UV illumination. Isolates that differed by one or more bands were considered to have different karyotypes (23, 26).
For REAG, digestion was carried out with SfiI at 37°C for 16 h or with BssHII at 50°C for 16 h. Electrophoresis for REAG-S was performed as for EK, except that a 1% agarose gel was used. Electrophoresis for REAG-B was for 20 h in a 1.0% agarose gel (SeaKem GTG agarose; FMC BioProducts) in 0.5× TBE buffer at 4 V/cm, with an initial switch time of 5 s and a final switch time of 50 s. The criteria proposed by Tenover et al. were used to analyze the REAG patterns (25). Briefly, strains with banding patterns identical in the sizes and numbers of bands were considered genetically indistinguishable and assigned to the same type, strains with banding patterns that differed by only three or fewer bands were considered closely related and described as subtypes (a or b) of a given clonal type, and strains with banding patterns that differed by four or more bands were considered different and assigned to separate types. When serial isolates from the same patient show a minor genotypic change (subtype) in the REAG pattern, it represents the occurrence of microevolution.
All isolates were analyzed at least twice (mean, three times; range, two to five times), repeating the procedure including subculturing of isolates from the original stock culture to Sabouraud dextrose agar, preparation of DNA, endonuclease digestion, and separation of the DNA by PFGE, to ascertain pattern relatedness and to ensure reproducibility.
RESULTS
Comparison of isolates from blood and other sites from the same patient.
During the study period, 35 patients whose blood was positive for C. albicans were identified at Chonnam National University Hospital. Of these, 15 patients met the inclusion criterion of catheter-related candidemia used in this study. Table 1 presents the sequence of isolates, their anatomical sites, the dates of isolation, and genotyping results for isolates from each of the 15 patients. The patients included seven adult patients (patients 1 to 7) and eight neonates admitted to the neonatal intensive care unit (NICU) (patients 8 to 15). In this study, all isolates obtained from sites other than blood were collected only after collection of the first blood isolate, with two exceptions (a catheter isolate of patient 1 and a urine isolate of patient 3). The interval between isolating C. albicans from the first positive blood culture and isolating it from another site in the same patient ranged from 1 to 37 days.
TABLE 1.
Genotyping results for C. albicans isolates from blood cultures and other body sites of 15 patients with catheter-related candidemiaa
| Patient | Ward | Isolate no. | Source | Isolation date (mo/ day/yr) | Microevolution detected by C1 fingerprintingb | PFGE
|
||
|---|---|---|---|---|---|---|---|---|
| EK | REAG-S type | REAG-B type | ||||||
| 1 | MICU | 1-C1 | Catheter | 05/21/2000 | 0 | K1 | S1 | B1 |
| 1-B1 | Blood | 05/26/2000 | 0 | K1 | S1 | B1 | ||
| 1-B2 | Blood | 06/14/2000 | 0 | K1 | S1 | B1 | ||
| 1-C2 | Catheter | 06/29/2000 | 0 | K2 | S1 | B1 | ||
| 2 | SICU | 2-C1 | Catheter | 07/31/2000 | 1+ | K2 | S2 | B2ac |
| 2-B1 | Blood | 07/31/2000 | 0 | K2 | S2 | B2b | ||
