Abstract
A commercially available repetitive-sequence-based PCR (rep-PCR) DNA fingerprinting assay adapted to an automated format, the DiversiLab system, enables rapid microbial identification and strain typing. We explored the performance of the DiversiLab system as a molecular typing tool for 69 Aspergillus isolates (38 A. fumigatus, 15 A. flavus, and 16 A. terreus isolates) had been previously characterized by morphological analysis. Initially, 27 Aspergillus isolates (10 A. fumigatus, 9 A. flavus, and 8 A. terreus isolates) were used as controls to create a rep-PCR-based DNA fingerprint library with the DiversiLab software. Then, 42 blinded Aspergillus isolates were typed using the system. The rep-PCR-based profile revealed 98% concordance with morphology-based identification. rep-PCR-based DNA fingerprints were reproducible and were consistent for DNA from both hyphae and conidia. DiversiLab dendrogram reports correctly identified all A. fumigatus (n = 28), A. terreus (n = 8), and A. flavus (n = 6) isolates in the 42 blinded Aspergillus isolates. rep-PCR-based identification of all isolates was 100% in agreement with the contiguous internal transcribed spacer (ITS) region (ITS1-5.8S-ITS2) sequence-based identification of the respective isolates. Additionally, the DiversiLab system could demonstrate strain-level differentiation of A. flavus and A. terreus. Automated rep-PCR may be a time-efficient, effective, easy-to-use, novel genotyping tool for identifying and determining the strain relatedness of fungi. This system may be useful for epidemiological studies, molecular typing, and surveillance of Aspergillus species.
Invasive aspergillosis (IA) has emerged as a major opportunistic mycosis, surpassing invasive candidiasis as the most frequent fungal infection found at autopsy in tertiary care oncology and bone marrow transplant centers (12, 23, 28). Despite the recent introduction of several new antifungals, mortality associated with IA remains high (12, 23). Among the more than 180 Aspergillus species, A. fumigatus, A. flavus, and A. terreus account for the majority of cases of IA (12). Since the pathogenic potential and the susceptibility profile of Aspergillus species differ, the rapid identification of Aspergillus at the species level is considered important (12). Such identification methods are currently based on a combination of macro- and micromorphological features that may take days to weeks for complete identification (22, 24). These methods are laborious, time-consuming, and require significant training (31) and expertise in laboratory mycology.
Since environmental factors play a key part in the acquisition of IA, infection control measures are crucial to reduce exposure to this opportunistic pathogen (23). Such measures have been shown to decrease the incidence of IA in epidemic and possibly in nonepidemic situations (2-4, 6, 7, 15, 23, 25, 29, 33). However, many controversies still exist regarding the epidemiology of IA (23). Among those controversies, what constitutes a hospital-acquired versus a community-acquired case of IA and what are the potential sources of nosocomial IA are major issues of uncertainty (23). In view of the tremendous genetic diversity among Aspergillus isolates (7, 9, 23, 25, 28, 35), a reliable, simple, and rapid molecular typing method system with high discriminatory power could greatly facilitate the investigation of the origin of IA cases.
Determining genetic sequence variation at the molecular level is an alternative to culturing for identification of Aspergillus species. For example, the ribosomal genes demonstrate conserved sequence regions ideal for primer targeting as well as regions of variability useful for species identification (38, 44). Amplification techniques, with subsequent species-specific probing of the amplicons, have been utilized to overcome the delays and lack of sensitivity and specificity that are encountered with conventional diagnostic mycology (12, 14, 39, 44). However, the use of species-specific probes is not always an efficient approach in fungal identification, given the large variety of potentially pathogenic fungi.
To address the problem of low resolution in Aspergillus species discrimination, various DNA fingerprinting systems, such as restriction fragment length polymorphism analysis with repetitive probes or with restriction endonuclease analysis, random amplified polymorphic DNA analysis, polymorphic microsatellite marker analysis, and microsatellite length polymorphism analysis, have been developed in order to delineate strains of Aspergillus species (6, 15, 23, 25, 37). Since each method has advantages and disadvantages (25), no single method has emerged as ideal for investigation of clustering of cases of IA or for routine surveillance (25). For example, random fragment length polymorphism analysis with Southern blotting is labor-intensive, polymorphic microsatellite marker analysis requires specialized equipment, random amplified polymorphic DNA analysis is prone to artifacts and lacks reproducibility, and restriction endonuclease analysis frequently entails subjective interpretation of the profiles (25). A combination of typing methods has been suggested in order to overcome the above issues (25).
