Abstract
Mycobacterial interspersed repetitive unit-variable number tandem repeat (MIRU-VNTR) typing of Mycobacterium tuberculosis complex isolates is portable, 100% reproducible, and highly discriminatory. Nondenaturing high-performance liquid chromatography (non-dHPLC) with use of a WAVE microbial analysis system is a promising method of PCR amplicon analysis as it is low cost and requires no preanalysis processing. The aims of this study were to validate the application of WAVE microbial analysis system technology to MIRU-VNTR typing. A collection of 70 strains were cultivated in liquid culture and extracted using the QIAamp DNA minikit. Novel primers were designed to target the 12 MIRU-VNTR loci (P. Supply et al., J. Clin. Microbiol. 39:3563-3571, 2001). After amplification, each PCR product was analyzed on a WAVE microbial analysis system. The fragment size was calculated from the chromatogram, and the number of tandem repeats at each locus was determined. For the collection of 70 strains 100% concordance was achieved when comparing MIRU-VNTR profiles obtained from agarose gel electrophoresis and PCRs analyzed on a WAVE microbial analysis system. The calculated fragment sizes, obtained from the WAVE microbial analysis system, were sufficiently accurate to ensure 100% confidence when assigning the number of tandem repeats to a MIRU-VNTR locus. This study is the first to report the successful use of non-dHPLC for screening for variations in the number of MIRU-VNTRs in mycobacterial DNA. Non-dHPLC analysis was demonstrated to be a rapid, low-labor input method for the detection and analysis of MIRU-VNTR amplicons. The combination with non-dHPLC further enhances the utility of MIRU-VNTR typing.
The global burden of tuberculosis remains substantial, mainly because of poor control in developing countries (7). Efficient local disease control depends on recognition and rapid treatment of infectious cases and on contact tracing and secondary prevention focused by epidemiological genotyping. On a broader scale, an international epidemiological surveillance scheme that can accurately monitor the dissemination of globally distributed clones will inform research on pathogenicity and transmission in different populations.
An ideal typing system must be accurate, discriminatory, reproducible, and at least semiautomated to ensure timeliness and cost-effectiveness. The generally accepted international “gold standard” for DNA fingerprinting of Mycobacterium tuberculosis has been IS6110 restriction fragment length polymorphism (RFLP) (27). This method has been proved to be extremely useful in population-based epidemiological studies (18). However, IS6110 RFLP typing is technically demanding, labor-intensive, and time-consuming. The analogue results are not easily reproducible between laboratories (1). The RFLP patterns of isolates containing fewer than six copies of IS6110 present a considerable challenge, as they exhibit a high rate of clustering by IS6110 RFLP (5). In a recent study in the United States, almost a quarter of isolates genotyped by IS6110 RFLP possessed less than six bands (9).
To combat the technical difficulties encountered by IS6110 RFLP typing, various rapid PCR-based methods have been developed (3) including mixed linker-PCR (2), multilocus sequence typing (19, 23), and spoligotyping (13, 17, 28). Variable number tandem repeat-PCR (VNTR-PCR), as applied to M. tuberculosis, was the first PCR method to analyze the VNTRs at specific genetic loci (10, 26, 29). These tandem repeat sequences are similar to the microsatellite DNA sequences used in human genomic fingerprinting (6). VNTR-PCR analyzes five distinct exact tandem repeat loci. Five-locus VNTR typing can define the geographic and evolutionary origin of strains (11) and has been useful in identifying laboratory cross-contamination incidents (12).
The range of loci used for typing that contain VNTRs has been extended to 12 mycobacterial interspersed repetitive unit (MIRU)-VNTR loci (24), which increased the discriminatory power of tandem repeat typing, such that it exhibits a performance comparable to that of IS6110 (15, 17, 20). VNTR and MIRU-VNTR typing have been shown to provide comparable clusters when applied to epidemiologically linked isolates (15).
