Abstract
Multiplex PCR assays were developed to identify Actinobacillus pleuropneumoniae serotypes 1, 2, and 8. Primers designed for the conserved capsular polysaccharide (CP) export region amplified a 489-bp DNA fragment from all serotypes. Primers specific to the CP biosynthesis regions of serotypes 1, 2, and 8 amplified fragments of 1.6 kb, 1.7 kb, and 970 bp from only their respective serotypes.
Actinobacillus pleuropneumoniae is the etiologic agent of swine pleuropneumonia, which is responsible for extensive economic losses each year to the swine industry. There are two biovars and 15 serotypes of A. pleuropneumoniae (1, 10, 22), and the capsular polysaccharide (CP) is the primary serotype-specific antigen, making the CP an important antigen for use in diagnostic assays (10). Early detection and identification of the causative serotype is important in controlling the spread of pleuropneumonia in the herd (22) and beginning proper treatment. However, serologic typing methods are problematic due to cross-reactivity between serotypes. PCR assays with specificity for different DNA regions have been used to identify and type A. pleuropneumoniae (5-8, 20, 21). Multiplex PCR assays to identify A. pleuropneumoniae serotype 5 (14) and serotypes 2, 5, and 6 (12) have been previously reported. The present work describes the partial characterization of the cps DNA region of serotype 8 and the development of three additional CP multiplex PCR assays for the identification of serotypes 1, 2, and 8.
The bacterial strains and plasmids used in this study are shown in Tables 1 and 2, respectively. All A. pleuropneumoniae strains were grown as previously described (14). The latex agglutination test was used to identify A. pleuropneumoniae field isolates of serotypes 1, 5, and 7 as previously described (11). A. pleuropneumoniae genomic DNA was isolated with the QIAamp DNA mini kit, following the manufacturer's recommendations (QIAGEN, Valencia, Calif.), and plasmid DNA was obtained by using the Qiaprep spin Miniprep kit (QIAGEN). DNA cloning and hybridizations were performed as described previously (19). DNA fragments to be used as probes were amplified by PCR, labeled with digoxigenin by the random primer method (Boehringer Mannheim Corp., Indianapolis, Ind.), and used for DNA hybridizations at 60°C (the cpxD probe), at 59°C (the cpsA probe), or at 49°C (the cps8CD probe) in solutions containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate).
TABLE 1.
Bacterial strains used in this study
| Species | Sero- type | Strain | Source |
|---|---|---|---|
| A. pleuropneumoniae | 1 | 4074 | ATCCa 27088 |
| A. pleuropneumoniae | 1 | 4045 | Brad Fenwick, Kansas State University, Manhattan, Kans. |
| A. pleuropneumoniae | 2 | 27089 | ATCC 27089 |
| A. pleuropneumoniae | 3 | 27090 | ATCC 27090 |
| A. pleuropneumoniae | 4 | 33378 | ATCC 33378 |
| A. pleuropneumoniae | 5 | J45 | B. Fenwick |
| A. pleuropneumoniae | 6 | Femø | ATCC 33590 |
| A. pleuropneumoniae | 7 | WF83 | Jacques Nicolet, University of Berne, Berne, Switzer- land |
| A. pleuropneumoniae | 8 | 405 | K. Mittal, Université de Montréal |
| A. pleuropneumoniae | 9 | 13261 | J. Nicolet |
| A. pleuropneumoniae | 10 | D13039 | K. Mittal |
| A. pleuropneumoniae | 11 | 56153 | B. Fenwick |
| A. pleuropneumoniae | 12 | 8329 | K. Mittal |
| A. pleuropneumoniae | Field isolates | Karen Post, Rollins Animal Disease Diagnostic Lab | |
| H. influenzae type b | Eagen | Porter Anderson, University of Rochester School of Medicine | |
| H. parasuis | Field isolate | K. Post | |
| H. paragallinarum | Field isolate | K. Post | |
| Pasteurella multocida | Field isolate | K. Post | |
| Histophilus somni | Field isolate | K. Post | |
| Escherichia coli chemi- cally competent DH5α | Life Technologies, Inc., Rockville, Md. |
American Type Culture Collection, Manassas, Va.
