Abstract
Sixty-three clinical isolates of charcoal-black-pigmented, gram-positive coryneform rods were received for identification by the Centers for Disease Control and Prevention (CDC) and were provisionally designated CDC fermentative coryneform group 4 (FCG4). Forty-five of these were characterized by morphological, physiologic, antimicrobial susceptibility, cellular fatty acids, 16S rRNA gene sequencing, and DNA-DNA hybridization analyses. Nitrate reduction, cellular fatty acid analysis, 16S rRNA gene sequencing, and DNA-DNA hybridization studies segregated these strains into two groups: FCG4a (8 strains) and FCG4b (37 strains). The FCG4a strains, only one of which was from a female genitourinary source, produced cellular fatty acid and biochemical profiles similar to those observed with reference strains of Rothia dentocariosa and Rothia mucilaginosa, while the FCG4b strains were similar to Corynebacterium species. DNA-DNA hybridization analysis demonstrated species-level relatedness among six FCG4a tested strains and showed that they were a charcoal-black-pigmented variant of R. dentocariosa. Sixteen isolates of the FCG4b group, mainly from female genitourinary tract specimens, as well as the type strains of two recently named species, Corynebacterium aurimucosum and Corynebacterium nigricans, were shown by DNA-DNA hybridization analysis and the sequencing of the 16S rRNA gene to be related at the species level and unrelated to the type strain of R. dentocariosa; therefore, the Corynebacterium-like strains were classified as a charcoal-black-pigmented variant of C. aurimucosum, because this name has nomenclatural priority over C. nigricans. These findings indicate that FCG4 represents a heterogeneous group that contains pigmented variants of both R. dentocariosa and C. aurimucosum; hence, the descriptions of both R. dentocariosa and C. aurimucosum have been amended to include charcoal-black-pigmented variants, and C. nigricans is a pro synonym of C. aurimucosum.
Between 1969 and 2004, the Special Bacteriology Reference Laboratory of the Centers for Disease Control and Prevention (CDC) received 63 unusual gram-positive, charcoal-black-pigmented coryneform isolates, recovered predominately from female genitourinary tract specimens. These isolates exhibited some phenotypic similarity to Rothia dentocariosa, an opportunistic pathogen usually isolated from the oral mucosa; however, their predominant sources of isolation, pigmentation, and variable reduction of nitrate were atypical. Provisionally, these organisms were designated CDC fermentative coryneform group 4 (FCG4).
Presented herein is a polyphasic study of 45 strains in this group, including morphological, biochemical, cellular fatty acid, antimicrobial susceptibility, DNA-DNA hybridization, and 16S rDNA analyses. This study indicates FCG4 to be a heterogeneous group that contains strains of charcoal-black-pigmented variants of R. dentocariosa isolated predominately from respiratory sources and of charcoal-black-pigmented variants of a recently described Corynebacterium species, Corynebacterium aurimucosum, isolated predominately from female urogenital sources.
MATERIALS AND METHODS
Bacterial strains.
Source, clinical information, geographic location, and year of receipt for 45 FCG4 clinical isolates are presented in Table 1. Reference strains Corynebacterium amycolatum ATCC 49368T, Corynebacterium matruchotii ATCC 14266T and ATCC 14265, Corynebacterium minutissimum ATCC 23348T, Corynebacterium nigricans ATCC 700975T, R. dentocariosa ATCC 17931T, and Rothia mucilaginosa (3) ATCC 25296T were obtained from the American Type Culture Collection (ATCC), Manassas, Va., C. aurimucosum NRRL B-24143T was obtained from the National Regional Research Laboratory (NRRL), Peoria, Ill., and Corynebacterium singulare CCUG 37330T was obtained from the Culture Collection of the University of Goteborg (CCUG), Goteborg, Sweden. All strains were cultured on heart infusion agar with 5% rabbit blood (rabbit blood agar; BBL, Becton Dickinson Microbiology Systems, Cockeysville, Md.) overnight at 35°C in a candle jar atmosphere before analysis. Some strains also were grown on trypticase soy agar with 5% sheep blood (BBL) to determine if differences in blood agar affected colonial morphology or pigmentation. For long-term storage, overnight cultures were suspended in defibrinated rabbit blood and frozen in liquid nitrogen.
TABLE 1.
CDC FCG4 strains studieda
| Strain no. | Yr received | Source, sex/age, and diagnosis | Geographic location | FCG4 subgroup |
|---|---|---|---|---|
| B4122 | 1969 | Mammary pustule, F/NG, NG | Michigan | Rothia-like a |
| B8899 | 1971 | Throat, NG/NG, NG | Colorado | Rothia-like a |
| C5560 (ATCC BAA-907, CCUG 48652) | 1973 | Sputum, M/NG, NG | Georgia | Rothia-like a |
| C9139 | 1974 | Left bronchus, M/NG, NG | Nevada | Rothia-like a |
| E3087 | 1978 | Urine, F/NG, NG | Maryland | Rothia-like a |
| E5063 | 1979 | Blood, F/NG, NG | North Carolina | Rothia-like a |
| G4818 | 1990 | Sputum, F/35, NG | Oregon | Rothia-like a |
| G8825 | 1993 | Blood, M/5, bacteremia | South Carolina | Rothia-like a |
| B8037 (ATCC 700540, CCUG 47034) | 1971 | Cervix, F/NG, NG | Pennsylvania | Corynebacterium-like b |
| C5610 | 1973 | Bartholin gland, F/NG, NG | Georgia | Corynebacterium-like b |
| C9693 | 1974 | Cervix, F/NG, NG | Illinois | Corynebacterium-like b |
| D6973 | 1976 | Vagina, F/NG, NG | New Jersey | Corynebacterium-like b |
| E7779 | 1980 | Cervix, F/NG, NG | Connecticut | Corynebacterium-like b |
| F1719 | 1981 | Vagina, F/NG, vaginal secretions | New York | Corynebacterium-like b |
| F4576 | 1983 | Cerebrospinal fluid, M/13 d, meningitis | Illinois | Corynebacterium-like b |
| F4578 | 1983 | Endometrium, F/17, delivery still born | California | Corynebacterium-like b |
| F4740 | 1983 | Placenta, F/NG, uterine infection | North Carolina | Corynebacterium-like b |
| F5348 | 1984 | Urine, F/36, NG | Arkansas | Corynebacterium-like b |
| G966 | 1988 | Cervix, F/NG, NG | Alabama | Corynebacterium-like b |