Although MIRU-VNTR has been applied to a DNA sequencer (25), we decided to evaluate denaturing high-performance liquid chromatography (dHPLC) because it has proved to be one of the most cost-effective methods for analytical applications to prokaryotic biology. The first of these applications was for the characterization of quinolone resistance in Staphylococcus aureus (14) and Salmonella enterica (8) and beta-lactam resistance in gram-negative organisms (21). The method has also been used for multilocus sequence typing-dHPLC typing of meningococci (22) and in the detection of mutations conferring antibiotic resistance in M. tuberculosis (4). However, the study described here is to our knowledge the first of its kind that utilizes the nondenaturing mode (non-dHPLC) for analysis of multiple fragments of double-stranded DNA on the WAVE microbial analysis system to type bacterial strains with use of tandem repeat sequences. Non-dHPLC employs reverse-phase ion pair chromatography in which DNA binds to the column, mediated by the ion-pairing reagent triethylammonium acetate; subsequently, this DNA bonding to the column is disrupted by a linear gradient of acetonitrile, which results in the movement of the DNA from the stationary phase into the mobile phase and progress through the column to the UV detector. This study describes the initial validation of a WAVE microbial analysis system for MIRU-VNTR typing of M. tuberculosis strains with use of 12 separate injections.
MATERIALS AND METHODS
Bacterial strains.
Seventy M. tuberculosis isolates were used to validate MIRU-VNTR typing on a WAVE microbial analysis system. These strains had been previously characterized by MIRU-VNTR typing via agarose gel electrophoresis to size amplified sequences and enumerate tandem repeats at each locus (24). Twenty isolates were selected to represent the entire range of repeats observed at each locus when previously typed. The remaining 50 isolates were randomly selected from the clinical isolates typed at the Birmingham Regional Centre for Mycobacteriology.
Growth conditions.
Strains were incubated in mycobacterial growth indicator tubes (Becton Dickinson Biosciences, Oxford, United Kingdom) at 37°C until a positive growth index was achieved.
DNA extraction.
A 1.5-ml portion of liquid culture was removed from the mycobacterial growth indicator tube and was centrifuged for 10 min at 8,000 × g. The supernatant was removed, and DNA was extracted from the deposit by using the QIAamp DNA minikit (Qiagen, Crawley, United Kingdom). Prior to amplification each extract was diluted 1:100 in Tris-EDTA buffer. DNA was stored at −20°C until used.
MIRU-VNTR typing by agarose gel electrophoresis.
MIRU-VNTR analysis to characterize the strains with use of agarose gels to size amplicons was performed by applying the primers and PCR conditions for the 12 MIRU-VNTR loci as initially published by Supply et al. (24). The amplification reaction was performed on an MBS Thermal Cycler (Thermohybaid, Ashford, United Kingdom) with the following program: an initial denaturation for 7 min at 95°C and 35 cycles of denaturation for 1 min at 95°C, annealing at 59°C for 1 min, and extension for 1 min 30 s at 72°C. A terminal extension step of 10 min at 72°C completed the program. The amplicons were analyzed on a 2% (wt/vol) Agarose-1000 gel (Invitrogen, Paisley, United Kingdom) for 3 h at 150 V with 50- and 100-bp ladder size standards (Promega, Southampton, United Kingdom).
MIRU-VNTR primer design.
A set of proprietary PCR oligonucleotides known as MIRU-SCAN were designed in conjunction with Transgenomic, Inc. (Omaha, Nebr.), to amplify the 12 MIRU-VNTR loci.
MIRU-VNTR typing by the nondenaturing program on a WAVE microbial analysis system.
The following reagents were added to each PCR mixture: 1 U of Optimase polymerase (Transgenomic), 500 nM (each) primer, 5 μl of 10× reaction buffer, 200 μM (each) four deoxynucleoside triphosphates, 1 mM MgSO4, 1 μl of template DNA diluted 1:100, and sterile H2O to a final reaction volume of 50 μl. We were able to optimize reaction conditions to use a single concentration of Mg2+ for each PCR.