TABLE 2.
Plasmids used in this study
| Plasmid | Source |
|---|---|
| pBluescript II SK(+/−) phagemid; cloning vector; | Promega Corp., Madison, Wis. |
| 2.9 kb; Ampr | |
| pJSAp81; 3.6-kb ClaI fragment of A. pleuropneumo- niae serotype 8 cloned into pBluescript II SK(+/−) Ampr | This work |
| pJSAp82; 1.8-kb EcoRV fragment of A. pleuropneu- moniae serotype 8 cloned into pBluescript II SK(+/−) Ampr | This work |
| pJSAp83; 2.0-kb EcoRV fragment of A. pleuropneu- moniae serotype 8 cloned into pBluescript II SK(+/−) Ampr | This work |
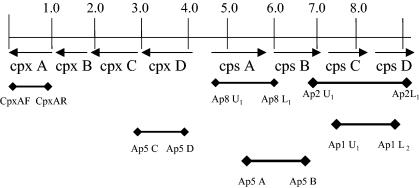
DNA for multiplex PCR was extracted as previously described (14). Five microliters of DNA template containing 1 to 2 ng of DNA was used for each reaction. Primers cpxAF, cpxAR, Ap5C, and Ap5D were designed from the conserved CP export region of A. pleuropneumoniae serotype 5. Forward and reverse primers, Ap1U1 and Ap1L2, Ap2U1 and Ap2L1, Ap5A and Ap5B, and Ap8U1 and Ap8L1, were designed from the serotype-specific CP biosynthesis regions of A. pleuropneumoniae serotypes 1, 2, 5, and 8, respectively (Fig. 1 and Table 3). The final volume of each master mix contained 1× PCR buffer (Fisher Scientific, Pittsburgh, Pa.), 200 μM concentrations of each deoxynucleoside triphosphate, and 2 U of Taq polymerase (Fisher Scientific). For identification of A. pleuropneumoniae serotype 1, the PCR mix contained a final concentration of 3 mM MgCl2, 20 pmol of each serotype-specific primer, and 10 pmol of each cpx primer. For identification of A. pleuropneumoniae serotype 2, the PCR mix contained a final concentration of 2 mM MgCl2 and 10 pmol of each of the cpx and cps primers. The A. pleuropneumoniae serotype 5 PCR mix contained a final concentration of 2 mM MgCl2 and 10 pmol of each cpx and cps primer. For identification of A. pleuropneumoniae serotype 8, the assay mix contained a final concentration of 3 mM MgCl2, 10 pmol of each serotype 8-specific cps primer, and 20 pmol of each cpx primer. Cycling parameters for each of the different PCRs are shown in Table 4. These parameters were found to be optimal for each of the primers used. However, modification of these times or temperatures may still be successful if the utilization of uniform cycling conditions is desired.
FIG. 1.
Map of the CP region of A. pleuropneumoniae and location of the conserved cpx primers and serotype-specific cps primers used for PCR.
TABLE 3.
Primer sequences used for multiplex PCR
| Primer and CP region | Name | Sequence | Primer size (bp) | Product size (bp) |
|---|---|---|---|---|
| Export region | ||||
| Forward primer | cpxAF | 5′-TAGAACCTTGTAAGCCTCGTCCATA-3′ | 25 | 489 |
| Reverse primer | cpxAR | 5′-CGTTTGTTAAGTGGTGTTGAGC-3′ | 22 | |
| Forward primer | Ap5C | 5′-TGGCGATACCGGAAACAGAGTC-3′ | 22 | 715 |
| Reverse primer | Ap5D | 5′-GCGAAAGGCTATGGTATGGGTATGG-3′ | 25 | |
| Biosynthesis region | ||||
| Forward primer | Ap1U1 | 5′-AGTGGCTGGATGAGACGAGAC-3′ | 21 | 1,603 |
| Reverse primer | Ap1L2 | 5′-TAGTTTGTTATGGTATTTCTGTA-3′ | 23 | |
| Forward primer | Ap2U1 | 5′-CGCAGCCGGACAAAAACAAATACACG-3′ | 26 | 1,725 |
| Reverse primer | Ap2L1 | 5′-CACCCCATGAATCGACTGATTGCCAT-3′ | 26 | |
| Forward primer | Ap5A | 5′-TTTATCACTATCACCGTCCACACCT-3′ | 25 | 1,100 |
| Reverse primer | Ap5B | 5′-CATTCGGGTCTTGTGGCTACTAA-3′ | 23 | |
| Forward primer | Ap8U1 | 5′-AACGGCTTTTGAACAACTTTATTTATTT-3′ | 28 | 977 |
| Reverse primer | Ap8L1 | 5′-TTCATTCCTAAACTCCGTATTGTCA-3′ | 25 |
TABLE 4.
