Abstract
Assessment of intrahepatic hepatitis B virus (HBV) DNA levels in patients with chronic hepatitis B is important in understanding the natural history of the disease and designing antiviral therapy regimens. However, there is no standardized method for the measurement of intrahepatic HBV DNA levels. We describe a convenient novel method for the measurement of intrahepatic HBV DNA levels based on a modified COBAS Amplicor HBV Monitor test for HBV DNA measurement and real-time PCR β-actin gene detection for human genomic DNA (hgDNA) quantitation. Fifteen hepatitis B e antigen (HBeAg)-positive patients, 26 patients positive for antibody to HBeAg (anti-HBe), and 8 control patients were recruited. The mean between-run coefficient of variation for the β-actin real-time PCR assay was 15.4%. All eight control patients had undetectable intrahepatic and serum HBV DNA levels. All chronic hepatitis B patients had detectable intrahepatic HBV DNA levels, and all but one anti-HBe-positive patient had detectable serum HBV DNA levels. HBeAg-positive patients had higher median intrahepatic and serum HBV DNA levels than anti-HBe-positive patients (6,950 versus 676 HBV DNA copies/ng of hgDNA, respectively [P < 0.001] and 184 × 106 versus 6.65 × 106 copies/ml, respectively [P < 0.001]). The intrahepatic HBV DNA levels correlated strongly with the serum HBV DNA levels (r = 0.842; P < 0.001) and with the degree of fibrosis (P = 0.014). We conclude that the method that we describe is reliable and convenient for the measurement of intrahepatic HBV DNA levels and has potential clinical significance.
Hepatitis B virus (HBV) is a major pathogen in the world, causing acute and chronic infections. The latter may lead to liver cirrhosis and/or hepatocellular carcinoma. Direct measurement of HBV DNA levels in serum has routinely been used as an indicator of the viral load during HBV infection. A comparison of several commercial assays for the measurement of serum HBV DNA levels has been reviewed by Yuen and Lai (18). Others have also developed various in-house PCR assays for the quantitation of HBV DNA in serum (1, 3, 17). However, there is no standardized method for the measurement of HBV DNA in liver biopsy specimens. Determination of intrahepatic HBV DNA levels is of importance in defining the natural history of chronic HBV infection as well as assessing the efficacy of antiviral therapy. The availability of potent nucleoside analogues, now and in the future, in the form of either monotherapy or combination therapy, makes standard methods for the determination of intrahepatic HBV DNA levels more important. With highly efficacious antiviral treatment, HBV DNA may become undetectable in serum. The HBV DNA content inside the liver may then become an important therapeutic end point. In this study, we describe a method for the quantitation of intrahepatic HBV DNA. We attempted to quantify intrahepatic HBV DNA using a modified protocol of the COBAS Amplicor HBV Monitor test. The intrahepatic HBV DNA levels were standardized with reference to the amount of human genomic DNA (hgDNA) in the sample, using the β-actin gene as an internal control.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Liver biopsy samples from 15 hepatitis B e antigen (HBeAg)-positive patients and 26 chronic hepatitis B patients positive for antibody to HBeAg (anti-HBe) were analyzed. Liver biopsy samples from six chronic hepatitis C patients and two nonhepatitis patients were used as controls. Of the two nonhepatitis patients, one had an unexplained derangement of liver function tests and one had suspected porphyria. Total DNA was extracted from frozen liver needle biopsy and serum samples with the Qiagen QIAamp DNA extraction kit, with final elution in 400 μl of deionized water. Serum specimens were collected within 1 week from the time of the liver biopsy.
HBV DNA detection.
The amount of total intrahepatic HBV DNA in the extract was directly measured by the COBAS Amplicor HBV Monitor test (Roche Diagnostics, Branchburg, N.J.) (lower limit of detection, 200 copies/ml). Instead of using 100 μl of serum for the test, 100 μl of diluted DNA extract was applied directly to the sample lysis buffer. The rest of the procedures were performed according to the instructions of the manufacturer. The final HBV DNA concentration was expressed as the number of copies of HBV DNA per milliliter of sample extract. Serum HBV DNA levels were measured by the COBAS Amplicor HBV Monitor test according to the instructions of the manufacturer.
hgDNA quantitation.