| 3 | 73W | 3-U1 | Urine | 07/14/2000 | 0 | K3 | S3 | B3 |
| 3-B1 | Blood | 08/01/2000 | 0 | K3 | S3 | B3 | ||
| 3-C1 | Catheter | 08/04/2000 | 0 | K3 | S3 | B3 | ||
| 4 | 11W | 4-B1 | Blood | 08/21/2000 | 0 | K4 | S4 | B4a |
| 4-C1 | Catheter | 08/21/2000 | 0 | K4 | S4 | B4a | ||
| 4-C2 | Catheter | 08/21/2000 | 1+, 1− | K4 | S4 | B4bc | ||
| 4-U1 | Urine | 08/29/2000 | 0 | K4 | S4 | B4a | ||
| 4-C3 | Blood | 09/08/2000 | 0 | K4 | S4 | B4a | ||
| 5 | SICU | 5-B1 | Blood | 07/14/2001 | 0 | K4 | S5 | B5 |
| 5-B2 | Blood | 07/16/2001 | 0 | K4 | S5 | B5 | ||
| 5-U1 | Urine | 07/16/2001 | 0 | K4 | S5 | B5 | ||
| 5-C1 | Catheter | 07/16/2001 | 0 | K4 | S5 | B5 | ||
| 5-B3 | Blood | 08/19/2001 | 0 | K4 | S5 | B5 | ||
| 6 | 72W | 6-B1 | Blood | 11/06/2001 | 0 | K5 | S6a | B6 |
| 6-C1 | Catheter | 11/06/2001 | NA | K6 | S6b | B7 | ||
| 7 | 10W | 7-B1 | Blood | 04/04/2002 | 0 | K2 | S7 | B8 |
| 7-R1 | TA | 04/09/2002 | 0 | K2 | S7 | B8 | ||
| 7-B2 | Blood | 04/12/2002 | 0 | K2 | S7 | B8 | ||
| 7-B3 | Blood | 04/13/2002 | 0 | K2 | S7 | B8 | ||
| 7-C1 | Catheter | 04/13/2002 | 0 | K2 | S7 | B8 | ||
| 8 | NICU | 8-C1 | Catheter | 03/10/2001 | 0 | K5 | S8ac | B9ac |
| 8-B1 | Blood | 03/11/2001 | 0 | K5 | S8b | B9b | ||
| 8-U1 | Urine | 03/11/2001 | 0 | K5 | S8b | B9b | ||
| 8-B2 | Blood | 03/15/2001 | 0 | K5 | S8b | B9b | ||
| 9 | NICU | 9-C1 | Catheter | 04/28/2001 | 0 | K5 | B8b | B9b |
| 9-B1 | Blood | 04/28/2001 | 0 | K5 | S8b | B9b | ||
| 9-U1 | Urine | 05/02/2001 | 0 | K5 | S8b | B9b | ||
| 9-U2 | Urine | 05/04/2001 | 0 | K5 | S8b | B9b | ||
| 9-C2 | Catheter | 05/04/2001 | 0 | K5 | S8b | B9b | ||
| 9-S1 | CSF | 05/07/2001 | 0 | K5 | S8b | B9b | ||
| 9-B2 | Blood | 05/09/2001 | 0 | K5 | S8b | B9b | ||
| 9-S2 | CSF | 05/15/2001 | 0 | K5 | S8b | B9b | ||
| 10 | NICU | 10-C1 | Catheter | 05/09/2001 | 0 | K5 | S8b | B9b |
| 10-B1 | Blood | 05/07/2001 | 0 | K5 | S8b | B9b | ||
| 11 | NICU | 11-C1 | Catheter | 01/21/2002 | 1+ | K6 | S9ac | B10ac |
| 11-B1 | Blood | 01/19/2002 | 0 | K6 | S9b | B10b | ||
| 11-U1 | Urine | 01/22/2002 | 0 | K6 | S9b | B10b | ||
| 11-B2 | Blood | 01/28/2002 | 0 | K6 | S9b | B10b | ||
| 11-U2 | Urine | 01/28/2002 | 0 | K6 | S9b | B10b | ||
| 12 | NICU | 12-C1 | Catheter | 04/08/2002 | 1− | K7 | S10ac | B11ac |
| 12-B1 | Blood | 04/08/2002 | 0 | K7 | S10b | B11b | ||
| 12-R1 | TA | 04/08/2002 | NA | K8 | S11 | B12 | ||
| 12-B2 | Blood | 04/12/2002 | 0 | K7 | S10b | B11b | ||
| 13 | NICU | 13-B1 | Blood | 05/21/2002 | 0 | K6 | S9b | B10b |
| 13-C1 | Catheter | 05/22/2002 | 0 | K6 | S9b | B10b | ||
| 13-B2 | Blood | 05/22/2002 | 0 | K6 | S9b | B10b | ||
| 14 | NICU | 14-B1 | Blood | 06/17/2002 | 0 | K6 | S9b | B10b |
| 14-C1 | Catheter | 06/18/2002 | 0 | K6 | S9b | B10b | ||
| 14-U1 | Urine | 06/22/2002 | 0 | K6 | S9b | B10b | ||
| 14-B2 | Blood | 06/26/2002 | 0 | K6 | S9b | B10b | ||
| 14-B3 | Blood | 06/28/2002 | 0 | K6 | S9b | B10b | ||
| 14-U2 | Urine | 06/29/2002 | 0 | K6 | S9b | B10b | ||
| 14-B4 | Blood | 06/29/2002 | 0 | K6 | S9b | B10b | ||
| 15 | NICU | 15-B1 | Blood | 08/05/2002 | 0 | K6 | S9b | B10b |
| 15-C1 | Catheter | 08/05/2002 | 0 | K6 | S9b | B10b | ||
MICU, medical intensive care unit; SICU, surgical intensive care unit; TA, tracheal aspirate.