Cycling times and temperatures for PCR
| PCR | Conditions |
|---|---|
| A. pleuropneumoniae serotype 1 specific | 1 cycle: 95°C for 3 min (denaturation); 33 cycles: 95°C for 1 min (denaturation), 56°C for 1 min (annealing), 72°C for 2 min (extension); 1 cycle: 72°C for 10 minute (final extension) |
| A. pleuropneumoniae serotype 2 specific | 1 cycle: 95°C for 3 min (denaturation); 35 cycles: 95°C for 1 min (denaturation), 60°C for 1 min (annealing), 72°C for 2 min (extension); 1 cycle: 72°C for 10 min (final extension) |
| A. pleuropneumoniae serotype 5 specific | 1 cycle: 95°C for 3 min (denaturation); 31 cycles: 95°C for 1 min (denaturation), 54°C for 1 min (annealing), 72°C for 1 min (extension); 1 cycle: 72°C for 10 min (final extension) |
| A. pleuropneumoniae serotype 8 specific | 1 cycle: 95°C for 3 min (denaturation); 29 cycles: 94°C for 30 s (denaturation), 60°C for 30 s (annealing), 72°C for 40 s (extension); 1 cycle: 72°C for 10 min (final extension) |
The cps sequence of serotypes 1 and 2 were previously determined (reference 24 and unpublished data). These sequences, combined with the current sequence of cps8, were used to design primers to expand the A. pleuropneumoniae serotype 5 multiplex PCR assay to include serotypes 1, 2, and 8. An additional set of primers was designed from the DNA sequence of the serotype 5 CP export region because the original primer sets Ap5C and Ap5D did not amplify the cpxCD fragment from serotype 4 (14) (Fig. 2). Primers cpx5AF and cpx5AR amplified a 489-bp DNA fragment from the cpxA gene of all serotypes, including serotype 4 (Fig. 3).
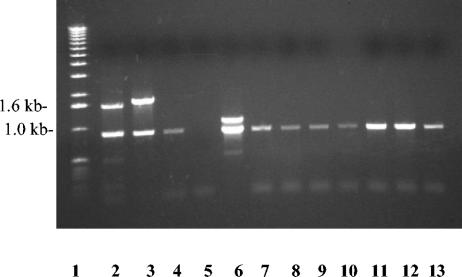
FIG. 2.
Agarose gel electrophoresis of DNA products amplified from A. pleuropneumoniae serotypes 1 to 12. Lane 1, 1-kb ladder; lanes 2, 4, 5, and 7 through 13, amplification of serotypes 1, 3, 4, and 6 through 12 with primers Ap1U1 and Ap1L2; lane 3, amplification of serotype 2 with primers Ap2U1 and Ap2L1; lane 6, amplification of serotype 5 DNA with primers Ap5A and Ap5B. Export primers Ap5C and Ap5D amplified a 715-bp fragment from all serotypes except serotype 4.
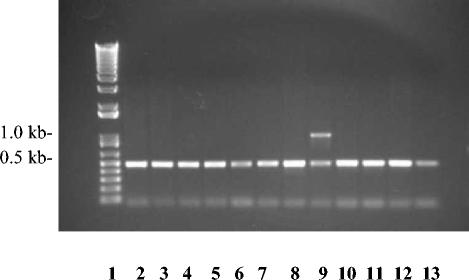
FIG. 3.