The β-actin gene was used as an internal control for the normalization of the amount of hgDNA in the DNA samples. A TaqMan PCR reagent kit (Applied Biosystems, Foster City, Calif.), which contained a specific forward primer (5′-2141-TCACCCACACTGTGCCCATCTACGA-2165-3′), a reverse primer (5′-2435-CAGCGGAACCGCTCATTGCCAATGG-2411-3′), and a fluorescence-labeled probe (5′-2171-FAM-ATGCCC-X-TAMRA-CCCCCATGCCATCCTGCGTp-2196-3′) for β-actin detection, was used to measure the amount of hgDNA in the samples. The primers and probe described above contain sequences homologous to exon 3 of the β-actin gene, and the numbers in the sequences refer to the nucleotide positions in a DNA sequence in GenBank (accession number M10277) (13). In the fluorescence-labeled probe, FAM is the fluorophore reporter dye 6-carboxyfluorescein, X is a nucleotide linker arm, TAMRA is the quencher dye 6-carboxytetramethylrhodamine, and p represents the phosphorylation site. The final concentrations of the primers and the probe in each reaction mixture were 300 and 200 μM, respectively, as recommended in the protocol of the manufacturer. hgDNA templates of known concentrations (supplied with the kit) were used as calibration standards. Real-time PCR amplification and detection were performed with an ABI Prism 7000 Sequence Detection system (Applied Biosystems, Foster City, Calif.). PCR amplification was done in a volume of 50 μl; and the PCR program was as follows: 50°C for 2 min and 95°C for 10 min, followed by 40 two-step cycles of 95°C for 15 s and 60°C for 1 min. The total HBV DNA concentration was standardized with reference to the amount (in nanograms) of hgDNA in the biopsy samples.
Mutational analysis.
The precore mutational status of HBV was determined by the INNO-LiPA HBV Precore assay (Innogenetics N.V., Ghent, Belgium), as described previously (20). The assay allows the detection of precore mutations when they occur in more than 5% of the total viral population.
Histology assessment.
Histologic sections of the liver biopsy specimens were stained with hematoxylin-eosin. The degree of necroinflammation was classified into the following categories on the basis of a modification of the scoring system of Knodell et al. (8): minimal, mild, moderate, and marked. The degree of fibrosis was classified into the following categories: none, mild, moderate, and cirrhosis.
Statistical analysis.
All statistical analyses were performed with the Statistical Program for Social Sciences (SPSS, version 10.0 for Windows; SPSS Inc., Chicago, Ill.). Continuous variables were tested for normality by the Kolmogorov-Smirnov test. The Student t test was used to test continuous variables with normal distributions, and the Mann-Whitney U test was used to test continuous variables with skewed distributions. After logarithmic transformation of the data with skewed distributions, correlation analyses were performed by Pearson's correlation analysis. Categorical variables were tested by the chi-square test or Fisher's exact test. Statistical significance was defined by a P value of less than 0.05.
RESULTS
Demographic, serological, and histologic data.
Table 1 and Table 2 show the demographic, serological and histologic data for the 15 HBeAg-positive and 26 anti-HBe-positive chronic hepatitis B patients. There was no significant difference between the HBeAg-positive and anti-HBe-positive patients in terms of age, median albumin levels, median alanine aminotransferase (ALT) levels, and median bilirubin levels. There was also no significant difference between the HBeAg-positive patients and the anti-HBe-positive patients in the degree of necroinflammation and fibrosis (for minimal or mild necroinflammation versus moderate or marked necroinflammation, P = 0.139; for no or mild fibrosis versus moderate fibrosis or cirrhosis, P = 0.742). There was no significant difference in the median serum HBV DNA levels between the patient group with minimal or mild necroinflammation and the patient group with moderate or marked necroinflammation (14.2 × 106 and 20.6 × 106 copies/ml, respectively; P = 0.905). The patient group whose fibrosis status was classified as moderate or cirrhosis had higher median serum HBV DNA levels than the patient group with no or mild fibrosis (18.2 × 106 and 3.61 × 106 copies/ml, respectively; P = 0.01).
TABLE 1.