| G1057 | 1988 | Urine, F/43, UTI | Illinois | Corynebacterium-like b |
| G1380 | 1988 | Vagina, F/17, endometriosis | Missouri | Corynebacterium-like b |
| G2504 | 1989 | Urine, F/67, NG | West Virginia | Corynebacterium-like b |
| G3305 | 1989 | Urine, F/27, UTI | South Carolina | Corynebacterium-like b |
| G3381 | 1989 | Urine, F/34, NG | Tennessee | Corynebacterium-like b |
| G3921 | 1989 | Vagina, F/26, vaginitis | Mississippi | Corynebacterium-like b |
| G4191 | 1990 | Placenta, F/NG, placental infection | Ohio | Corynebacterium-like b |
| G5798 | 1990 | Vagina X2, F/NG, NG | Mississippi | Corynebacterium-like b |
| G6250 | 1991 | Vulva, F/16, vulval ulcer | Canada | Corynebacterium-like b |
| G6373 | 1991 | Cervix, F/18, NG | New York | Corynebacterium-like b |
| G6414 | 1991 | Vagina, F/21, NG | Pennsylvania | Corynebacterium-like b |
| G7852 | 1992 | Vulva, F/60, NG | New York | Corynebacterium-like b |
| G8234 | 1993 | Cervix, F/28, abdominal pain and G1 bleeding | Michigan | Corynebacterium-like b |
| G8236 | 1993 | Cervix, F/21, NG | Michigan | Corynebacterium-like b |
| G9202 | 1994 | Vagina, F/37, pain in vaginal incision | Louisiana | Corynebacterium-like b |
| G9230 | 1994 | Cervix, F/63, NG | New Jersey | Corynebacterium-like b |
| G9277 | 1994 | Amniotic fluid, F/31, twin fatal demise | Louisiana | Corynebacterium-like b |
| G9297 | 1994 | Vagina, F/child unknown age, NG | California | Corynebacterium-like b |
| G9328 | 1994 | Vagina, F/24, NG | Canada | Corynebacterium-like b |
| G9366 | 1995 | Cervix, F/28, IUP | New Jersey | Corynebacterium-like b |
| G9460 | 1995 | Urine, F/40, NG | California | Corynebacterium-like b |
| G9487 | 1995 | Cervix, F/78, NG | New Jersey | Corynebacterium-like b |
| G9488 | 1995 | Genital, F/55, NG | New Jersey | Corynebacterium-like b |
| G9585 | 1995 | Cervix, F/37, vaginitis | Hawaii | Corynebacterium-like b |
| G9631 | 1995 | Cervix, F/54, NG | New Jersey | Corynebacterium-like b |
| G9766 | 1995 | Cervix, F/61, NG | New Jersey | Corynebacterium-like b |
Abbreviations: F, female; M, male; NG, not given; UTI, urinary tract infection; GI, gastrointestinal; IUP, intrauterine pregnancy.
Chemical and phenotypic analysis.
Biochemical characteristics and Gram stain morphology were determined by using methods described previously (20). Biochemical tests were incubated aerobically at 35°C. Gelatin digestion tests were incubated for 14 days. Catalase and growth temperature tests were read at 1 day of incubation. All other biochemical tests were read at 2 and 7 days. Carbohydrate reactions were performed in enteric fermentation base broth (20). Cells for cellular fatty acid (CFA) determination were saponified, and the liberated fatty acids were methylated and analyzed by capillary gas-liquid chromatography (8, 20). CFA profiles were identified using a commercially available system (MIDI, Newark, Del.) utilized with a CDC library created using LGS software (20).
DNA relatedness.
Total DNA relatedness studies were performed on a set of 22 FCG4 strains which included all phenotypic variants. The methods used to cultivate cells and to prepare, isolate, and purify labeled and unlabeled DNA, as well as the methods used for DNA reassociation and the separation of single-stranded and double-stranded DNA on hydroxyapatite, have been described elsewhere (1). DNA relatedness was determined at optimal (60°C) and, on selected strains, stringent (75°C) reassociation criteria. Percent divergence was calculated to the nearest 0.5%.
16S ribosomal DNA (rDNA) sequencing.
The 16S rRNA gene of strains Rothia-like FCG4a ATCC BAA-907 (C5560) and Corynebacterium-like FCG4b ATCC 700540 (B8037) along with the type strains of C. aurimucosum and C. nigricans were amplified by PCR utilizing the primers fD1 and rD1 originally described by Weisburg et al. (18). The PCR product was then cycle sequenced with several subunit primers as previously described (2). Related sequences were identified in a BLAST search against the GenBank database. Similarity searches were performed with Clustal W, and a distance matrix was created. In Treecon, the phylogenetic tree of aligned sequences was constructed with the neighbor-joining method and bootstrapped based on 1,000 replications (2).
Antimicrobial susceptibility testing.
Antimicrobial MICs were determined using the broth microdilution method in accordance with the National Committee for Clinical Laboratory Standards (NCCLS) (9). The bacteria were initially subcultured onto trypticase soy agar plates containing 5% defibrinated sheep blood and incubated at 35°C in ambient air supplemented with 5% CO2 for 24 h. Inocula were prepared by direct colony suspension method or by the broth method if a uniform suspension was not possible by direct colony suspension because of clumping. The broth microdilution plates were prepared in-house with cation-adjusted Mueller-Hinton broth (Difco, BD) containing 5% lysed horse blood (Lampire Biological Laboratories, Pipersville, Pa.). After inoculation, broth microdilution plates were incubated at 35°C in ambient air. MICs were read at 24 and 48 h. The antimicrobial agents and the concentrations tested were as follows: cefotaxime (Sigma, St. Louis, Mo.), 0.008 to 8 μg/ml; ceftriaxone (Sigma), 0.008 to 8 μg/ml; cefepime (Bristol Myers Squibb, Syracuse, N.Y.), 0.008 to 8 μg/ml; chloramphenicol (Sigma), 0.25 to 32 μg/ml; clindamycin (USP, Rockville, Md.), 0.015 to 2 μg/ml; erythromycin (Eli Lilly and Co., Indianapolis, Ind.), 0.015 to 2 μg/ml; levofloxacin (Johnson and Johnson, Spring House, Pa.), 0.12 to 16 μg/ml; penicillin (Sigma), 0.008 to 16 μg/ml; tetracycline (Sigma), 0.06 to 8 μg/ml; trimethoprim-sulfamethoxazole (SXT; Sigma), 0.006 to 8 μg/ml; and vancomycin (Eli Lilly and Co.), 0.015 to 2 μg/ml. Susceptibility testing performance was monitored by testing Streptococcus pneumoniae ATCC 49619 (10) daily during the study.
Nucleotide sequence accession number.
The 16S rRNA sequences were submitted to GenBank, and the assigned accession numbers are listed below in Table 3.
TABLE 3.