Agarose gel electrophoresis of A. pleuropneumoniae DNA products. Lane 1, 1-kb ladder; lanes 2 through 13, PCR products from serotypes 1 through 12 amplified with cpxAF, cpxAR, Ap8U1, and Ap8L1. Export primers cpxAF and cpxAR amplified a band of 489 bp from all A. pleuropneumoniae serotypes.
In order to develop a serotype-specific PCR assay for the identification of serotype 8, the CP biosynthesis (cps) genes were sequenced. Templates for DNA sequencing were constructed by subcloning CP cps fragments generated from specific restriction sites within the serotype 8 genome into pBluescript II SK(+). Templates were sequenced on an ABI 3100 capillary sequencer, and sequence analysis was performed using DNA Star (Madison, Wis.) software.
The conserved cpx8D gene and part of the predicted adjacent serotype-specific cps DNA were identified from ClaI-digested genomic DNA of A. pleuropneumoniae serotype 8 by use of a 480-bp DNA probe to the cpxD gene of serotype 5. A 3.7-kb fragment of serotype 8 genomic DNA hybridized with the probe at 60°C. This fragment was cloned into pBluescript, and the clone was sequenced and designated pJSAp81. DNA sequencing revealed that the pJSAp81 insert contained 843 bp of the cpx8C gene, the entire cpx8D gene, and two open reading frames (ORFs), one complete and one incomplete, that were transcribed in the opposite direction. These two ORFs were putatively designated cps8A and cps8B. A PCR was used to generate DNA fragments of cps8B that were labeled and used as probes to clone additional downstream sequences, which included the remainder of cps8B, complete ORFs designated cps8C and cps8D, and 624 bp of additional sequence.
DNA sequencing revealed that cpsA was 1,143 bp long, cps8B was 429 bp long, cps8C was 1,152 bp long, and cps8D was 999 bp long. The majority of the cps genes from A. pleuropneumoniae serotype 8 did not reveal any substantial homology at the nucleotide level with other sequences in the nucleotide databases at the National Center for Biotechnology Information (NCBI). The exception was cps8B, which had 82% identity at the nucleotide level with tagD from the teichoic acid biosynthesis locus in Bacillus subtilis and at the amino acid level demonstrated 70% and 68% identity with glycerol 3-phosphate cytidyltransferases from B. subtilis and Listeria monocytogenes, respectively. Of interest is that cps2B from serotype 2 also had a high degree of homology with B. subtilis tagD, and both A. pleuropneumoniae serotypes 2 and 8 contain glycerol teichoic acids in their CPs (16). The lack of homology observed at the nucleotide level between the ORFs cps8A, cps8C, and cps8D from A. pleuropneumoniae serotype 8 and sequences in the NCBI databases was not surprising. Little homology appears to exist between some genes that encode glycosyltransferases involved in the biosynthesis of unique CPs (4, 17). A lack of substantial homology at the nucleotide level was also noticed with most genes from the CP regions of A. pleuropneumoniae serotype 2 (unpublished data) and serotype 5 (24). These findings reflect the unique structure of the CP of each serotype.
The use of A. pleuropneumoniae serotype-specific primers Ap1U1 and Ap1L2, Ap2U1 and Ap2L1, Ap5A and Ap5B, and Ap8U1 and Ap8L1 amplified fragments of 1.6, 1.7, and 1.1 kb and 970 bp from only serotypes 1, 2, 5, and 8, respectively (Fig. 2 and 3). Neither the 880-bp or 489-bp cpx fragments nor any of the cps fragments were amplified from controls lacking bacterial DNA or from Histophilus somni (“Histophilus ovis”), Haemophilus influenzae, Haemophilus parasuis, Haemophilus paragallinarum, or Pasteurella multocida (data not shown). The optimum annealing temperature for the serotype 1 PCR was 56°C, whereas the optimum annealing temperature for the serotype 2 and serotype 8 PCRs was 60°C (Table 4).