The amplification program was performed on an MBS Thermal Cycler with the following conditions: initial denaturation stage of 5 min at 95°C; 35 cycles of 1 min at 95°C, annealing for 1 min at 62°C, and extension for 2 min at 72°C; and a final extension step of 72°C for 5 min. Five microliters of each PCR mixture was loaded onto a DNASep cartridge in a WAVE microbial analysis system (Transgenomic, Inc.) with use of the double-stranded multiple fragment DNA sizing analysis program. The gradient for separation was an 8.4-min gradient at a flow rate of 0.9 ml min−1 at 50°C. The gradient size range was set at 50 to 900 bp at a rate of 0.5 min/100 bp. Peaks that exhibited an absorbance of greater than 1 mV were accepted as true peaks representing PCR fragments. The gradient parameters were controlled by WAVEMAKER software (Transgenomic, Inc.). The principle behind fragment sizing by dHPLC operating in a nondenaturing mode is the differential retention of DNA primarily depending on the length of the sequence, which reflects the number of repeats in the fragment. A sizing standard with GC content comparable to that of the analyzed DNA must be used when sizing PCR fragments on a WAVE microbial analysis system (16). The MIRU-VNTR loci of M. tuberculosis all have a high GC content (65 to 70%). Therefore, to ensure that an appropriate standard was used, a DNA ladder was constructed from combined PCR products of MIRU-VNTR locus 23 that contained known alleles that ranged from 1 to 10 repeats. With use of this DNA standard, no offset effects of different sequence composition were observed.
With use of a Microsoft Excel spreadsheet, the retention time of each PCR fragment was compared against the nearest standard bands and the size in base pairs was calculated. From this the number of repeats at each locus was determined.
Cluster analysis.
MIRU-VNTR results were entered into BioNumerics (Applied Maths, St.-Martin-Latem, Belgium), analyzed by using the categorical coefficient, and displayed via the unweighted pair group method with arithmetic averages (UPGMA).
Statistical analysis of results.
From the MIRU-VNTR results obtained, the minimum, mean, and maximum observed repeat values and fragment size (base pairs) were calculated. These results were then compared against a defined range of allowable results.
Validation of duplex MIRU-VNTR typing with use of a WAVE microbial analysis system.
MIRU-VNTR results obtained from 12 single injections were analyzed to evaluate if each single MIRU locus could be duplexed into six paired PCRs of MIRU loci (MIRU 2 and 10, 4 and 20, 16 and 23, 24 and 40, 26 and 39, and 27 and 31).
RESULTS
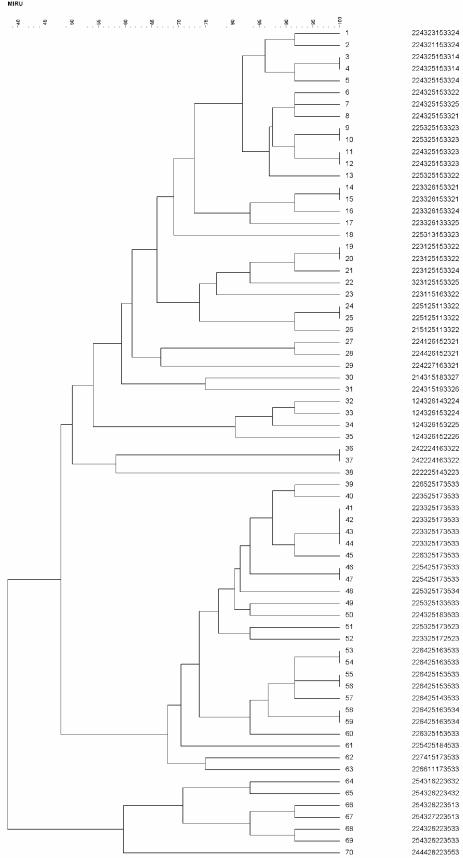
Figure 1 displays the 70 strains analyzed by agarose gel electrophoresis and with use of the WAVE microbial analysis system as single amplifications. A concordance level of 100% between MIRU-VNTR profiles generated by agarose gel electrophoresis and by the non-dHPLC method was observed when results from the two methods were compared. The strains were selected from the Midlands Regional Centre for Mycobacteriology culture collection and represented the entire spectrum of alleles observed when typing clinical strains in the Midlands to date. Due to the confined geographical origins of strains analyzed in this collection the spectrum of alleles is not as wide as that observed previously (25), when globally originating strains were analyzed. There were 56 distinct MIRU-VNTR profiles with a total of 26 isolates indistinguishable by MIRU-VNTR in 12 clusters. Forty-four isolates possessed a unique MIRU-VNTR profile compared to other strains in the collection. The largest cluster of four isolates (223325173533) represented a group of four epidemiologically linked patients from one sending location. Of the other 11 clusters each containing a pair of isolates, only two clusters contained isolates received from the same sending laboratory. The other nine clusters containing pairs of isolates from different sending laboratories could be confirmed by IS6110 RFLP typing.
FIG. 1.