Percent similarity of 16S rRNA gene sequence of Corynebacterium-like FCG4b strain B8037 (ATCC 700540) to sequences in Rothia-like FCG4a and related species of Rothia and Corynebacterium
| 16S rRNA gene sequence source | Strain number | GenBank accession no. | % Similarity of Corynebacterium-like FCG4b strain B8037 |
|---|---|---|---|
| R. dentocariosa | ATCC 17931T | M59055 | 89.5 |
| Rothia-like FCG4ab | C5560 (ATCC BAA-907) | AY546095 | 89.3 |
| C. aurimucosum | NRRL B-24143T | AY536427 | 99.7 |
| Corynebacterium-like FCG4b | B8037 (ATCC 700540) | AY546096 | 100.0 |
| C. nigricans | ATCC 700975T | AY536426 | 99.9 |
| C. amycolatum | ATCC 49368T | X82057 | 92.8 |
| C. argentoratense | ATCC 51927T | X83955 | 95.9 |
| C. confusum | DMMZ 2439T | Y15886 | 95.0 |
| C. coyleae | DSM 44184T | X96497 | 94.6 |
| C. diphtheriae | NCTC 11397T | X84248 | 94.8 |
| C. durum | CCUG 37331T | Z97069 | 92.3 |
| C. falsenii | CCUG 33651T | Y13024 | 94.9 |
| C. glucuronolyticum | ATCC 51860T | X86688 | 91.4 |
| C. imitans | CCUG 36877T | Y09044 | 93.8 |
| C. minutissimum | NCTC 10288T | X82064 | 99.0 |
| C. pseudotuberculosis | ATCC 19410 | D38578 | 94.1 |
| C. riegelii | CCUG 38180T | Y14651 | 94.6 |
| C. singulare | CCUG 37330T | Y10999 | 98.9 |
| Corynebacterium sp. (uncultured) MTcorylP | NAc | AF115934 | 99.5 |
| Corynebacterium sp. (uncultured) MTcory16R | NA | AF115941 | 98.5 |
| C. striatum | NCTC 764T | X84442 | 97.0 |
| C. thomssenii | DSM 44276T | AF010474 | 94.0 |
| C. ulcerans | NCTC 7910 | X84256 | 94.9 |
| C. xerosis | ATCC 373T | X84446 | 93.3 |
Abbreviations: DMMZ, Department of Medical Microbiology, University of Zurich, Zurich, Switzerland; DSM = DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany; NCTC, National Collection of Type Cultures, London, United Kingdom.
Rothia-like FCG4a is 100% similar to the type strain of R. dentocariosa (see text).
NA, not applicable.
RESULTS
Of 45 FCG4 clinical isolates studied, 37 (82%) were from female genitourinary tract specimens (Table 1). The other eight isolates were from a variety of sources, including mammary pustule, throat, sputum, bronchus, blood, and spinal fluid; of these, three were obtained from females, three from males, and the sex of one was unknown. The spinal fluid isolate was of interest because it was obtained from a neonate, suggesting the possibility of maternal transmission. All but two isolates were submitted from U.S. state health departments, representing 25 states; the remaining two were from Canada.
Cells of FCG4, grown on heart infusion agar at 35°C for 18 to 24 h, were small coccoid to short coryneform rods. The organisms were gram positive and generally stained uniformly, although some strains produced a beaded reaction. When grown on rabbit or sheep blood agar, either aerobically or in a candle jar atmosphere for 18 to 24 h, colonies ranged in size from punctate to 1.5 mm in diameter, and they were circular, convex, entire, opaque, and charcoal-black in color. Colonies of most strains were smooth, but some strains produced rough and adherent colonies that appeared to grow into the agar. Some strains produced a mixture of smooth, rough, and very rough colony types with variable gray to black pigmentation (4). Subculture of the individual colony types produced the same mixture, indicating that individual strains were capable of colonial variation. For most strains, no hemolytic reaction was observed on rabbit blood agar after overnight incubation at 35°C; however, 12 strains produced green discoloration. Three other strains produced a dark brown to black discoloration of the medium.
CFA analysis of these strains revealed the presence of two distinct groups: FCG4a, which was Rothia-like, and FCG4b, which was Corynebacterium-like. The Rothia-like FCG4a group included eight strains (B4122, B8899, C5560, C9139, E5063, E3087, G4818, and G8825), all of which produced a CFA profile similar to the profile of R. dentocariosa and R. mucilaginosa reference strains. This profile was characterized by the presence of major amounts of a-C15:0 (22 to 43%), i-C16:0 (7 to 14%), C16:0 (9 to 23%), a-C17:0 (11 to 20%), C18:2 (6 to 11%), and C18:1T9c (4 to 12%). The remaining 37 strains (Table 1) (Corynebacterium-like FCG4b) produced a CFA profile similar to the profile of C. matruchotii, C. amycolatum, C. minutissimum, C. aurimucosum (21), C. singulare (11), and C. nigricans (13). This profile was characterized by major amounts of C16:0 (33 to 47%), C18:1T9c (37 to 54%), and C18:0 (4 to 12%). A small amount (≤6%) of tuberculostearic acid (10-CH3-C18:0) was detected in 23 Corynebacterium-like FCG4b strains, of which all but one contained ≤3%, while strain G6250 contained 6%. CFA profiles of strains that contained tuberculostearic acid were more closely related to C. minutissimum, C. aurimucosum (21), or C. nigricans (13), whereas strains that did not contain tuberculostearic acid had CFA profiles more closely related to those of C. matruchotii, C. amycolatum, or C. singulare (11).
The phenotypic characteristics of the Rothia-like FCG4a and Corynebacterium-like FCG4b isolates and the reference strains are listed in Table 2. All FCG4 strains fermented d-glucose, sucrose, and maltose. Other positive characteristics for all strains included growth at 35°C and growth in nutrient broth without NaCl. Negative characteristics shared by all strains included motility, fermentation of d-xylose and lactose, growth on MacConkey agar, citrate alkalinization, reduction of nitrate to gas, indole production, and H2S production in the butt of the triple sugar iron agar slant. In general, these strains were also d-mannitol, urease, and oxidase negative; however, one Corynebacterium-like FCG4b strain produced a weak and late urease reaction, three Corynebacterium-like FCG4b strains fermented d-mannitol, and one strain each of Rothia-like FCG4a and Corynebacterium-like FCG4b produced a positive oxidase reaction.
TABLE 2.