Field isolates of A. pleuropneumoniae from the United States were assayed by the latex agglutination test and by PCR using serotype 1- and serotype 5-specific primers (Fig. 4). Out of 72 field isolates from pigs with respiratory disease originally identified by phenotypic properties as A. pleuropneumoniae, 58 isolates were typed as A. pleuropneumoniae serotypes 1, 5, or 7 by latex agglutination and 14 isolates were other serotypes or nontypeable. All isolates identified as serotype 1 or serotype 5 by latex agglutination were also identified as serotype 1 or 5 by amplification of the cpx band and the 1.6-kb and 1.1-kb cps bands, respectively.
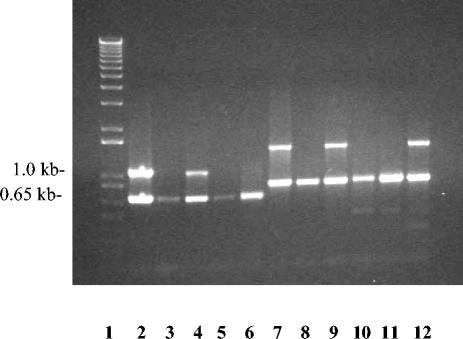
FIG. 4.
Agarose gel electrophoresis of DNA products amplified from A. pleuropneumoniae field isolates by using primers for serotypes 1 and 5. Lane 1, 1-kb ladder; lanes 2 through 6, PCR DNA products from field isolates of A. pleuropneumoniae serotypes 5, 7, 5, 1, and 7, respectively, amplified with Ap5C and Ap5D and serotype 5-specific primers Ap5A and Ap5B; lanes 7 through 12, PCR DNA products from field isolates of A. pleuropneumoniae serotypes 1, 5, 1, 5, 7, and 1, respectively, amplified with cpxU1 and cpxL1 and serotype 1-specific primers Ap1U1 and Ap1L2.
Due to the prevalence of A. pleuropneumoniae serotype 8 in Denmark, primers designed specifically for the serotype 8 cps region were tested at the Danish Veterinary Institute in Copenhagen, Denmark. The primers were used in a PCR assay to screen 41 field isolates previously identified as A. pleuropneumoniae serotype 8 by gel immunodiffusion as well as 2 confirmed A. pleuropneumoniae serotype 6 field isolates and the A. pleuropneumoniae serotype 6 reference strain Femø. Thirty-nine of the 41 A. pleuropneumoniae serotype 8 field isolates were confirmed to be A. pleuropneumoniae serotype 8 by amplification of the 970-bp fragment. The 970-bp fragment was not amplified from the A. pleuropneumoniae serotype 6 field isolates or the serotype 6 reference strain Femø (data not shown).
As determined by sequencing, the serotype 8 cps locus was located upstream from the cpx8DCBA gene cluster, which encodes proteins involved in CP export. The location of the A. pleuropneumoniae serotype 8-specific DNA upstream from the cpx8D gene was consistent with the location of the CP biosynthesis genes from serotypes 1, 2, and 5 (23, 24) and other bacterial species that express type II/III CP (2, 4, 13, 17, 18). These findings provide further evidence that the genetic organizations of the A. pleuropneumoniae CP locus are similar between serotypes and very similar to the organization of the CP loci of encapsulated H. influenzae and Neisseria meningitidis.
Primers Ap1U1 and Ap1L2, designed specifically for the A. pleuropneumoniae serotype 1 cps region, amplified a 1.6-kb fragment by PCR. However, a longer extension time was required with these primers for amplification of the 1.6-kb fragment than for the amplification of other fragments. An increase in the concentration of the A. pleuropneumoniae serotype 1 cps primers compared to cpx primers was also important for amplification of the 1.6-kb fragment. These modifications were necessary due to preferential amplification of shorter fragments, possible competition for binding sites between the cps and cpx primers, or simply the requirement for more cps primers to amplify a larger fragment (9). A disadvantage of the previous multiplex PCR assay described by Lo et al. (14) was that the cpx5D primers Ap5C and Ap5D did not amplify a product from serotype 4. The absence of the cpx DNA fragment from serotype 4 indicated that areas of nonhomology or low homology may be present even in the conserved export region. Therefore, primers were designed for the cpxA gene, which encodes a portion of an ATP-binding cassette that is essential for the export of CP (3). As expected, these primers amplified a DNA fragment from all serotypes of A. pleuropneumoniae.