Demographic, serological, and histologic data for the patients
| Characteristic | HBeAg positive (n = 15) | Anti-HBe positive (n = 26) | P |
|---|---|---|---|
| Sex ratio (no. of males:no. of females) | 11:4 | 23:3 | |
| Mean age at biopsy ± SD (yr) | 37.0 ± 10.0 | 43.2 ± 10.6 | 0.072 |
| Median albumin level (g/liter [range]) | 44 (38-50) | 44 (36-49) | 0.785 |
| Median ALT level (U/liter [range]) | 74 (38-357) | 74.5 (21-275) | 0.626 |
| Median bilirubin level (μmol/liter [range]) | 15 (4-77) | 12 (5-22) | 0.238 |
| Histologic data (no. of patients) | |||
| Necroinflammation | 0.139 | ||
| Minimal | 3 | 12 | |
| Mild | 12 | 9 | |
| Moderate | 0 | 5 | |
| Marked | 0 | 0 | |
| Fibrosis | 0.742 | ||
| None | 0 | 6 | |
| Mild | 5 | 5 | |
| Moderate | 9 | 14 | |
| Cirrhosis | 1 | 1 | |
| Serum HBV DNA level (no. of copies [106]/ml [range]) | 184 (1.86-3,640) | 6.65 (<0.0002-1,430) | 0.001 |
TABLE 2.
Patients' serological and histologic data
| Patient group and no. | Intrahepatic HBV DNA level (no. of copies/ng of hgDNA) | Serum HBV DNA level (log10 copies/ml) | ALT concn (U/liter) | Necroinflammation statusa | Fibrosis statusa |
|---|---|---|---|---|---|
| HBeAg positive | |||||
| 1 | 511 | 7.82 | 74 | Minimal | Mild |
| 2 | 1,800 | 6.62 | 53 | Mild | Moderate |
| 3 | 2,360 | 8.26 | 175 | Minimal | Cirrhosis |
| 4 | 4,210 | 8.76 | 71 | Mild | Moderate |
| 5 | 4,310 | 7.05 | 43 | Mild | Mild |
| 6 | 5,460 | 8.49 | 124 | Mild | Moderate |
| 7 | 6,790 | 9.37 | 38 | Mild | Mild |
| 8 | 6,950 | 7.21 | 212 | Mild | Moderate |
| 9 | 9,460 | 8.75 | 57 | Mild | Moderate |
| 10 | 10,700 | 8.71 | 70 | Mild | Mild |
| 11 | 12,700 | 6.27 | 85 | Mild | Mild |
| 12 | 15,100 | 7.25 | 72 | Mild | Moderate |
| 13 | 21,300 | 8.94 | 83 | Mild | Moderate |
| 14 | 22,700 | 8.00 | 357 | Mild | Moderate |
| 15 | 55,500 | 9.56 | 95 | Minimal | Moderate |
| Anti-HBe positive | |||||
| 16 | 0.158 | 4.07 | 21 | Minimal | None |
| 17 | 0.31 | <2.30b | 28 | Minimal | Mild |
| 18 | 1.14 | 4.29 | 38 | Minimal | None |
| 19 | 3.31 | 5.26 | 22 | Minimal | None |
| 20 | 5.10 | 4.57 | 25 | Minimal | None |
| 21 | 43.6 | 5.99 | 190 | Mild | Moderate |
| 22 | 79.2 | 3.22 | 28 | Mild | Mild |
| 23 | 101 | 4.71 | 32 | Minimal | None |
| 24 | 166 | 6.67 | 130 | Mild | Moderate |
| 25 | 254 | 6.79 | 41 | Mild | Moderate |
| 26 | 404 | 7.19 | 73 | Minimal | Moderate |
| 27 | 585 | 6.25 | 51 | Moderate | Moderate |
| 28 | 610 | 7.11 | 170 | Mild | Mild |
| 29 | 742 | 6.54 | 43 | Moderate | Moderate |
| 30 | 782 | 6.73 | 275 | Minimal | Mild |
| 31 | 1,050 | 7.70 | 216 | Moderate | Mild |
| 32 | 1,200 | 7.31 | 76 | Moderate | Moderate |
| 33 | 1,510 | 7.26 | 67 | Mild | Moderate |
| 34 | 2,230 | 7.10 | 85 | Mild | Moderate |
| 35 | 2,410 | 6.85 | 223 | Minimal | None |
| 36 | 3,250 | 9.16 | 154 | Minimal | Cirrhosis |
| 37 | 3,310 | 7.23 | 70 | Mild | Moderate |
| 38 | 5,120 | 6.86 | 153 | Mild | Moderate |
| 39 | 5,250 | 8.11 | 87 | Minimal | Moderate |
| 40 | 5,360 | 7.54 | 248 | Moderate | Moderate |
| 41 | 7,310 | 7.58 | 134 | Minimal | Moderate |
The histologic classification are described in the Materials and Methods section.