MIRU typing results obtained for each of the 70 M. tuberculosis strains analyzed. The dendrogram was constructed with use of the categorical coefficient algorithm and produced via UPGMA.
Table 1 is a comparison of our results with those of Supply et al. For nine of the MIRU loci, isolates from a global collection displayed a greater range of alleles than did our collection. For two MIRU loci the allele range was the same for the two strain collections. For one locus (MIRU 26) the collection from Birmingham Regional Centre for Mycobacteriology exhibited a greater range of alleles than did the global collection.
TABLE 1.
Allelic diversity of typed strains from West Midlands (England) compared to that of global strains
| Locus | Allelic diversity
|
|
|---|---|---|
| Midlands | Global | |
| 2 | 1-3 | 1-3 |
| 4 | 1-5 | 1-9 |
| 10 | 2-7 | 2-8 |
| 16 | 1-6 | 2-8 |
| 20 | 1-2 | 1-2 |
| 23 | 1-7 | 1-11 |
| 24 | 1-2 | 1-6 |
| 26 | 1-9 | 1-6 |
| 27 | 2-4 | 2-5 |
| 31 | 2-5 | 1-6 |
| 39 | 1-3 | 1-4 |
| 40 | 1-7 | 1-8 |
Table 2 displays the statistical analysis of peaks obtained over multiple runs when the group of 70 strains were typed on the WAVE microbial analysis system. Three separate groups of MIRU repeats were analyzed, derived from clinical strains that have been previously typed by agarose gel electrophoresis. The minimum, median, and maximum numbers of repeats observed in clinical strains previously typed in the West Midlands were statistically analyzed.
TABLE 2.
Analysis of product sizes calculated from WAVE microbial analysis system DNA fragment analysis chromatograms
| Value type and locus | Value for product containing expected no. of repeats
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Expected no. of repeats | No. of results | Size (bp)
|
SD (bp) | 95% CI (bp) | Mean variance (bp) | ||||
| Expected | Observed
|
||||||||
| Minimum | Mean | Maximum | |||||||
| Minimum | |||||||||
| 2 | 1 | 10 | 168 | 155 | 160 | 164 | 3 | 158-162 | −8 |
| 4 | 1 | 11 | 196 | 182 | 195 | 211 | 10 | 189-201 | −1 |
| 10 | 2 | 14 | 195 | 183 | 190 | 198 | 5 | 187-193 | −5 |
| 16 | 1 | 39 | 107 | 95 | 103 | 108 | 4 | 102-104 | −4 |
| 20 | 1 | 12 | 156 | 144 | 150 | 156 | 4 | 148-152 | −6 |
| 23 | 1 | 10 | 134 | 121 | 133 | 138 | 5 | 132-138 | −1 |
| 24 | 1 | 59 | 179 | 166 | 173 | 178 | 3 | 172-174 | −6 |
| 26 | 1 | 17 | 113 | 100 | 108 | 115 | 5 | 106-110 | −5 |
| 27 | 2 | 10 | 210 | 193 | 201 | 208 | 4 | 198-204 | −9 |
| 31 | 2 | 13 | 183 | 175 | 177 | 183 | 3 | 175-179 | −6 |
| 39 | 1 | 17 | 187 | 178 | 182 | 187 | 3 | 181-183 | −5 |
| 40 | 1 | 16 | 206 | 197 | 200 | 203 | 2 | 199-201 | −6 |
| Median | |||||||||
| 2 | 2 | 67 | 221 | 209 | 216 | 222 | 3 | 215-217 | −5 |
| 4 | 2 | 59 | 273 | 261 | 270 | 275 | 4 | 269-271 | −3 |
| 10 | 4 | 29 | 301 | 287 | 293 | 301 | 4 | 292-294 | −8 |
| 16 | 3 | 39 | 213 | 201 | 209 | 214 | 4 | 208-210 | −4 |
| 20 | 2 | 63 | 233 | 221 | 228 | 235 | 3 | 227-229 | −5 |
| 23 | 5 | 45 | 346 | 338 | 347 | 359 | 6 | 345-349 | 1 |
| 24 | 1 | 59 | 179 | 166 | 173 | 178 | 3 | 172-174 | −6 |
| 26 | 5 | 30 | 317 | 305 | 316 | 323 | 4 | 315-317 | −1 |
| 27 | 3 | 64 | 263 | 247 | 255 | 261 | 3 | 254-256 | −8 |