Microevolution by C1 fingerprinting is assessed according to the number of bands differing from those of the blood isolate from each patient and whether there is a band addition (1+), a band loss (1−), or no microevolution (0). NA, not applicable because the isolate showed a C1 fingerprinting pattern different from that of the blood isolate from the same patient.
A minor change in REAG patterns in serial isolates from the same patient suggests microevolution.
The blood isolates showed EK patterns identical to those from the catheter (14 of 15 patients), urine (7 of 7 patients), CSF (1 patient), and respiratory (1 of 2 patients) specimens from the same patient. Only two isolates of two patients (a catheter isolate in patient 6 and a tracheal isolate in patient 12) showed EK patterns different from those of the corresponding blood isolates. In these two cases, both the REAG and C1 fingerprinting disclosed different patterns. All other sequential isolates from each patient, which had the same EK pattern, showed the same or similar REAG patterns, suggesting that they were clonal in origin.
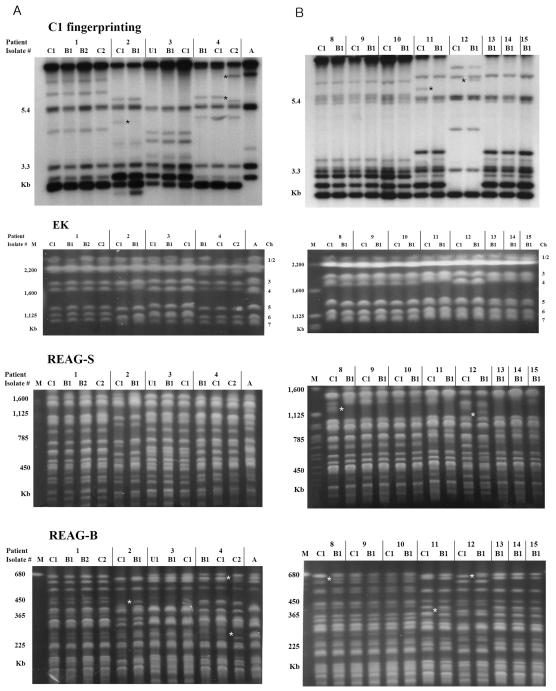
For 14 of 15 patients, catheter isolates from each patient had an EK pattern identical to that of the corresponding blood isolate. Of these, the catheter isolates from each of nine patients had REAG and C1 fingerprinting patterns identical to those of the corresponding blood isolates, while those of five patients (36%) showed minor genetic differences (one or two bands) by REAG-B (n = 5), REAG-S (n = 3), or C1 fingerprinting (n = 4), suggesting that microevolution had occurred (Table 1; Fig. 1). Microevolution was detected by both REAG and C1 fingerprinting for four patients (patients 2, 4, 11, and 12) but was detected by REAG for only one patient (patient 8). For patient 4, two isolates were obtained from catheters on the same day; one isolate showed the same genotype as the blood isolate, but the other showed microevolution that was detected by both REAG-B and C1 fingerprinting. The changes observed in the C1 fingerprinting and REAG patterns were usually restricted to one or two high-molecular-weight bands. In five cases of candidemia in which the catheter isolate showed microevolution in comparison with the blood isolate, the time interval between admission and the first positive blood culture ranged from 14 to 34 days.