Undetectable.
All non-hepatitis B control patients had undetectable serum HBV DNA levels. All 15 HBeAg-positive patients and 25 of 26 (96%) of the anti-HBe-positive patients had detectable serum HBV DNA levels. HBeAg-positive patients had a median serum HBV DNA level of 184 × 106 copies/ml (range, 1.86 × 106 to 3.64 × 109 copies/ml), and anti-HBe-positive patients had a median serum HBV DNA level of 6.65 × 106 copies/ml (range, <200 to 1.43 × 109 copies/ml). Compared to the HBeAg-positive patients, the anti-HBe-positive patients had lower median serum HBV DNA levels (P = 0.001).
Reproducibilities of the tests.
The reproducibility of the COBAS Amplicor HBV Monitor test has been documented previously (11, 14). Four liver biopsy samples were analyzed three times by the COBAS Amplicor HBV Monitor test for the determination of the mean between-run coefficient of variation (CV), which was determined to be 22.3%. Three samples were retested in triplicate in the same COBAS Amplicor HBV Monitor test run, and the mean within-run CV was 10.8%. These CVs were consistent with those reported previously (11, 14).
For the determination of hgDNA levels by β-actin gene real-time PCR quantitation, each test was done in duplicate. According to the instructions of the manufacturer, it is suggested that samples be retested if the within-run CV is greater than 10%, and 5 of the 49 samples (10.2%) tested required retesting. Sixteen randomly chosen samples were subjected to three repeat tests. The mean between-run CV was 15.4%, indicating that the test is reproducible.
Quantitation of intrahepatic HBV DNA in HBeAg- and anti-HBe-positive patients.
All 15 HBeAg-positive patients and 26 anti-HBe-positive patients had detectable intrahepatic HBV DNA levels. The median HBV DNA level in the liver biopsy specimen extract measured by this method was 1.50 × 107 copies/ml of DNA extract (range, 472 to 3.09 × 108 copies/ml of DNA extract). All eight non-hepatitis B control patients had undetectable intrahepatic HBV DNA levels.
Since each biopsy sample contained different amount of liver tissue, the concentration of HBV DNA was calculated with reference to the amount of hgDNA in the extracts by using the β-actin gene as an internal control. The amount of hgDNA ranged from 0.228 to 29.8 ng/μl of DNA extract, and the amount of HBV DNA per nanogram of hgDNA was calculated. The mean hgDNA contents in the liver tissue extracts were comparable between the HBeAg-positive and anti-HBe-positive patients (11.0 and 7.76 ng/μl, respectively [P = 0.141]; range, 2.75 to 23.2 ng/μl and 0.228 to 29.8 ng/μl, respectively). The median intrahepatic HBV DNA level in the HBeAg-positive patients was 6,950 HBV DNA copies/ng of hgDNA (range, 511 to 55,500 HBV DNA copies/ng of hgDNA), and that for the anti-HBe patients was 676 HBV DNA copies/ng of hgDNA (range, 0.158 to 7,310 copies/ng of hgDNA). Anti-HBe-positive patients had lower median intrahepatic HBV DNA levels than HBeAg-positive patients (P < 0.001). The anti-HBe-positive patient in whose serum HBV DNA was undetectable had an intrahepatic HBV DNA level of 0.31 DNA copies/ng of hgDNA.
Correlation of serum and intrahepatic HBV DNA levels.
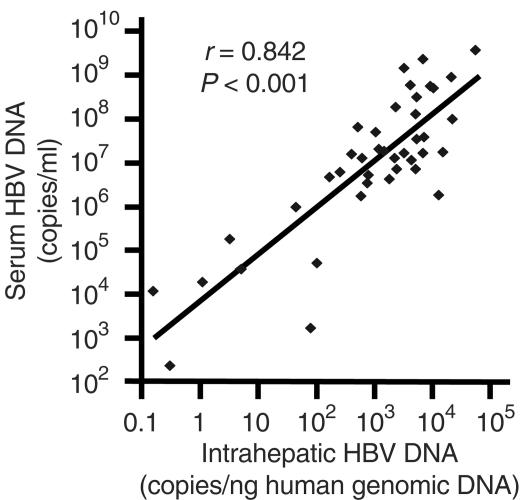
There was a strong correlation between the serum and the intrahepatic HBV DNA levels of the samples tested in this study (r = 0.842; P < 0.001) (Fig. 1). As determined by the line with the best fit in Fig. 1, for patients with serum HBV DNA levels of 105, 104, and 103 copies/ml, the intrahepatic HBV DNA levels were approximately 11, 1.2, and 0.15 copies/ng of hgDNA respectively.