| 31 | 3 | 37 | 236 | 225 | 231 | 235 | 3 | 230-232 | −5 |
| 39 | 2 | 38 | 240 | 228 | 233 | 237 | 2 | 232-234 | −7 |
| 40 | 3 | 31 | 314 | 301 | 308 | 314 | 3 | 307-309 | −6 |
| Maximum | |||||||||
| 2 | 2 | 67 | 221 | 209 | 216 | 222 | 3 | 215-217 | −5 |
| 4 | 5 | 10 | 504 | 481 | 490 | 503 | 8 | 485-495 | −14 |
| 10 | 7 | 10 | 460 | 439 | 450 | 460 | 6 | 446-454 | −10 |
| 16 | 6 | 14 | 372 | 365 | 368 | 374 | 2 | 367-369 | −4 |
| 20 | 2 | 63 | 233 | 221 | 228 | 235 | 3 | 227-229 | −5 |
| 23 | 7 | 18 | 452 | 441 | 451 | 465 | 6 | 448-454 | −1 |
| 24 | 2 | 11 | 233 | 226 | 228 | 231 | 2 | 227-229 | −5 |
| 26 | 9 | 14 | 521 | 512 | 517 | 522 | 3 | 515-519 | −4 |
| 27 | 4 | 10 | 316 | 303 | 306 | 308 | 2 | 305-307 | −10 |
| 31 | 5 | 28 | 342 | 332 | 340 | 347 | 4 | 338-342 | −2 |
| 39 | 3 | 27 | 293 | 281 | 286 | 290 | 3 | 285-287 | −7 |
| 40 | 7 | 10 | 530 | 505 | 515 | 524 | 5 | 512-518 | −15 |
For the products containing the lowest number of repeats observed at each locus (112111112211), the standard deviation (SD) ranged from 2 to 10 bp (±0.04 to 0.13 repeats) with an average from the 12 MIRU loci of ±4 bp (±0.07 repeats). The 95% confidence interval (CI) ranged from 2 to 12 bp (0.04 to 0.16 repeats) with a mean interval of 5 bp (0.07 repeats). The variance of the mean calculated sizes from the expected product sizes ranged from 1 to 9 bp (0.01 to 0.16 repeats) less than the expected product size, with an average of −5 bp (−0.09 repeats) across the 12 MIRU loci.
The median number of repeats for each locus was 224325153323. For these PCR products the SD range was ±2 to 6 bp (0.04 to 0.11 repeats) with a mean of ±3 bp (±0.06 repeats) for all 12 MIRU loci combined. The 95% CI varied from 2 to 4 bp (0.02 to 0.06 repeats) with an average interval of 2 bp (0.04 repeats). The mean of the calculated product sizes varied from the expected size by −1 to −8 bp (−0.01 to −0.13 repeats) with an average of −5 bp (−0.09 repeats) for all 12 MIRU loci.
Two hundred eighty-two PCR products were analyzed from isolates that possessed the maximum number of repeats observed at each locus in clinical strains (257627294537); the SD for each locus ranged from a minimum of ±2 bp to a maximum of 8 bp (0.04 to 0.12 repeats) with an average SD of ±4 bp (±0.07 repeats) for all 12 MIRU loci combined. The 95% CI ranged from 2 to 10 bp (0.02 to 0.14 repeats), with a mean interval of 4 bp (0.07 repeats). The variance of the mean calculated size from the expected product size ranged from −1 to −14 bp (−0.02 to −0.27 repeats) with an average variance from the expected size of −7 bp (−0.12 repeats).
Of 840 PCR fragments analyzed, 828 (99%) were sized accurately within a repeat integer range of ±0.25 repeats. For all 840 PCR fragments analyzed within this study, the WAVE microbial analysis system exhibited a mean accuracy of +4.8 bp or +0.09 repeats. The 12 PCR products that were not within this range had an average variance of −0.29 repeats from the expected repeat number. Upon repeat analysis, all of the 12 PCR products that did not initially group within this interval were sized accurately to within ±0.25 repeats.