FIG. 1.
Genotyping patterns of C. albicans obtained by C1 fingerprinting, EK, REAG-S, and REAG-B. The sequential C. albicans isolates were obtained from catheter (C), blood (B), and urine (U) cultures of four adult patients (patients 1 to 4) (A) and eight NICU patients (patients 8 to 15) (B) with catheter-related fungemia. See Table 1 for detailed information on each isolate. Sequential isolates from each patient had the same karyotype and showed the same or similar REAG and C1 fingerprinting patterns, suggesting that they were clonal in origin. Stars indicate the positions of added or deleted bands in the minor patterns for clonal strains from the same patient. The catheter isolates from five patients (patients 2, 4, 8, 11, and 12), in comparison with the first blood isolates from the patients, showed the same EK pattern but had one- or two-band differences in the C1 fingerprinting and REAG banding patterns, suggesting that microevolution had occurred. Microevolution of isolates from patient 8 was detected with both REAG-S and REAG-B but not with C1 fingerprinting. Two DNA types were shared by isolates from seven patients (type I, patients 8 to 10; type II, patients 11 and 13 to 15), which suggested two nosocomial clusters. None of these type I and type II isolates, except for two catheter isolates from index patients (patients 8 and 11), showed any microevolution during cross-infection among three and four patients, respectively. A, C. albicans ATCC 90028. M, Saccharomyces cerevisiae DNA concatemers as a molecular size marker. Ch, chromosomal assignments on the EK patterns.
For nine patients for whom two or three blood culture isolates were available, the sequential blood isolates obtained from each patient had the same karyotype, REAG-B, and REAG-S, and C1 fingerprinting patterns. All other sequential isolates from urine (7 patients), CSF (1 patient), and respiratory (1 patient) specimens of each of these patients had the same EK, REAG, and C1 fingerprinting patterns. Overall, 36% of catheter isolates (5 of 14) showed microevolution when compared with their blood isolates, while none of the other sequential blood, urinary, or CSF isolates from patients with catheter-related candidemia showed any microevolution.
Comparison among isolates from different patients.
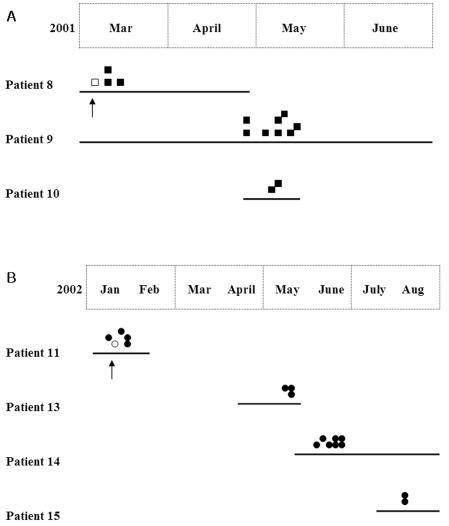
When analyzed by using three PFGE typing methods, the 61 isolates from the 15 patients yielded 8 different EK patterns, 11 different patterns after digestion with SfiI, and 12 different patterns after digestion with BssHII. Overall, the combination of the three PFGE methods and C1 fingerprinting identified 12 distinct DNA types. Two DNA types (types I and II) were shared among 31 isolates from seven neonatal patients in the NICU, which suggested two nosocomial clusters (Table 1 and Fig. 1B). Type I was detected in isolates of three NICU patients (patients 8, 9, and 10) and persisted over a period of 2 months (between 10 March and 19 May 2001). Another identical DNA pattern, type II, was first recovered from patient 11 in January 2002 and then recovered from three other NICU patients (patients 13 to 15) from May to August 2002 (Fig. 2). During each cluster of C. albicans candidemia, most of the patient hospitalizations overlapped, except that of patient 11, and the same health care workers cared for the patients. Although the first two catheter isolates from the two index patients (patients 8 and 11) showed microevolution when compared with each type isolate, none of the other isolates from each of the two clusters showed any microevolution during the 2- or 7-month duration of the cluster.