FIG. 1.
Correlation between serum and intrahepatic HBV DNA levels in chronic hepatitis B patients.
Liver histology and intrahepatic and serum HBV DNA levels.
The degree of necroinflammation showed no significant correlation with the median intrahepatic HBV DNA levels in the patients studied (for patients with minimal or mild necroinflammation versus patients with moderate or marked necroinflammation, 2,380 versus 1,050 copies/ng of hgDNA, respectively; P = 0.633). However, the patients with no or mild fibrosis had lower median intrahepatic HBV DNA levels than the group with moderate fibrosis or cirrhosis (560 versus 3,310 copies/ng of hgDNA, respectively; P = 0.014).
Precore mutations in anti-HBe-positive patients.
The INNO-LiPA HBV precore mutation assay was performed for 25 of 26 of the anti-HBe-positive patients. No PCR-amplified product could be detected in the remaining patient with the 5′-biotinylated primers used. Ten of 25 anti-HBe-positive patients (38.5%) harbored mutants with precore mutations. The median serum HBV DNA levels did not differ significantly between anti-HBe-positive patients with and without precore mutations (5.04 × 106 versus 7.17 × 106 copies/ml, respectively; P = 0.683), nor did the median intrahepatic HBV DNA levels (676 versus 585 copies/ng of hgDNA respectively; P = 0.683). The degree of necroinflammation and fibrosis also did not differ significantly between the anti-HBe patients with and without precore mutations (for minimal or mild versus moderate or marked necroinflammation, P = 0.358; for no or mild fibrosis versus moderate fibrosis or cirrhosis, P = 0.122).
DISCUSSION
Despite the availability of a variety of commercial tests for the measurement of serum HBV DNA levels, standardized methods for the measurement of intrahepatic HBV DNA levels in chronic hepatitis B patients are lacking. This study demonstrated a convenient method for the quantitation of intrahepatic HBV DNA. Cacciola et al. (2) attempted to quantify intrahepatic HBV DNA using spectrophotometry for the determination of the liver DNA concentration and the COBAS Amplicor HBV Monitor test for the quantitation of HBV DNA in the sample extract. However, physical methods of quantitating the DNA concentration by standard optical spectrophotometry invariably introduce errors due to optical interference by other contaminating materials, such as rRNA and tRNA. In this study, the amount of liver DNA was measured by using the β-actin gene as an internal control. This allows the HBV DNA content to be standardized. β-Actin mRNA quantitation has been widely used as an internal mRNA control for mammalian gene expression studies because of its relatively stable expression under various conditions (15, 16). Since β-actin is a single-copy gene, it can be used as an internal DNA control to adjust for differences in the amounts of genomic DNA in test samples. Josefsson et al. (7) have used β-actin as a housekeeping gene for the measurement of the human papillomavirus type 16 viral load. This study makes use of this property of β-actin to adjust for the amount of hgDNA in liver tissue for the determination of intrahepatic HBV DNA levels.
The assay described here is designed to measure total intrahepatic HBV DNA only. In the future, a separate analysis should also be performed to assay the proportion of covalently closed circular (ccc) HBV DNA since ccc HBV DNA is resistant to nucleoside analogue therapy for chronic hepatitis B (12) and is probably responsible for the rebound of HBV DNA following the cessation of therapy (9). However, it should be noted that when the total intrahepatic HBV DNA load decreases, the ccc HBV DNA load would be expected to decrease, too.
Intrahepatic HBV DNA was found to be of clinical significance in several aspects. First, the intrahepatic HBV DNA concentration was found to correlate significantly with the degree of fibrosis. Second, in at least one patient, intrahepatic HBV DNA was still present even when serum HBV DNA was no longer detectable. It may be significant that the biopsy sample from this patient showed a mild degree of fibrosis, even though necroinflammation was minimal. This supports the previous findings that for Asian HBV carriers, there is no cutoff HBV DNA value below which disease progression would not occur (4, 21).