Biochemical characteristics of Rothia-like FCG4a, Corynebacterium-like FCG4b, R. dentocariosa, R. mucilaginosa, C. aurimucosum, C. nigricans, C. matruchotii, C. minutissimum, C. amycolatum, and C. singularea
| Test performed |
Rothia-like FCG4a (n = 8)
|
R. dentocariosa ATCC 17931T | R. mucilaginosa ATCC 25296T |
Corynebacterium-like FCG4b (n = 37)
|
C. aurimucosum NRRL B-24143T | C. nigricansb ATCC 700975T |
C. matruchotii ATCC 14266T and ATCC 1265 (n = 2)
|
C. minutissimum ATCC 23348T | C. amycolatum ATCC 49368T | C. singularec CCUG 37330T | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. positive/no. tested | % Positive | No. of positive/no. tested | % Positive | No. positive/no. tested | % Positive | ||||||||
| Acid from: | |||||||||||||
| d-Glucose | 8/8 | 100 | + | + | 36(1)/37 | 97(3) | + | + | 2/2 | 100 | + | + | + |
| d-Mannitol | 0/8 | 0 | − | − | 1(2)/37 | 3(5) | − | − | 0/2 | 0 | − | − | − |
| Lactose | 0/8 | 0 | − | − | 0/37 | 0 | − | − | 0/2 | 0 | − | − | − |
| Sucrose | 7(1)/8 | 87 (13) | + | + | 25 (12)/37 | 68 (32) | w | + | 2/2 | 100 | + | − | + |
| Maltose | 8/8 | 100 | + | + | 35 (2)/37 | 95 (5) | + | + | (2)/2 | (100) | + | + | + |
| Catalase | 4, 1w/8 | 50, 13w | + | − | 37/37 | 100 | + | + | 2/2 | 100 | + | + | + |
| Oxidase | 1/8 | 13 | − | − | 1/37 | 3 | − | − | 0/2 | 0 | − | − | − |
| Urea, Christensen's | 0/8 | 0 | − | − | (1)/37 | (3) | − | − | 0/2 | 0 | − | + | + |
| Nitrate reduction | 8/8 | 100 | + | + | 2/37 | 5 | − | − | 2/2 | 100 | − | − | − |
| TSI slant, acid | 7 (1)/8 | 87 (13) | + | (+) | 29 (5)/37 | 78 (14) | + | ND | 2/2 | 100 | + | − | ND |
| TSI butt, acid | 5 (3)/8 | 62 (38) | + | (+) | 24 (9)/37 | 65 (24) | w | ND | 2/2 | 100 | + | + | ND |
| H2S (Pb Ac paper) | 6, 2w/8 | 75, 25w | + | + | 10 (12)/37 | 27 (32) | − | ND | 2/2 | 100 | + | + | ND |
| Gelatin hydrolysis | 2/5 | 40 | + | − | 1 (3)/37 | 3(8) | − | − | 0/2 | 0 | − | − | − |
| Charcoal growth pigment | 8/8 | 100 | − | − | 37/37 | 100 | −d | − | 0/2 | 0 | − | − | − |
| Growth at: | |||||||||||||
| 25°C | 4w/6 | 67w | − | − | 21/37 | 57 | − | ND | 1w/2 | 50w | w | + | ND |
| 35°C | 8/8 | 100 | + | + | 37/37 | 100 | + | + | 2/2 | 100 | + | + | + |
| 42°C | 1w/6 | 17w | − | w | 20/37 | 54 | + | ND | 1w/2 | 50w | w | w | ND |
| Esculin hydrolysis | 8/8 | 100 | + | + | 12 (4)/37 | 32 (11) | − | − | (2)/2 | (100) | − | − | − |
| Nutrient broth, 0% NaCl | 7/8 | 87 | + | (+) | 37/37 | 100 | + | ND | 2/2 | 100 | + | + | ND |
| Nutrient broth, 6% NaCl | 1/8 | 13 | − | − | 29/37 | 78 | + | ND | 0/2 | 0 | w | + | ND |
Symbols and abbreviations: n, the number of strains tested; IR, indicator reduction; +, positive; −, negative; w, weak positive; TSI, triple sugar iron; Pb Ac, lead acetate; ND, not determined. All strains were negative for growth on MacConkey agar, Simmons citrate, gas from nitrate, indole, H2S in TSI butt, motility, and acid from xylose. Values were obtained after 2 days’ incubation. The numbers of weakly positive strains are indiated by a lowercase w following the value. Values in parentheses were obtained after 3 to 7 days’ incubation.
Data from Shukla et al. (13).
Data from Riegel et al. (11).
Colonies on blood agar were sticky and slightly yellow; on trypticase soy agar they were colorless and slimy.
CFA analysis as well as the nitrate test were useful in differentiating between Rothia-like FCG4a and Corynebacterium-like FCG4b strains. All Rothia-like FCG4a strains reduced nitrate, whereas 95% of Corynebacterium-like FCG4b strains were nitrate negative.
The reference strains most similar to the Rothia-like FCG4a group were R. dentocariosa and R. mucilaginosa. The Rothia-like FCG4a group shared a common CFA profile and many common biochemical reactions. The charcoal water-insoluble pigmentation differentiated the Rothia-like FCG4a strains from all the reference strains. The morphology and sometimes catalase activity differed from that of the type strains of R. dentocariosa and R. mucilaginosa.
The reference strains most similar to the Corynebacterium-like FCG4b strains were C. amycolatum, C. aurimucosum, C. matruchotii, C. minutissimum, C. nigricans, and C. singulare. In addition to the charcoal pigmentation, Corynebacterium-like FCG4b strains could also be differentiated from some of the reference strains by nitrate reduction and urea hydrolysis.
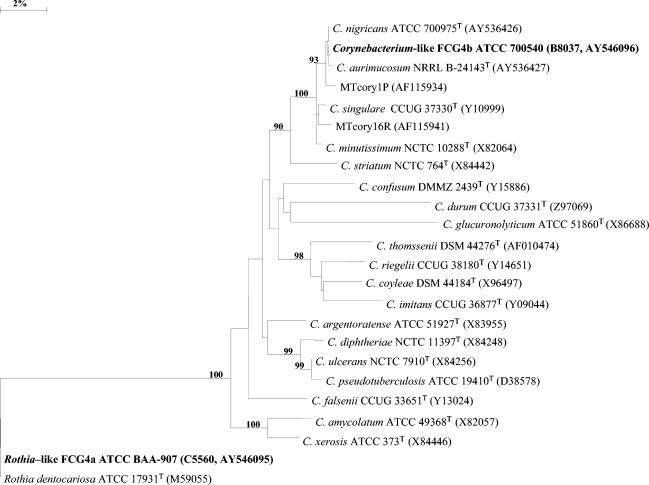
The 16S rDNA from a Rothia-like FCG4a strain (ATCC BAA-907, C5560), a Corynebacterium-like FCG4b strain (ATCC 700540, B8037), and the type strains of C. aurimucosum and C. nigricans were amplified and sequenced. The resulting sequences were aligned with several relevant Corynebacterium 16S rDNA sequences from GenBank, as well as the 16S rDNA sequences of two uncultured Corynebacterium-like organisms and R. dentocariosa. A phylogenetic tree was generated from an edited alignment and demonstrated the close relationships among most of the species in the genus Corynebacterium (Fig. 1). Rothia-like FCG4a ATCC BAA-907 was positioned with R. dentocariosa with 100% similarity. Corynebacterium-like FCG4b strain ATCC 700540 (CDC B8037) was positioned in a group with C. aurimucosum, C. nigricans, C. singulare, C. minutissimum, and two sequences from uncultured Corynebacterium-like organisms, strains MTcory1P and MTcory16R, which were similar to those given by Yassin et al. (21) and Shukla et al. (13). A similarity matrix (Table 3) generated from the edited alignment indicated that the Corynebacterium-like FCG4b strain ATCC 700540 sequence was 99.9% similar to that of C. nigricans, 99.7% to that of C. aurimucosum, 99.5% to that of MTcory1P, 99.0% to that of C. minutissimum, 98.9% to that of C. singulare, and 98.5% to that of MTcory16R. The similarity values between Corynebacterium-like FCG4b ATCC 700540 and the remaining Corynebacterium sequences ranged from 91.4% for C. glucuronolyticum to 97.0% for C. striatum.
FIG. 1.