FIG. 2.
Schematic representation of the epidemiological situation and genetic relatedness of isolates in two nosocomial clusters. The duration of hospitalization (from the date of admission to discharge) of each patient is represented by a thick line. Identical symbols indicate isolates with identical REAG and C1 patterns. Two identical isolates of type I and II strains were obtained from three (A) and four (B) NICU patients, respectively. Only the first two catheter isolates (indicated by arrows) from the two index patients (patients 8 and 11) showed microevolution when compared with each type of isolate.
DISCUSSION
The phenomenon of microevolution in BSIs caused by Candida species has not been completely explored. We examined microevolution among C. albicans isolates from catheter-related candidemia, in relation to the transition from catheter colonization to hematogenous dissemination or nosocomial transmission. We showed that in catheter-related candidemia, microevolution may occur in some C. albicans strains during catheter colonization. In addition, the findings suggest the utility of the PFGE methods in detecting microevolution within individual patient isolates by comparison with the results of C1 fingerprinting.
The 11-kb complex DNA fingerprinting probe Ca3 is effective both for cluster analyses of C. albicans isolates and for identifying microevolutionary changes in the sizes of hypervariable genomic fragments (10, 11, 16, 21, 24). The microevolutionary changes identified by Ca3 are due exclusively to the reorganization of genomic sequences identified by the 0.98-kb C1 fragment (16). Lockhart et al. (11) demonstrated that C1 fingerprinting is an excellent indicator of microevolution, although it has some limitations. Using C1 fingerprinting, they assessed C. albicans populations colonizing the oral cavities and vaginal canals of healthy individuals and vaginitis patients and found that each population of the same clone contained minor variants, demonstrating microevolution.
We used both C1 fingerprinting and PFGE methods to detect microevolution in C. albicans strains. The PFGE typing performed in this study consisted of EK, REAG-B, and REAG-S. Barton et al. (4) reported the stability of karyotypes in serial isolates of C. albicans from neutropenic patients. Using additional typing methods, they demonstrated that a change in the karyotype of a group of serial clinical isolates of C. albicans represents colonization by a new strain. Our results also indicate that different karyotypes of C. albicans isolates can occur only with different REAG patterns, which represent the occurrence of a new strain. In addition, we found that sequential isolates that had the same karyotype had the same or similar REAG patterns in the same patient. Therefore, without the karyotype information, a minor change (a one- or two-band difference) in the REAG patterns, in serial isolates of C. albicans from the same patient, indicates microevolution in the same strain.
Comparing the REAG patterns of isolates from the same patient, microevolution was detected in isolates from 5 out of 15 patients in this study. These results agree with the results obtained by using C1 fingerprinting, except for the isolates from one patient: microevolution was detected in isolates from patient 8 only by REAG. These findings suggest that PFGE methods and C1 fingerprinting provide a powerful means of detecting microevolution among sequential isolates of C. albicans from catheter-related BSI. In addition, since the microevolution in isolates from patient 8 was not detected by C1 fingerprinting, microevolution may occur without a change in the size of the major repeat sequence. Forche et al. (7) pointed out that genetic changes, such as mitotic recombination and gene conversion, occur at a sufficiently high frequency to be important in the transition of C. albicans from a commensal to a pathogenic organism.
When we compared isolates obtained from catheters with blood isolates from the same individuals, the C. albicans strain recovered from the catheter was identical or similar to the blood isolate in 14 of 15 patients. These findings suggest that the portal of entry of the organism in most patients is transvenous, via the catheter. The genetic identity of sequential isolates from blood (9 of 9 patients), urine (6 of 6 patients), respiratory (1 of 2 patients), and CSF (1 patient) specimens from the same patient was confirmed by using the PFGE and C1 fingerprinting methods. In this study of catheter-related candidemia, because most of the isolates were obtained after collecting the first blood isolate from each patient and because catheter colonization frequently follows catheter-related BSI (26), we postulate that in these patients, most of the blood, urine, or CSF strains arose recently from the catheter or BSI strain.