This study has shown that anti-HBe-positive patients had lower serum and intrahepatic HBV DNA levels than HBeAg-positive patients. Even among the anti-HBe-positive patients, however, 96% (25 of 26) of them had detectable levels of HBV DNA in both serum and the intrahepatic location. This may account for the mild to moderate necroinflammation and the moderate fibrosis to cirrhosis in liver biopsy samples from more than 50% of our anti-HBe-positive patients.
It is believed that precore mutations emerge under the selection of the host's immune pressure during the process of HBeAg seroconversion (5). A previous study (20) demonstrated that there is no significant difference in serum HBV DNA levels between patients with and patients without precore mutations. The present study further showed that intrahepatic HBV DNA levels and the degrees of necroinflammation and fibrosis in our anti-HBe-positive patients with and without precore mutations were comparable. These findings, together with the findings from the previous study (20), suggest that Asian anti-HBe-positive patients with precore mutations and wild-type virus have the same viral loads and disease activities.
There is a strong correlation between serum and intrahepatic HBV DNA levels, suggesting that serum HBV DNA levels can be used as an indication of intrahepatic HBV DNA levels. However, the measurement of intrahepatic HBV DNA levels is still of importance, especially in patients with low serum HBV DNA levels, as demonstrated by the presence of detectable intrahepatic HBV DNA in the anti-HBe-positive subject who had undetectable serum HBV DNA levels by the COBAS Amplicor HBV Monitor test. Although it has been proposed at a recent National Institutes of Health workshop that a serum HBV DNA level below 105 copies/ml may represent an inactive hepatitis B surface antigen (HBsAg) carrier state (10), the present study has shown that intrahepatic HBV DNA is detectable in patients with serum HBV DNA levels below 105 copies/ml. The estimated intrahepatic HBV DNA level for patients with serum HBV DNA levels of 105 copies/ml was approximately 11 copies/ng of hgDNA, and this value is well above the minimum intrahepatic HBV DNA level associated with fibrosis estimated in this study (0.31 copies/ng of hgDNA). This further confirms that, at least in Chinese patients, cirrhosis or complications of cirrhosis can still occur with HBV DNA levels below 105 copies/ml (6, 20).
Chinese patients with chronic hepatitis B infection can develop complications of cirrhosis and/or hepatocellular carcinoma after HBeAg seroconversion and even after they lose HBsAg (19). Quantitative measurement of intrahepatic HBV DNA correlates with the degree of fibrosis and may give some insight and prognostic guide for Asian patients. It may also provide a guide for the ideal time for the cessation of antiviral therapy. The method described in this study has provided a convenient tool for intrahepatic HBV DNA quantitation.
REFERENCES
- 1.Brechtbuehl, K., S. A. Whalley, G. M. Dusheiko, and N. A. Saunders. 2001. A rapid real-time quantitative polymerase chain reaction for hepatitis B virus. J. Virol. Methods 93:105-113. [DOI] [PubMed] [Google Scholar]
- 2.Cacciola, I., T. Pollicino, G. Squadrito, G. Cerenzia, D. Villari, R. de Franchis, T. Santantonio, S. Brancatelli, G. Colucci, and G. Raimondo. 2000. Quantification of intrahepatic hepatitis B virus (HBV) DNA in patients with chronic HBV infection. Hepatology 31:507-512. [DOI] [PubMed] [Google Scholar]
- 3.Chen, R. W., H. Piiparinen, M. Seppanen, P. Koskela, S. Sarna, and M. Lappalainen. 2001. Real-time PCR for detection and quantitation of hepatitis B virus DNA. J. Med. Virol. 65:250-256. [DOI] [PubMed] [Google Scholar]
- 4.Chu, C. J., M. Hussain, and A. S. Lok. 2002. Quantitative serum HBV DNA levels during different stages of chronic hepatitis B infection. Hepatology 36:1408-1415. [DOI] [PubMed] [Google Scholar]
- 5.Gunther, S., L. Fischer, I. Pult, M. Sterneck, and H. Will. 1999. Naturally occurring variants of hepatitis B virus. Adv. Virus Res. 52:25-137. [DOI] [PubMed] [Google Scholar]