The successful application of the multiplex PCR assay to type field isolates provided a quick and simple method to identify A. pleuropneumoniae serotypes 1, 2, 5, and 8. The preparation and identification of the isolates was relatively simple, and PCR amplification was completed in less than 4 h. The A. pleuropneumoniae cpx and cps fragments were specific to A. pleuropneumoniae, and DNA was not amplified from other species of the Pasteurellaceae tested. The A. pleuropneumoniae serotype 8-specific primers were successfully used to confirm the identity of 39 serotype 8 field isolates previously determined to be serotype 8. Serological cross-reactivity between serotypes 6 and 8 has been problematic (15), but no cross-reactions were seen with the limited number of serotype 6 isolates tested using the CP-specific multiplex PCR.
In conclusion, the multiplex PCR assay described was effective in detecting A. pleuropneumoniae and identifying serotypes 1, 2, 5, and 8 from purified DNA and bacterial cells. The assay was relatively rapid, easy to perform, and highly sensitive and specific compared to serological assays.
Nucleotide sequence accession number.
The sequence of cps8ABC genes has been deposited in the GenBank database under accession number AY356527.
Acknowledgments
We thank Gretchen Glindemann for technical assistance, Nicole Kauffman for DNA sequencing, Abey Bandara for providing DNA sequences, Stephen Boyle for valuable advice, and Jane Duncan for advice and review of the manuscript.
This work was supported by USDA/CSREES NRI grant 98-35204-6811 from the Cooperative State Research, Education, and Experiment Service of the U.S. Department of Agriculture and by HATCH formula funds to the Virginia Agricultural Experiment Station.
REFERENCES
- 1.Blackall, P. J., H. B. L. M. Klaasen, H. van den Bosch, P. Kuhnert, and J. Frey. 2002. Proposal of a new serovar of Actinobacillus pleuropneumoniae: serovar 15. Vet. Microbiol. 84:47-52. [DOI] [PubMed] [Google Scholar]
- 2.Clarke, B. R., R. Pearce, and L. S. Roberts. 1999. Genetic organization of the Escherichia coli K10 capsule gene cluster: identification and characterization of two conserved regions in group III capsule gene clusters encoding polysaccharide transport functions. J. Bacteriol. 181:2279-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fath, M. J., and R. Kolter. 1993. ABC transporters: bacterial exporters. Microbiol. Rev. 57:995-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frosch, M., U. Edwards, K. Bousset, B. Krausse, and C. Weisgerber. 1991. Evidence for a common molecular origin of the capsule gene loci in gram-negative bacteria expressing group II capsular polysaccharides. Mol. Microbiol. 5:1251-1263. [DOI] [PubMed] [Google Scholar]
- 5.Gram, T., P. Ahrens, and J. P. Nielsen. 1996. Evaluation of a PCR for detection of Actinobacillus pleuropneumoniae in mixed bacterial cultures from tonsils. Vet. Microbiol. 51:95-104. [DOI] [PubMed] [Google Scholar]
- 6.Gram, T., and P. Ahrens. 1998. Improved diagnostic PCR assay for Actinobacillus pleuropneumoniae based on the nucleotide sequence of an outer membrane lipoprotein. J. Clin. Microbiol. 36:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gram, T., P. Ahrens, M. Andreasen, and J. P. Nielsen. 2000. An Actinobacillus pleuropneumoniae PCR typing system based on the apx and omlA genes—evaluation of isolates from lungs and tonsils of pigs. Vet. Microbiol. 75:43-57. [DOI] [PubMed] [Google Scholar]
- 8.Hennessy, K. J., J. J. Iandolo, and B. W. Fenwick. 1993. Serotype identification of Actinobacillus pleuropneumoniae by arbitrarily primed polymerase chain reaction. J. Clin. Microbiol. 31:1155-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Innis, M. A., D. H. Gelfand, and J. J. Sninsky. 1995. PCR strategies. Academic Press, Inc., San Diego, Calif.