Phylogenetic tree, including selected relevant Rothia-like, Corynebacterium-like, and Corynebacterium species organisms. The tree was rooted using R. dentocariosa as the outgroup. Bootstrap analysis was done with 1,000 resamplings; bootstrap values are indicated at some branch points. The scale bar represents a 2% difference in DNA sequence. The strain numbers and GenBank accession number for the sequences used in the study are shown. DMMZ, Department of Medical Microbiology, University of Zurich; DSM = DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen; NCTC, National Collection of Type Cultures.
DNA relatedness among FCG4 strains was determined (Table 4) with radiolabeled DNA from Corynebacterium-like FCG4b strain B8037 (ATCC 700540) and type strains of C. aurimucosum (NRRL B-24143T), C. nigricans (ATCC 700975T), and R. dentocariosa (ATCC 17931T). Two DNA relatedness groups were evident. One group comprised six strains of Rothia-like FCG4a and the type strain of R. dentocariosa. Relatedness among these strains was 79 to 85%, with 1.5 to 3.5% divergence at 60°C and 69 to 81% at 75°C. R. dentocariosa ATCC 17931T was <20% related to R. mucilaginosa, <5% related to C. minutissimum, C. matruchotii, and C. amycolatum, and <30% related to 16 strains of Corynebacterium-like FCG4b (relatedness to all but one strain was <7%). The second relatedness group comprised 16 strains of Corynebacterium-like FCG4b, the type strain of C. aurimucosum, and the type strain of C. nigricans. Relatedness among these strains was 73 to 93%, with 1.5 to 4.0% divergence at 60°C and 62 to 90% at 75°C. Corynebacterium-like FCG4b strain B8037 (ATCC 700540) was 39% related to C. minutissimum ATCC 23348T and less than 7% to the type strains of other Corynebacterium species, R. dentocariosa, and R. mucilaginosa.
TABLE 4.
DNA relatedness of Rothia-like FCG4a; Corynebacterium-like FCG4b, R. dentocariosa, C. aurimucosum, C. nigricans, C. minutissimum, C. matruchotii, C. amycolatum, and R. mucilaginosa strainsa
| Source of unlabeled DNA |
|
Relatedness of labeled DNA from:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
R. dentocariosa
|
Corynebacterium-like FCG4b
|
C. aurimucosum
|
|||||||
| ATCC 17931T
|
B8037
|
NRRL B-24143T
|
|||||||
| RBRb at 60°C | % Dc | RBR at 75°C | RBR at 60°C | % D | RBR at 75°C | RBR at 60°C | % D | RBR at 75°C | |
| R. dentocariosa ATCC 17931T | 100 | 0.0 | 100 | 5 | |||||
| Rothia-like FCG4a | |||||||||
| C5560 | 85 | 1.5 | 81 | ||||||
| G4818 | 85 | 2.0 | 75 | ||||||
| G8825 | 84 | 2.5 | 76 | ||||||
| E5063 | 83 | 1.5 | 77 | 0 | |||||
| B8099 | 83 | 2.0 | 80 | ||||||
| E3087 | 79 | 3.5 | 69 | ||||||
| C. aurimucosum NRRL B-24143T | 100 | 0.0 | 100 | ||||||
| Corynebacterium-like FCG4b | |||||||||
| B8037 | 2 | 100 | 0.0 | 100 | 85 | 4.0 | 87 | ||
| F4740 | 3 | 93 | 1.5 | 90 | |||||
| G6414 | 6 | 90 | 3.0 | 82 | |||||
| G8234 | 1 | 90 | |||||||
| G6250 | 3 | 86 | 3.0 | 69 | |||||
| G9230 | 4 | 85 | 2.0 | 68 | |||||
| G8236 | 2 | 85 | 2.5 | 75 | |||||
| D6973 | 26 | 84 | 2.0 | 73 | |||||
| G9631 | 4 | 84 | 2.0 | 68 | |||||
| G9277 | 4 | 82 | 2.5 | 69 | |||||
| G1057 | 4 | 80 | 3.0 | 67 | |||||
| F4578 | 3 | 79 | 2.0 | 67 | |||||
| F5348 | 6 | 78 | 3.0 | 67 | |||||
| G9488 | 3 | 74 | 3.0 | 64 | |||||
| F4576 | 5 | 73 | 2.5 | 62 | |||||
| C5610 | 3 | 73 | 3.5 | 67 | |||||
| C. nigricans ATCC 700975T | 97 | 0.5 | 100 | ||||||
| C. minutissimum ATCC 23348T | 3 | 39 | |||||||
| C. matruchotii ATCC 14266T | 2 | 7 | |||||||
| C. amycolatum ATCC 49368T | 2 | 5 | |||||||
| R. mucilaginosa ATCC 25296T | |||||||||
| Colony type 1 | 18 | 0 | |||||||
| Colony type 2 | 12 | 1 | |||||||
Blank space in the table indicates that the reaction was not done.
RBR, relative binding ratio [(percent heterologous DNA bound to hydroxyapatite/percent homologous DNA bound to hydroxyapatite) × 100]. All reactions were done twice:
D, divergence, the decrease in thermal stability (°C) of heterologous DNA duplexes compared with those of homologous duplexes.
Isolates of the coryneform group Corynebacterium-like FCG4b grew well in the broth microdilution plates, and so antimicrobial MICs could be read after 24 h without difficulty. Growth was not as good for isolates of the Rothia-like FCG4a group, but there was sufficient growth to read MICs at 24 h, and rarely did a 48-h reading result in a significant change in the MIC (i.e., greater than a single doubling dilution increase in the MIC).
Results for the 11 antimicrobial agents are shown in Table 5. At present there are no interpretive criteria for the determination of breakpoints recommended by NCCLS for the coryneform group of organisms. As a conservative approach, it has been suggested that the NCCLS interpretive criteria for streptococci be used when reporting erythromycin, penicillin, and vancomycin results (19). Using NCCLS breakpoints for streptococci (10), the Rothia-like FCG4a isolates were susceptible to cefepime, cefotaxime, ceftriaxone, chloramphenicol, levofloxacin, penicillin, tetracycline, and vancomycin. One of the five isolates was resistant to erythromycin (>4 μg/ml), and three of the five isolates were resistant to clindamycin (1 and >2 μg/ml). There are no interpretive breakpoints for SXT established for streptococci. However, for the Rothia-like FCG4a isolates, MICs of SXT were low.
TABLE 5.