Marco et al. (12) compared C. albicans isolates from urine, stool, and other sites (not including catheter isolates) with blood isolates from the same individuals. They found that urine and stool isolates were similar or identical to the blood isolates in approximately 90% of the patients and that in one-third of the patients (33%), microevolution had occurred in the colonizing strain. This value is below the values of 66 and 55% that they had previously observed for collections of commensal isolates and isolates causing vaginitis, respectively (10, 11). The authors suggested that the apparent differences in incidence of microevolution were due to the difference in the time frames of the studies. In the latter studies, subjects may carry the same commensal strains or the strains causing vaginitis for very long periods, up to several years. The lower figure for microevolution of C. albicans in hospitalized patients suggests that the strains that colonized patients in the intensive care units had insufficient time to diversify, supporting the idea that they had recently colonized their present hosts (11).
In our study, catheter strains from 36% of patients (5 of 14) showed microevolution in comparison with the corresponding blood isolates. This 36% incidence of microevolution of catheter strains is similar to the 33% incidence of microevolution in urine and stool isolates compared to blood isolates from candidemic patients observed by Marco et al. (12). Two isolates simultaneously isolated from catheter cultures of a patient (patient 4) on the same day showed minor band differences between the two strains, which suggested that microevolution occurred at the site of the catheter. In contrast to catheter strains, none of the sequential blood, urine, or CSF isolates showed microevolution in our study. This suggests that in catheter-related fungemia, microevolution of the C. albicans isolates often occurs during catheter colonization but that consecutive blood or urinary isolates collected during catheter-related fungemia appear to be genetically stable in the same patient.
Our study showed that two genotypes (types I and II) were common among isolates from the seven NICU patients, demonstrating a high rate of nosocomial clusters of catheter-related BSI caused by C. albicans in the NICU. Pfaller et al. (13) demonstrated that particular BSI strains of C. albicans were more highly concentrated in particular geographic locales and that established BSI strains were endemic in some, but not all, hospitals in the study and underwent microevolution in the hospital setting. Marco et al. (12) found that an endemic strain had cross-contaminated the hands of hospital coworkers over a period of approximately 1 year and that this strain had undergone significant microevolution. They reported that in the majority of cases, health care workers were contaminated by isolates from colonized patients but that in a significant minority, the reverse was true. In our study, interestingly, catheter strains from the index patients, patients 8 and 11, of each of the two nosocomial clusters showed microevolution in comparison with the type I or II strains, respectively. However, genotyping of other strains causing nosocomial clusters revealed that the nosocomial transmission of C. albicans BSI may occur among individuals without microevolution of the strain for several months.
In five cases of candidemia in which the catheter isolate showed microevolution in comparison with the corresponding blood isolate, the time interval between admission and the first positive blood culture ranged from 14 to 34 days, whereas identical isolates of type I and II strains were obtained repeatedly, without evidence of microevolution, for the 2- or 7-month duration of each of the two clusters, respectively. These data suggest that the occurrence of microevolution depends not only on the duration of colonization or infection but also on the genetic stability of the strain in a given host environment. This was also supported by our finding that microevolution occurred frequently in catheter strains but occurred in no consecutive strains obtained from blood, urine, or CSF isolates collected after the first blood isolates. Therefore, it is possible that the microevolution of C. albicans strains takes place during colonization of catheters or mucosal surfaces of the patient, on the hands of health care workers, or in the hospital environment. However, once a genetically stable strain is established in an individual patient, it may be responsible for invasion of the bloodstream and then spread to urine or CSF in the same patient or by nosocomial transmission among the different patients over time without significant microevolutionary changes.
The pathogenesis of catheter-related BSI is complex and incompletely understood. The occurrence and rate of BSI are dependent on microbial virulence factors, host factors, and catheter characteristics (6, 17, 22). The pathogenic process probably begins with attachment to and colonization of either the outer or inner surface of the catheter by the infecting microorganisms. While most catheters are colonized with organisms embedded in biofilms after placement, only a few of these organisms cause BSI (2, 17). The ratio of BSI incidence to catheter colonization was highest for Staphylococcus aureus, followed by C. albicans and then coagulase-negative staphylococci (6). This probably reflects the relative virulences of these organisms as pathogens on intravascular devices (6). In C. albicans, in addition to the capacity to form biofilms, a number of protein receptors, including fibronectin (9), fibrinogen (5), and vitronectin (8), may facilitate adhesion to epithelial, endothelial, and foreign body surfaces.