The MIRU-VNTR results from the 70 isolates analyzed were compared in six putative paired MIRU loci (MIRU 2 and 10, 4 and 20, 16 and 23, 24 and 40, 26 and 39, and 27 and 31). From this analysis, PCR amplicons amplified from all of the MIRU loci are allocated to their correct locus. The paired amplicons from 815 of 840 MIRU loci can be correctly allocated to their respective locus compared to previously calculated profiles. The remaining 25 amplicons from the 840 MIRU loci can be allocated to their respective locus on an exclusion basis. After the analysis of each amplicon for calculating its number of repeats to two decimal places, the first amplicon of the paired MIRU loci that falls within the range of ±0.25 repeats is assigned to its locus. Then the second amplicon of the paired MIRU loci is assigned to the only possible remaining MIRU locus.
DISCUSSION
The results of this study show that non-dHPLC with use of a WAVE microbial analysis system produces accurate and reproducible sizing of MIRU-VNTR amplicons when utilized for single PCR MIRU-VNTR typing of M. tuberculosis strains. The highest number of repeats analyzed in this study is nine repeats (521 bp) for MIRU-VNTR locus 26. For this locus the total range of calculated amplicon sizes was 512 to 522 bp. The SD was ±3 bp, the 95% CI was 515 to 519 bp, and the mean variance of calculated product sizes from expected product size was −4 bp. However, the largest product size analyzed was 530 bp for MIRU 40 (seven repeats). For this locus the calculated amplicon size ranged from 505 to 524 bp, with an SD of ±5 bp, a 95% CI of 512 to 518 bp, and a mean variance from calculated to expected amplicon size of −15 bp. The relatively narrow SD and 95% CI values indicate that the majority of PCR products are grouped together. When sizing amplicons for tandem repeat typing, very high levels of accuracy of sizing are not necessary as MIRU-VNTR units are all longer than 50 bp. Therefore, it is necessary only to have sufficient accuracy to unambiguously determine the number of repeat sequences at each locus. Combining the 281 amplicons analyzed for the maximum number of repeats category, the mean variance for all samples compared to the expected amplicon size was −6 bp. For quality control purposes, certain ranges of allowable repeat numbers must be implemented. For single amplicon analysis, a “buffer zone” of 50% of the repeat size between predicted PCR fragment sizes can be used. This means that any PCR product calculated as ±0.25 repeats within a repeat integer can be accepted. On a WAVE microbial analysis system 828 of 840 (99%) of single PCR fragments analyzed were within this interval. The 12 PCR products that were not within this range had an average variance of −0.29 repeats (i.e., only −0.04 repeats outside the limit) from the expected repeat number. This variation probably occurred as a result of systematic calculation because retention times are recorded to only two decimal places. They are then manipulated and then rounded to two decimal places before rounding up or down to a repeat integer value.
In a previous study (25), sizing of MIRU-VNTR PCR amplicons with use of a DNA sequencer was found to be reproducible, with within-run and between-run average precisions of ±0.5 and ±0.6 bp, respectively. Mean errors for the sizing accuracy were 1.1 bp for fragments below 500 bp and 0.8 bp for fragments from 500 to up to 971 bp. The WAVE microbial analysis system exhibits a mean error of 4.8 bp for all PCR fragments analyzed within this study.
We have calculated that, to perform MIRU-VNTR typing with 12 single PCRs, postamplification analysis by non-dHPLC on a WAVE microbial analysis system would cost $6 per isolate. Using a duplex method would approximately halve these costs.
After PCR amplification for non-dHPLC MIRU-VNTR typing then the next step is straightforward uncapping and loading of tubes into the autosampler unit of the WAVE microbial analysis system. No intermediary manipulation of the PCR product such as addition of loading buffer, denaturation, and prerunning, which is required on a 96-well ABI 377 automatic sequencer (Bio-Whittaker Molecular Applications, Rockland, Maine), is required (24).
Pairing of the 12 MIRU-VNTR loci into six duplex PCRs would increase the throughput of isolates. Without using fluorescent dyes, this would be possible, as the WAVE microbial analysis system exhibits a mean accuracy of 4.8 bp. This would enable accurate identification of PCR products to the correct MIRU-VNTR locus.
WAVE DNA fragment analysis equipment can be supplemented with various units to further enhance the speed of analysis such as a fluorescence detector, an accelerator, and the potential for 2- by 96-well plate capacity. The potential capacity is 78 strains per week with performance of daily runs with duplex PCRs and a dual-plate autosampler. By using the exclusion method described previously to correctly assign amplicons to paired MIRU-VNTR loci, it is possible to perform duplex MIRU-VNTR typing on a WAVE microbial analysis system.