MICs of 11 antimicrobial agents for Rothia-like FCG4a and Corynebacterium-like FCG4b
| Strain | MIC (μg/ml)a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CHL | CRO | CTX | ERY | FEP | LEV | PEN | SXT | TET | VAN | |
| Rothia-like FCG4a | |||||||||||
| C5560 | 1 | 1 | ≤0.008 | ≤0.008 | 0.6 | ≤0.015 | 0.25 | ≤0.008 | 0.12/2.28 | 0.5 | 1 |
| E3087 | 0.25 | 1 | ≤0.008 | ≤0.008 | 0.03 | ≤0.015 | 0.25 | ≤0.008 | 0.25/4.75 | 0.5 | 1 |
| E5063 | 1 | 0.5 | ≤0.008 | ≤0.008 | 0.03 | ≤0.015 | 0.25 | ≤0.008 | 0.25/4.75 | 0.5 | 1 |
| G4818 | >2 | 1 | ≤0.008 | ≤0.008 | >2 | 0.03 | 2 | ≤0.008 | 0.25/4.75 | 0.5 | 1 |
| G8825 | 0.25 | 0.5 | 0.03 | 0.03 | 0.03 | 0.06 | 0.25 | ≤0.008 | 0.12/2.28 | 0.5 | 1 |
| Corynebacterium-like FCG4b | |||||||||||
| B8037 | 2 | 4 | 1 | 1 | 0.06 | 0.25 | ≤0.12 | 0.25 | 4/76 | 0.5 | 0.25 |
| C5610 | >2 | 4 | 1 | 0.5 | 0.06 | 0.25 | ≤0.12 | 0.25 | 2/38 | 0.5 | 0.25 |
| D6973 | 2 | 4 | 1 | 0.5 | 0.06 | 0.25 | ≤0.12 | 0.25 | 2/38 | 0.5 | 0.25 |
| F4576 | 2 | 32 | 2 | 1 | 0.06 | 0.5 | ≤0.12 | 0.5 | >8/152 | 4 | 0.25 |
| F4578 | 1 | 32 | 1 | 0.5 | 0.06 | 0.25 | ≤0.12 | 0.25 | 2/38 | 2 | 0.25 |
| F4740 | 1 | 2 | 1 | 1 | 0.03 | 0.25 | ≤0.12 | 0.25 | 1/19 | 0.25 | 0.25 |
| F5348 | 1 | 4 | 2 | 1 | 0.06 | 0.5 | ≤0.12 | 0.25 | 1/19 | 0.5 | 0.25 |
| G1057 | 1 | 4 | 1 | 1 | 0.06 | 0.25 | ≤0.12 | 0.25 | 1/19 | 0.25 | 0.25 |
| G6250 | 2 | 2 | 1 | 0.5 | 0.06 | 0.25 | ≤0.12 | 0.06 | 1/19 | 0.5 | 0.25 |
| G6414 | 2 | 4 | 1 | 0.5 | 0.06 | 0.25 | ≤0.12 | 0.12 | 2/38 | 0.5 | 0.25 |
| G8234 | >2 | 4 | 1 | 0.5 | >2 | 0.25 | ≤0.12 | 0.25 | 2/38 | >8 | 0.25 |
| G8236 | >2 | 4 | 1 | 0.5 | >2 | 0.25 | ≤0.12 | 0.25 | 1/19 | 0.5 | 0.25 |
| G9230 | 2 | 4 | 0.5 | 0.25 | 0.03 | 0.12 | ≤0.12 | 0.03 | 1/19 | 1 | 0.25 |
| G9277 | >2 | 4 | 1 | 1 | >2 | 0.25 | ≤0.12 | 0.25 | 1/19 | 0.25 | 0.25 |
| G9488 | >2 | 4 | 1 | 0.5 | >2 | 0.25 | ≤0.12 | 0.25 | 2/38 | 1 | 0.25 |
| G9631 | 1 | 4 | 1 | 0.5 | 0.06 | 0.25 | ≤0.12 | 0.25 | 1/19 | 0.5 | 0.25 |
Abbreviation: CC, clindamycin; CHL, chloramphenicol; CRO, ceftriaxone; CTX, cefotaxime; ERY, erythromycin; FEP, cefepime; LEV, levofloxacin; PEN, penicillin; SXT, trimethoprim-sulfamethoxazole; TET, tetracycline; VAN, vancomycin. Where two values are given, they are the MICs of the individual drugs.
All of the Corynebacterium-like FCG4b isolates were susceptible to cefotaxime, cefepime, vancomycin, and levofoxacin. Of the 16 isolates, two demonstrated decreased susceptibility to ceftriaxone (MIC of 2 μg/ml; intermediate) and 13 isolates showed decreased susceptibility to penicillin (MICs of 0.25 to 0.5 μg/ml; intermediate). Two isolates were resistant to chloramphenicol (32 μg/ml), four isolates were resistant to erythromycin (MICs of >2 μg/ml), and all of the isolates were resistant to clindamycin (MIC of ≥1 μg/ml). Two isolates demonstrated decreased susceptibility to tetracycline (MICs of 4 μg/ml [intermediate] and >8 μg/ml [resistant]). The Corynebacterium-like FCG4b isolates consistently demonstrated decreased susceptibility to SXT.
DISCUSSION
The CDC Special Bacteriology Reference Laboratory began studying unusual clinical bacterial isolates in 1950. In 1969, the first of a group of unusual charcoal-black-pigmented coryneform isolates was submitted from a mammary pustule specimen. Between 1969 and 2004, 62 additional isolates were received, the majority of which had been submitted after 1989 and were predominately from female genitourinary sources.
The results of this study indicate that FCG4 is a heterogeneous group. Using biochemical methods and CFA analysis, we identified two distinct subgroups: Rothia-like FCG4a and Corynebacterium-like FCG4b. These subgroups differ in their CFA profiles and nitrate reduction. The clinical sources of most of the Rothia-like FCG4a strains (mammary pustule, throat, sputum, left bronchus, urine, and blood) also differed from the sources of the larger Corynebacterium-like FCG4b subgroup, which were predominately isolated from the female genitourinary tract.
Phenotypically, except for pigmentation, the Rothia-like strains generally agreed with the type strain of R. dentocariosa. The phenotypic characteristics of the Corynebacterium-like strains, C. aurimucosum, and C. nigricans were similar except on pigmentation and sometimes esculin hydrolysis, which may be a strain variation.
The similarity values derived from the 16S rDNA sequence alignment of Rothia-like FCG4a ATCC BAA-907, Corynebacterium-like FCG4b ATCC 700540, and 21 Corynebacterium or Corynebacterium-like organisms with R. dentocariosa indicated that Rothia-like FCG4a ATCC BAA-907 was most closely related to R. dentocariosa and that Corynebacterium-like FCG4b CDC B8037 (ATCC 700540) was most closely related to C. aurimucosum, C. nigricans, C. singulare, C. minutissimum, and two uncultured organisms. This close phylogenetic relationship between CDC B8037 and C. aurimucosum was shown previously by Yassin et al. (21), and that between CDC B8037 and C. aurimucosum and C. nigricans, which was investigated by Shukla et al. (13). In the former report, DNA hybridization studies did not include the CDC isolate, and in the latter report of Shukla et al. (13) no DNA hybridization studies were included. It should be emphasized that the description of a new species based on 16S rDNA sequencing and production of charcoal pigment requires caution. In addition, the relationships with uncultured bacteria should be viewed with caution. Sequencing 16S rDNA from clinical specimens can be problematic; there is a distinct possibility that an apparently new species is actually a hybrid sequence from two or more organisms (16).