- 6.Hsu, Y. S., R. N. Chien, C. T. Yeh, I. S. Sheen, H. Y. Chiou, C. M. Chu, and Y. F. Liaw. 2002. Long-term outcome after spontaneous HBeAg seroconversion in patients with chronic hepatitis B. Hepatology 35:1522-1527. [DOI] [PubMed] [Google Scholar]
- 7.Josefsson, A. M., P. K. Magnusson, N. Ylitalo, P. Sorensen, P. Qwarforth-Tubbin, P. K. Andersen, M. Melbye, H. O. Adami, and U. B. Gyllensten. 2000. Viral load of human papilloma virus 16 as a determinant for development of cervical carcinoma in situ: a nested case-control study. Lancet 355:2189-2193. [DOI] [PubMed] [Google Scholar]
- 8.Knodell, R. G., K. G. Ishak, W. C. Black, T. S. Chen, R. Craig, N. Kaplowitz, T. W. Kiernan, and J. Wollman. 1981. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology 1:431-435. [DOI] [PubMed] [Google Scholar]
- 9.Lai, C. L., C. K. Ching, A. K. Tung, E. Li, J. Young, A. Hill, B. C. Wong, J. Dent, and P. C. Wu. 1997. Lamivudine is effective in suppressing hepatitis B virus DNA in Chinese hepatitis B surface antigen carriers: a placebo-controlled trial. Hepatology 25:241-244. [DOI] [PubMed] [Google Scholar]
- 10.Lok, A. S., E. J. Heathcote, and J. H. Hoofnagle. 2001. Management of hepatitis B: 2000—summary of a workshop. Gastroenterology 120:1828-1853. [DOI] [PubMed] [Google Scholar]
- 11.Lopez, V. A., E. J. Bourne, M. W. Lutz, and L. D. Condreay. 2002. Assessment of the COBAS Amplicor HBV Monitor test for quantitation of serum hepatitis B virus DNA levels. J. Clin. Microbiol. 40:1972-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moraleda, G., J. Saputelli, C. E. Aldrich, D. Averett, L. Condreay, and W. S. Mason. 1997. Lack of effect of antiviral therapy in nondividing hepatocyte cultures on the closed circular DNA of woodchuck hepatitis virus. J. Virol. 71:9392-9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakajima-Iijima, S., H. Hamada, P. Reddy, and T. Kakunaga. 1985. Molecular structure of the human cytoplasmic beta-actin gene: interspecies homology of sequences in the introns. Proc. Natl. Acad. Sci. USA 82:6133-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noborg, U., A. Gusdal, E. K. Pisa, A. Hedrum, and M. Lindh. 1999. Automated quantitative analysis of hepatitis B virus DNA by using the Cobas Amplicor HBV Monitor test. J. Clin. Microbiol. 37:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sturzenbaum, S. R., and P. Kille. 2001. Control genes in quantitative molecular biological techniques: the variability of invariance. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 130:281-289. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki, T., P. J. Higgins, and D. R. Crawford. 2000. Control selection for RNA quantitation. BioTechniques 29:332-337. [DOI] [PubMed] [Google Scholar]
- 17.Weinberger, K. M., E. Wiedenmann, S. Bohm, and W. Jilg. 2000. Sensitive and accurate quantitation of hepatitis B virus DNA using a kinetic fluorescence detection system (TaqMan PCR). J. Virol. Methods 85:75-82. [DOI] [PubMed] [Google Scholar]
- 18.Yuen, M. F., and C. L. Lai. 1999. Debates in hepatitis: how to assess HBV DNA reductions in association with therapy. Viral Hepatitis Rev. 5:159-175. [Google Scholar]
- 19.Yuen, M. F., and C. L. Lai. 2000. Natural history of chronic hepatitis B virus infection. J. Gastroenterol. Hepatol. 15(Suppl.):E20-E24. [DOI] [PubMed] [Google Scholar]
- 20.Yuen, M. F., E. Sablon, H. J. Yuan, C. K. Hui, D. K.-H. Wong, J. Doutreloigne, B. C.-Y. Wong, A. O.-O. Chan, and C. L. Lai. 2002. Relationship between the development of precore and core promoter mutations and hepatitis B e antigen seroconversion in patients with chronic hepatitis B virus. J. Infect. Dis. 186:1335-1338. [DOI] [PubMed] [Google Scholar]
- 21.Yuen, M. F., H. J. Yuan, C. K. Hui, D. K.-H. Wong, W. M. Wong, A. O.-O. Chan, B. C.-Y. Wong, and C. L. Lai. 2003. A large population study of spontaneous HBeAg seroconversion and acute exacerbation of chronic hepatitis B infection: implications for antiviral therapy. Gut 52:416-419. [DOI] [PMC free article] [PubMed] [Google Scholar]