- 10.Inzana, T. J., and B. Mathison. 1987. Serotype specificity and immunogenicity of the capsular polymer of Haemophilus pleuropneumoniae. Infect. Immun. 55:1580-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inzana, T. J. 1995. Simplified procedure for preparation of sensitized latex particles to detect capsular polysaccharides: application to typing and diagnosis of Actinobacillus pleuropneumoniae. J. Clin. Microbiol. 33:2297-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jessing, S. G., Ø. Angen, and T. J. Inzana. 2003. Evaluation of a multiplex PCR test for simultaneous identification and serotyping of Actinobacillus pleuropneumoniae serotypes 2, 5, and 6. J. Clin. Microbiol. 41:4095-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo, R. Y. C., L. J. McKerral, T. L. Hills, and M. Kostrzynska. 2001. Analysis of the capsule biosynthesis locus of Mannheimia (Pasteurella) haemolytica A1 and proposal of a nomenclature system. Infect. Immun. 69:4458-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo, T. M., C. K. Ward, and T. J. Inzana. 1998. Detection and identification of Actinobacillus pleuropneumoniae serotype 5 by multiplex PCR. J. Clin. Microbiol. 36:1704-1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicolet, J. 1988. Taxonomy and serological identification of Actinobacillus pleuropneumoniae. Can. Vet. J. 29:578-579. [PMC free article] [PubMed] [Google Scholar]
- 16.Perry, M. B., E Altman, J. R. Brisson, L. M. Beynon, and J. C. Richards. 1990. Structural characteristics of the antigenic capsular polysaccharide and lipopolysaccharide involved in serological classification of Actinobacillus pleuropneumoniae strains. Serodiagn. Immunother. Infect. Dis. 4:299-308. [Google Scholar]
- 17.Roberts, I. S., R. Mountford, N. High, D. Bitter-Suerman, K. Jann, K. N. Timmis, and G. J. Boulnois. 1986. Molecular cloning and analysis of the genes for production of the K5, K7, K12, and K92 capsular polysaccharides of Escherichia coli. J. Bacteriol. 168:1228-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Russo, T. A., S. Wenderoth, U. B. Carlino, J. M. Merrick, and A. J. Lesse. 1998. Identification, genomic organization, and analysis of the group III capsular polysaccharide genes kpsD, kpsM, kpsT, and kpsE from an extraintestinal isolate of Escherichia coli (CP9, O/4/K54/H5). J. Bacteriol. 180:338-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 1-3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Savoye, C., J. L. Jobert, F. Berthelot-Herault, A. M. Keribin, R. Cariolet, H. Morvan, F. Madec, and M. Kobish. 2000. A PCR assay used to study aerosol transmission of Actinobacillus pleuropneumoniae from samples of live pigs under experimental conditions. Vet. Microbiol. 73:337-347. [DOI] [PubMed] [Google Scholar]
- 21.Schaller, A., S. P. Djordjevic, G. J. Eamens, W. A. Forbes, R. Kuhn, P. Kuhnert, M. Gottschalk, J. Nicolet, and J. Frey. 2001. Identification and detection of Actinobacillus pleuropneumoniae by PCR based on the gene apxIVA. Vet. Microbiol. 79:47-62. [DOI] [PubMed] [Google Scholar]
- 22.Taylor, D. J. 1999. Actinobacillus pleuropneumoniae, p. 343-354. In B. E. Straw, S. D'Allaire, W. L. Mengeling, and D. J. Taylor (ed.), Diseases of swine. Iowa State University Press, Ames, Iowa.
- 23.Ward, C. K., and T. J. Inzana. 1997. Identification and characterization of a DNA region involved in the export of capsular polysaccharide by Actinobacillus pleuropneumoniae serotype 5a. Infect. Immun. 65:2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward, C. K., M. L. Lawerence, H. P. Veit, and T. J. Inzana. 1998. Cloning and mutagenesis of a serotype-specific DNA region involved in encapsulation and virulence of Actinobacillus pleuropneumoniae serotype 5a: concomitant expression of serotype 5a and 1 capsular polysaccharides in recombinant A. pleuropneumoniae serotype 1. Infect. Immun. 66:3326-3336. [DOI] [PMC free article] [PubMed] [Google Scholar]