The DNA-DNA hybridization results in this study were consistent with the morphological and CFA results. This analysis showed the Rothia-like FCG4a subgroup to be a charcoal-black-pigmented variant of R. dentocariosa. It shares a common CFA profile with R. dentocariosa. Rothia-like FCG4a strains and the type and reference strains of R. dentocariosa form a highly related DNA hybridization group. Even though the 16S rDNA sequence similarity values between Corynebacterium-like FCG4b ATCC 700540, C. aurimucosum, and C. nigricans were high (99.7 and 99.9%, respectively), ATCC 700540 produced charcoal-black-pigmented colonies, suggesting that it is physiologically different from C. aurimucosum, an organism with slightly yellow-pigmented colonies that has only recently been described based on two clinical isolates, one from blood and one from an unknown source (21), but it is similar to C. nigricans, a more recently named black-pigmented Corynebacterium species recovered from the human female urogenital tract (13). Although no rigid criterion exists for defining species based on close 16S rDNA sequence similarity, sequencing of 16S rDNA has been recommended by Stackebrandt et al. (15) as one of the minimal characteristics in species description along with DNA-DNA reassociation. Because of the existing high sequence similarity values between the three organisms (Corynebacterium-like FCG4b ATCC 700540, C. aurimucosum, and C. nigricans), DNA-DNA hybridization was investigated. DNA relatedness studies determined that the Corynebacterium-like FCG4b subgroup, C. aurimucosum, and C. nigricans belong to the same species (Table 4). The strains are within the limits of the same species that include strains with approximately 70% or greater DNA-DNA relatedness and with less than 5°C divergence (17). Both C. aurimucosum and C. nigricans were recently proposed, but the name C. aurimucosum was published first (21) and would have nomenclature priority. Similarly, high levels of sequence similarities were observed when the Corynebacterium-like FCG4b ATCC 700540 sequence was aligned with the C. singulare (98.9%) and C. minutissimum (99.0%) sequences. Corynebacterium-like FCG4b strain B8037 (ATCC 700540) can be differentiated from C. singulare based on phenotypic characteristics. Unlike C. singulare, Corynebacterium-like FCG4b strain B8037 (ATCC 700540) is a urease-negative organism with charcoal-black-pigmented colonies. Further, DNA-DNA hybridization data indicated that Corynebacterium-like FCG4b strain B8037 (ATCC 700540) was not the same species as C. minutissimum (39% relatedness at 60EC). Therefore, as previously reported by Yassin et al. (21), high 16S rDNA sequence similarity values between strains belonging to the same genus do not invariably indicate membership of the same species.
For clinically significant infections caused by organisms from the coryneform group, antimicrobial susceptibility testing of organisms from the coryneform group is often warranted, because resistance may be found in many classes of antimicrobial agents (5). However, because of the lack of a recommended method and interpretive criteria, only a dilution method should be used, in which case MICs can be reported without an interpretation. Although the isolates in the Rothia-like FCG4a group were all extremely susceptible to penicillin, the Corynebacterium-like FCG4b group MICs were more likely to be in the range of 0.25 to 0.5 μg/ml, and it is unknown whether penicillin would be effective. Data for 10 strains of R. dentocariosa tested with the disk diffusion method have been published (7). Although clindamycin was not included (7), results with other antimicrobial agents were susceptible and comparable to our results. Data given for five C. nigricans isolates based on results using the Etest (13) were in agreement with the present study of 16 isolates of Corynebacterium-like FCG4b.
Although R. dentocariosa opportunistic infection of the oral cavity and pharynx is well established, serious infections such as endocarditis and bacteremia are rare. In a recent review of R. dentocariosa bacteremia and endocarditis cases, the authors suggested this rarity may be attributed to lack of awareness and inadequate identification methods (12). In the present study, two of eight of charcoal-black-pigmented Rothia-like isolates were from blood specimens, which emphasizes the pathogenic potential of R. dentocariosa.
The observation that these charcoal-black-pigmented Corynebacterium-like isolates have been predominantly isolated from specimens associated with female genitourinary tract infections merits further investigation. Limited clinical information was available in this study; however, based on recent reports by Shukla et al. (13, 14) and the number of strains that have been submitted to us, this organism does appear to be clinically significant. The awareness of these charcoal-black-pigmented isolates may result in increased recognition of these organisms by clinical microbiologists and physicians and, thereby, a better understanding of their pathogenic potential.
On the basis of the results in this study, C. nigricans is a later synonym of C. aurimucosum, and an amended description of R. dentocariosa and C. aurimucosum is given.
Amended description of R. dentocariosa Georg and Brown 1967 emend. Daneshvar et al. 2004.
The description below is based on the studies of the type strain ATCC 17931 by Georg and Brown (6) as well as the results of this study. Mycelia are composed of branched filaments, often with clavate ends (1 μm or less in diameter) which fragment readily into bacillary and coccoid forms. Occasionally cultures are isolated in diplococcoid or coccoid form; however, filaments can be demonstrated in subcultures either on the surface of agar plates or in liquid medium. Microcolonies on agar surfaces may be completely filamentous or may present a filamentous border only, with the center of colony a mass of rods and cocci. Mature colonies may be raised and highly convoluted, reflecting a filamentous micromorphology, or they may be round, convex, and smooth surfaced, reflecting a bacillary or coccoid micromorphology. All mature colonies, whether rough or smooth, are creamy white or sometimes charcoal-black, entire, and of a soft texture. No aerial hyphae or spores are produced. In broth there is usually a mixture of soft, turbid, and finely granular or floccose growth. Requires organic nitrogen for growth. Poor growth on pyruvate, lactate, or acetate. Gram positive, non-acid fast, nonmotile. Aerobic to microaerophilic. Very poor growth or none anaerobically. CO2 not required for growth. Cell walls composed of galactose, alanine, glutamic acid, lysine, and possibly ornithine. Ferments fructose, d-glucose, maltose, sucrose, and glycerol. Lactic acid is the major product produced from glucose, together with acetic acid and a small amount of succinic acid. Voges-Proskauer test positive. Gelatin usually liquefied. Starch, albumin, and casein not hydrolyzed. Milk unchanged or slowly peptonized. Nitrate and nitrite reduced. Indole not produced. Nonhemolytic on human and sheep blood, variable on rabbit blood. Catalase is variable.
Amended description of C. aurimucosum Yassin et al. 2002 emend. Daneshvar et al. 2004.