The total size range of amplicons amplified by our novel set of primers is 107 to 521 bp (one to nine repeats). This optimized range further keeps the analysis time to 8.4 min per injection, as the primer-binding flanking regions are relatively small (the largest is MIRU-VNTR 40 at 152 bp).
This study is the first to report the successful use of non-dHPLC for screening for variations in the number of MIRU-VNTRs in mycobacterial DNA. The fragments of various retention times detected by agarose gel analysis could be rapidly and reproducibly identified in single PCR products by non-dHPLC, producing specific DNA WAVE patterns that correlate with MIRU-VNTR genotypes.
Non-dHPLC analysis was demonstrated to be a rapid, low-labor input method for the detection and analysis of PCR fragments containing various numbers of tandem repeats in M. tuberculosis. VNTR and MIRU-VNTR typing have recently been described as valuable in investigating clusters of tuberculosis in a large health care region in the United Kingdom (15). The combination with non-dHPLC further enhances the utility of this typing method.
REFERENCES
- 1.Braden, C. R., J. T. Crawford, and B. A. Schable. 2002. Quality assessment of Mycobacterium tuberculosis genotyping in a large laboratory network. Emerg. Infect. Dis. 8:1210-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler, W. R., W. H. Haas, and J. T. Crawford. 1996. Automated DNA fingerprinting analysis of M. tuberculosis using fluorescent detection of PCR products. J. Clin. Microbiol. 34:1801-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collyns, T. A., D. M. Gascoyne-Binzi, and P. M. Hawkey. 2002. Molecular fingerprinting of Mycobacterium tuberculosis: does it help in understanding the epidemiology of tuberculosis? Rev. Med. Microbiol. 13:119-127. [Google Scholar]
- 4.Cooksey, R. C., G. P. Morlock, B. P. Holloway, J. Limor, and M. Hepburn. 2002. Temperature-mediated heteroduplex analysis performed by using denaturing high-performance liquid chromatography to identify sequence polymorphisms in Mycobacterium tuberculosis complex organisms. J. Clin. Microbiol. 40:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowan, L. S., L. Mosher, L. Diem, J. P. Massey, and J. T. Crawford. 2002. Variable-number tandem repeat typing of Mycobacterium tuberculosis isolates with low copy numbers of IS6110 by using mycobacterial interspersed repetitive units. J. Clin. Microbiol. 40:1592-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devaney, J. M., J. E. Girard, and M. A. Marino. 2000. DNA microsatellite analysis using ion-pair reversed-phase high-performance liquid chromatography. Anal. Chem. 72:858-864. [DOI] [PubMed] [Google Scholar]
- 7.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. WHO Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 8.Eaves, D. J., E. Liebana, M. J. Woodward, and L. J. V. Piddock. 2002. Detection of gyrA mutations in quinolone-resistant Salmonella enterica by denaturing high-performance liquid chromatography. J. Clin. Microbiol. 40:4121-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis, B. A., J. T. Crawford, C. R. Braden, S. J. McNabb, M. Moore, and S. Kammerer. 2002. Molecular epidemiology of tuberculosis in a sentinel surveillance population. Emerg. Infect. Dis. 8:1197-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frothingham R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 11.Gascoyne-Binzi, D. M., R. E. Barlow, A. Essex, R. Gelletlie, M. A. Khan, S. Hafiz, T. A. Collyns, R. Frizzell, and P. M. Hawkey. 2002. Predominant VNTR family of strains of Mycobacterium tuberculosis isolated from South Asian patients. Int. J. Tuberc. Lung Dis. 6:492-496. [DOI] [PubMed] [Google Scholar]
- 12.Gascoyne-Binzi, D. M., R. E. Barlow, R. Frothingham, G. Robinson, T. A. Collyns, R. Gelletlie, and P. M. Hawkey. 2001. Rapid identification of laboratory contamination with Mycobacterium tuberculosis using variable number tandem repeat analysis. J. Clin. Microbiol. 39:69-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groenen, P. M., A. E. Bunschoten, D. van Soolingen, and J. D. van Embden. 1993. Nature of DNA polymorphism in the direct repeat cluster of Mycobacterium tuberculosis; application for strain differentiation by a novel typing method. Mol. Microbiol. 10:1057-1065. [DOI] [PubMed] [Google Scholar]