Although bacterial microevolution is responsible for the rapid emergence of variants with novel virulence and resistance properties (27), the significance of the microevolution of C. albicans in the pathogenesis of catheter-related BSI is completely unknown. Perhaps it is one of the various adaptation mechanisms that permitted C. albicans to become a successful human pathogen, although further investigation is needed to confirm this speculation. In cases of catheter-related BSI, these organisms may evolve genetically and overcome host defenses, resulting in catheter colonization and eventual invasion of the fungus into the bloodstream. Such a role for microevolution is supported by our observations that microevolution frequently occurred at catheter sites but rarely occurred among consecutive blood, urine, or CSF isolates in the same patient or in clonal isolates from two nosocomial clusters. Our findings suggest that in catheter-related candidemia, some C. albicans strains undergo microevolution during catheter colonization, producing genetically stable variants and thereby allowing the establishment BSI in the patient and nosocomial transmission among patients.
Acknowledgments
This work was supported by a grant (R04-2002-000-00036-0) from the Basic Research Program of the Korea Science & Engineering Foundation.
REFERENCES
- 1.Abi-Said, D., E. Anaissie, O. Uzun, I. Raad, H. Pinzcowski, and S. Vartivarian. 1997. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin. Infect. Dis. 24:1122-1128. [DOI] [PubMed] [Google Scholar]
- 2.Anaissie, E., G. Samonis, D. Kontoyiannis, J. Costerton, U. Sabharwal, G. Bodey, and I. Raad. 1995. Role of catheter colonization and infrequent hematogenous seeding in catheter-related infections. Eur. J. Clin. Microbiol. Infect. Dis. 14:134-137. [DOI] [PubMed] [Google Scholar]
- 3.Banerjee, S., T. Emori, D. Culver, R. Gaynes, W. Jarvis, T. Horan, J. Edwards, J. Tolson, T. Henderson, and W. Martone. 1991. Secular trends in nosocomial primary bloodstream infections in the United States, 1980-1989. Am. J. Med. 91:86S-89S. [DOI] [PubMed] [Google Scholar]
- 4.Barton, R. C., A. van Belkum, and S. Scherer. 1995. Stability of karyotype in serial isolates of Candida albicans from neutropenic patients. J. Clin. Microbiol. 33:794-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casanova, M., J. L. Lopez-Ribot, C. Monteagudo, A. Llombart-Bosch, R. Sentandreu, and J. P. Martinez. 1992. Identification of a 58-kilodalton cell surface fibrinogen-binding mannoprotein from Candida albicans. Infect. Immun. 60:4221-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crump, J. A., and P. J. Collignon. 2000. Intravascular catheter-associated infections. Eur. J. Clin. Microbiol. Infect. Dis. 19:1-8. [DOI] [PubMed] [Google Scholar]
- 7.Forche, A., G. May, J. Beckerman, S. Kauffman, J. Becker, and P. T. Magee. 2003. A system for studying genetic changes in Candida albicans during infection. Fungal Genet. Biol. 39:38-50. [DOI] [PubMed] [Google Scholar]
- 8.Jakab, E., M. Paulsson, F. Ascencio, and A. Ljungh. 1993. Expression of vitronectin and fibronectin binding by Candida albicans yeast cells. APMIS 101:187-193. [PubMed] [Google Scholar]
- 9.Klotz, S. A., R. C. Hein, R. L. Smith, and J. B. Rouse. 1994. The fibronectin adhesin of Candida albicans. Infect. Immun. 62:4679-4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lockhart, S. R., B. D. Reed, C. L. Pierson, and D. R. Soll. 1996. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “substrain shuffling”: demonstration by sequential DNA fingerprinting with probes Ca3, C1, and CARE2. J. Clin. Microbiol. 34:767-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockhart, S. R., J. J. Fritch, A. S. Meier, K. Schroppel, T. Srikantha, R. Galask, and D. R. Soll. 1995. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J. Clin. Microbiol. 33:1501-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marco, F., S. R. Lockhart, M. A. Pfaller, C. Pujol, M. S. Rangel-Frausto, T. Wiblin, H. M. Blumberg, J. E. Edwards, W. Jarvis, L. Saiman, J. E. Patterson, M. G. Rinaldi, R. P. Wenzel, D. R. Soll, et al. 1999. Elucidating the origins of nosocomial infections with Candida albicans by DNA fingerprinting with the complex probe Ca3. J. Clin. Microbiol. 37:2817-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfaller, M. A., S. R. Lockhart, C. Pujol, J. A. Swails-Wenger, S. A. Messer, M. B. Edmond, R. N. Jones, R. P. Wenzel, and D. R. Soll. 1998. Hospital specificity, region specificity, and fluconazole resistance of Candida albicans bloodstream isolates. J. Clin. Microbiol. 36:1518-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaller, M. 1996. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clin. Infect. Dis. 22(Suppl.):589-594. [DOI] [PubMed] [Google Scholar]
- 15.Pfaller, M. 1995. Epidemiology of candidiasis. J. Hosp. Infect. 30(Suppl.):329-338. [DOI] [PubMed] [Google Scholar]
- 16.Pujol, C., S. Joly, B. Nolan, T. Srikantha, and D. R. Soll. 1999. Microevolutionary changes in Candida albicans identified by the complex Ca3 fingerprinting probe involve insertions and deletions of the full-length repetitive sequence RPS at specific genomic sites. Microbiology 145:2635-2646. [DOI] [PubMed] [Google Scholar]
- 17.Raad, I. 1998. Intravascular-catheter-related infections. Lancet 351:893-898. [DOI] [PubMed] [Google Scholar]
- 18.Reagan, D. R., M. A. Pfaller, R. J. Hollis, and R. P. Wenzel. 1990. Characterization of the sequence of colonization and nosocomial candidemia using DNA fingerprinting and a DNA probe. J. Clin. Microbiol. 28:2733-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richard, B., J. R. Thomson, and J. M. Miller. 2003. Specimen collection, transport, and processing: bacteriology, p. 286-330. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 20.Scherer, S., and D. A. Stevens. 1987. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J. Clin. Microbiol. 25:675-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schroppel, K., M. Rotman, R. Galask, K. Mac, and D. R. Soll. 1994. Evolution and replacement of Candida albicans strains during recurrent vaginitis demonstrated by DNA fingerprinting. J. Clin. Microbiol. 32:2646-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin, J. H., S. J. Kee, M. G. Shin, S. H. Kim, D. H. Shin, S. K. Lee, S. P. Suh, and D. W. Ryang. 2002. Biofilm production by isolates of Candida species recovered from nonneutropenic patients: comparison of bloodstream isolates with isolates from other sources. J. Clin. Microbiol. 40:1244-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin, J. H., D. H. Shin, J. W. Song, S. J. Kee, S. P. Suh, and D. W. Ryang. 2001. Electrophoretic karyotype analysis of sequential Candida parapsilosis isolates from patients with persistent or recurrent fungemia. J. Clin. Microbiol. 39:1258-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soll, D., R. Galask, J. Schmid, C. Hanna, K. Mac, and B. Morrow. 1991. Genetic dissimilarity of commensal strains of Candida spp. carried in different anatomical locations of the same healthy women. J. Clin. Microbiol. 29:1702-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voss, A., R. J. Hollis, M. A. Pfaller, R. P. Wenzel, and B. N. Doebbeling. 1994. Investigation of the sequence of colonization and candidemia in nonneutropenic patients. J. Clin. Microbiol. 32:975-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziebuhr, W., K. Ohlsen, H. Karch, T. Korhonen, and J. Hacker. 1999. Evolution of bacterial pathogenesis. Cell. Mol. Life Sci. 30:719-728. [DOI] [PMC free article] [PubMed] [Google Scholar]