The description below is based on the studies of the type strain DSM 20651 by Yassin et al. (21) as well as the results of this study. Cells are gram positive and non-acid fast. They are thin, nonmotile, non-spore-forming, and pleomorphic coryneform. On Columbia blood agar supplemented with 5% sheep blood, the colonies are sticky and slightly yellow or charcoal black in color. On trypticase soy agar they appear colorless or charcoal black and slimy. On brain heart infusion agar supplemented with 1% Tween 80, some strains are able to form a coralloid precipitin in agar when grown in the presence of CO2. Growth is facultatively anaerobic. Catalase positive. It contains meso-diaminopimelic acid as wall diamino acid in addition to galactose and arabinose in whole-cell hydrolysates (i.e., the cell wall chemotype is chemotype IV). Short-chain corynemycolic acids are present. The fatty acid profile contains saturated, unsaturated, and tuberculostearic acids. It has type PI phospholipid pattern with no nitrogen-containing compounds. Acid is produced from fructose, d-glucose, maltose, sucrose, and occasionally d-mannitol but not from adonitol, amygdalin, arabinose, cellobiose, glycerol, glycogen, inulin, lactose, mannose, melezitose, raffinose, rhamnose, ribose, salicin, sorbitol, trehalose, or xylose. It hydrolyzed hippurate and occasionally gelatin and urea but not starch. Esculin hydrolysis is variable, and nitrate reductase is rare. Positive for alkaline phosphatase, leucine arylamidase, and pyrazinamidase activities, but negative for acid phosphatase, arginine dihydrolase, esterase (C4), ester lipase (C8), lipase(C14), α-glucosidase, β-glucosidase, α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, α-mannosidase, α-fucosidase, naphthol-AS-bi-phosphohydrolase, pyrrolidonyl arylamidase, valine arylamidase, cystine arylamidase, trypsin, and chemotrypsin. Acetoin positive, indole negative. Lactate is the major product of glucose fermentation. The GC content of the type species is 63.7 mol%. The type strain of C. aurimucosum is strain IMMIB D-1488T (DSM 44532T).
Acknowledgments
We thank Deanna Jannat-Khah for her assistance with the chemical analysis and Jean Euzeby for taxonomic clarity.
REFERENCES
- 1.Brenner, D. J., A. C. McWhorter, J. K. Leete Knutson, and A. G. Steigerwalt. 1982. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J. Clin. Microbiol. 15:1133-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carvalho, M. G. S., A. G. Steigerwalt, R. E. Morey, P. L. Shewmaker, L. M. Teixeira, and R. R. Facklam. 2004. Characterization of three new enterococcal species, Enterococcus sp. nov. CDC PNS-E1, Enterococcus sp. nov. CDC PNS-E2, and Enterococcus sp. nov. CDC PNS-E3, isolated from human clinical specimens. J. Clin. Microbiol. 42:1192-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins, M. D., R. A. Hutson, V. Baverud, and E. Falsen. 2000. Characterization of a Rothia-like organism from a mouse: description of Rothia nasimurium sp. nov. and reclassification of Stomatococcus mucilaginosus as Rothia mucilaginosa comb. nov. Int. J. Syst. Evol. Microbiol. 50:1247-1251. [DOI] [PubMed] [Google Scholar]
- 4.Funke, G., and K. A. Bernard. 2003. Coryneform gram-positive rods, p. 472-501. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. ASM Press, Washington, D.C.
- 5.Funke, G., V. P'unter, and A. von Graevenitz. 1996. Antimicrobial susceptibility patterns of some recently established coryneform bacteria. Antimicrob. Agents Chemother. 40:2874-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Georg, L. K., and J. M. Brown. 1967. Rothia gen. nov. an aerobic genus of the family Actinomycetaceae. Int. J. Syst. Bacteriol. 17:79-88. [Google Scholar]
- 7.Kronvall, G., M. Lanner-Sjoberg, L. V. von Stedingk, H. S. Hanson, B. Pettersson, and E. Falsen. 1998. Whole cell protein and partial 16S rRNA gene sequence analysis suggest the existence of a second Rothia species. Clin. Microbiol. Infect. 4:255-263. [DOI] [PubMed] [Google Scholar]
- 8.Moss, C. W., and M. A. Lambert-Fair. 1989. Location of double bonds in monounsaturated fatty acids of Campylobacter cryaerophila with dimethyl disulfide derivatives and combined gas chromatography-mass spectrometry. J. Clin. Microbiol. 27:1467-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Committee for Clinical Laboratory Standards. 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 6th ed. Approved standard M7-A6. NCCLS, Wayne, Pa.
- 10.National Committee for Clinical Laboratory Standards. 2003. Performance standards for antimicrobial susceptibility testing. 13th informational supplement, M100-S13. NCCLS, Wayne, Pa.
- 11.Riegel, P., R. Ruimy, F. N. R. Renaud, J. Freney, G. Prevost, F. Jehl, R. Christen, and H. Monteil. 1997. Corynebacterium singulare sp. nov., a new species for urease-positive strains related to Corynebacterium minutissimum. Int. J. Syst. Bacteriol. 47:1092-1096. [DOI] [PubMed] [Google Scholar]
- 12.Salamon, S. A., and J. Prag. 2002. Three cases of Rothia dentocariosa bacteraemia: frequency in Denmark and a review. Scand. J. Infect. Dis. 34:153-157. [DOI] [PubMed] [Google Scholar]
- 13.Shukla, S. K., K. A. Bernard, M. Harney, D. N. Frank, and K. D. Reed. 2003. Corynebacterium nigricans sp. nov.: proposed name for a black-pigmented Corynebacterium species recovered from the human female urogenital tract. J. Clin. Microbiol. 41:4353-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shukla, S. K., M. Harney, B. Jhaveri, K, Andrews, and K. D. Reed. 2003. Is a black-pigmented Corynebacterium species an opportunistic pathogen during pregnancy? Literature review and report of 3 new cases. Clin. Infect. Dis. 37:834-837. [DOI] [PubMed] [Google Scholar]
- 15.Stackebrandt, E., W. Frederiksen, G. M. Garrity, P. A. D. Grimont, P. Kampfer, M. C. J. Maiden, X. Nesme, R. Rossella-Mora, J. Swings, H. G. Truper, L. Vauterin, A. C. Ward, and W. B. Whitman. 2002. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 52:1043-1047. [DOI] [PubMed] [Google Scholar]
- 16.Wang, G. C., and Y. Wang. 1997. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63:4645-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wayne, L. G., D. J. Brenner, R. R. Colwell, P. A. D. Grimont, O. Kandler, M. I. Krichevsky, L. H. Moore, W. E. C. Moore, R. G. E. Murray, E. Stackebrandt, M. P. Starr, and H. G. Truper. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37:463-464. [Google Scholar]
- 18.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss, K., M. Laverdiere, and R. Rivest. 1996. Comparison of antimicrobial susceptibilities of Corynebacterium species by broth microdilution and disk diffusion methods. Antimicrob. Agents Chemother. 40:930-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weyant, R. S., C. W. Moss, R. E. Weaver, D. G. Hollis, J. J. Jordan, E. C. Cook, and M. I. Daneshvar. 1996. Identification of unusual pathogenic gram-negative aerobic and facultatively anaerobic bacteria, 2nd ed. Williams & Wilkins, Baltimore, Md.
- 21.Yassin, A. F., U. Steiner, and W. Ludwig. 2002. Corynebacterium aurimucosum sp. nov. and emended description of Corynebacterium minutissimum Collins and Jones (1983). Int. J. Syst. Evol. Microbiol. 52:1001-1005. [DOI] [PubMed] [Google Scholar]