- 14.Hannachi-M'Zali, F., J. E. Ambler, C. F. Taylor, and P. M. Hawkey. 2002. Examination of single and multiple mutations involved in resistance to quinolones in Staphylococcus aureus by a combination of PCR and denaturing high-performance liquid chromatography (dHPLC). J. Antimicrob. Chem. 50:649-655. [DOI] [PubMed] [Google Scholar]
- 15.Hawkey, P. M., E. G. Smith, J. T. Evans, P. Monk, G. Bryan, H. H. Mohamed, M. Bardhan, and R. N. Pugh. 2003. Mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis compared to IS6110-based restriction fragment length polymorphism analysis for investigation of apparently clustered cases of tuberculosis. J. Clin. Microbiol. 8:3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber, C. G. 1998. Micropellicular stationary phases for high-performance liquid chromatography of double-stranded DNA. J. Chromatogr. A 806:3-30. [DOI] [PubMed] [Google Scholar]
- 17.Kremer, K., D. van Soolingen, R. Frothingham, W. H. Haas, P. W. Hermans, C. Martin, P. Palittapongarnpim, P. B. Plikaytis, L. W. Riley, M. A. Yakrus, J. M. Musser, and J. D. van Embden. 1999. Comparison of methods based on different molecular epidemiological markers for typing of Mycobacterium tuberculosis complex strains: interlaboratory study of discriminatory power and reproducibility. J. Clin. Microbiol. 37:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maguire, H., J. W. Dale, T. D. McHugh, P. D. Butcher, S. H. Gillespie, A. Costetsos, H. Al-Ghusein, R. Holland, A. Dickens, L. Marston, P. Wilson, R. Pitman, D. Strachan, F. A. Drobniewski, and D. K. Banerjee. 2002. Molecular epidemiology of tuberculosis in London 1995-7 showing low rate of active transmission. Thorax 57:617-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of M. tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pérez-Pérez, F. J., and N. D. Hanson. 2002. Detection of plasmid-mediated AmpC β-lactamase genes in clinical isolates by using multiplex PCR. J. Clin. Microbiol. 40:2153-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shlush, L. I., D. M. Behar, A. Zelazny, N. Keller, J. R. Lupski, A. L. Beaudet, and D. Bercovich. 2002. Molecular epidemiological analysis of the changing nature of a meningococcal outbreak following a vaccination campaign. J. Clin. Microbiol. 40:3565-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sreevatsan, S., X. Pan, K. E. Stockbauer, N. D. Connell, B. N. Kreiswirth, T. S. Whittam, and J. M. Musser. 1997. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 94:9869-9874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 25.Supply, P., S. Lesjean, E. Savine, K. Kremer, D. van Soolingen, and C. Locht. 2001. Automated high-throughput genotyping for study of global epidemiology of Mycobacterium tuberculosis based on mycobacterial interspersed repetitive units. J. Clin. Microbiol. 39:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Belkum, A., W. van Leeuwen, S. Scherer, and H. Verbrugh. 1999. Occurrence and structure-function relationship of pentameric short sequence repeats in microbial genomes. Res. Microbiol. 150:617-626. [DOI] [PubMed] [Google Scholar]
- 27.van Embden, J. D., M. D. Cave, J. T. Crawford, J. W. Dale, K. D. Eisenach, B. Gicquel, P. Hermans, C. Martin, R. McAdam, and T. M. Shinnick. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J. Clin. Microbiol. 31:406-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warren, R. M., E. M. Streicher, S. Charalambous, G. Churchyard, G. D. van der Spuy, A. D. Grant, P. D. van Helden, and T. C. Victor. 2002. Use of spoligotyping for accurate classification of recurrent tuberculosis. J. Clin. Microbiol. 40:3851-3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaganehdoost, A., E. A. Graviss, M. W. Ross, G. J. Adams, S. Ramaswamy, A. Wanger, R. Frothingham, H. Soini, and J. M. Musser. 1999. Complex transmission dynamics of clonally related virulent Mycobacterium tuberculosis associated with barhopping by predominantly human immunodeficiency virus-positive gay men. J. Infect. Dis. 180:1245-1251. [DOI] [PubMed] [Google Scholar]