In this work, we present a new method of molecular typing for Aspergillus species by using automated repetitive-sequence-based PCR (rep-PCR), an approach that has been established for subspecies identification and strain delineation for bacteria (10, 11, 42, 43) and yeasts (8, 34). We also present a comparison of rep-PCR-based DNA fingerprint analysis with an internal transcribed spacer (ITS) region, specifically the ITS1-5.8S-ITS2 region, and sequence variations for Aspergillus species identification, and we demonstrate the potential of the DiversiLab system in Aspergillus strain discrimination.
(A part of this work was presented at the 103rd General Meeting of the American Society for Microbiology, Washington, D.C., May 2003 [Abstr. 103rd Gen. Meet. Am. Soc. Microbiol., abstr. M-1034, 2003].)
MATERIALS AND METHODS
Identification of Aspergillus isolates by morphological and cultural characteristics.
Clinical isolates (n = 69) for the three most common pathogenic Aspergillus species, A. fumigatus (n = 38), A. flavus (n = 15), and A. terreus (n = 16), were identified by review of culture reports (1998 to 2002) from the Clinical Microbiology Laboratory at M.D. Anderson Cancer Center. All fungal samples submitted to the Clinical Microbiology Laboratory were routinely plated on Sabouraud dextrose plates, Mycosel agar (BBL, Cockeysville, Md.), Sabouraud dextrose slants, and brain heart infusion-10% sheep blood agar with chloramphenicol and gentamicin. The media were incubated at 25°C without CO2 for a total of 4 weeks and read twice per week. Aspergillus species were identified using standard morphological criteria (24). Fungal strains were subsequently cultured from frozen stocks on yeast maltose broth or potato dextrose agar plates at 37°C for at least 1 day for molecular analysis.
Clinical data.
Corresponding medical records were retrieved and reviewed retrospectively in order to determine whether the positive culture for Aspergillus species represented colonization or infection (definite or probable IA) as defined according to the National Institute of Allergy and Infectious Diseases Mycoses Study Group-European Oncology Research Cooperative criteria (1).
DNA extraction.
DNA was extracted from approximately 10 μl of hyphae or conidia of each Aspergillus isolate by using the UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Solano Beach, Calif.). Manufacturer's instructions were followed directly for conidia. For the hyphae, a heating (65°C for 15 min) step before extraction was added, and the bead beating was extended from 10 to 30 min. Sample DNA was standardized to approximately 25 ng/μl, and genomic integrity was visualized using agarose gel electrophoresis.
rep-PCR DNA fingerprinting.
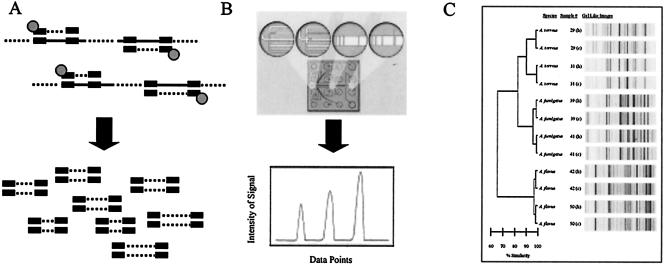
The extracted DNA was amplified using the DiversiLab Aspergillus DNA fingerprinting kit (Bacterial Barcodes, Inc., Houston, Tex.) following the manufacturer's instructions. Briefly, 50 ng of genomic DNA, 2.5 U of AmpliTaq, and 1.5 μl of 10× PCR buffer (Applied Biosystems, Foster City, Calif.) were added to the rep-PCR master mix for a total of 25 μl per reaction mixture. Thermal cycling parameters were the following: initial denaturation at 94°C for 2 min, 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, extension at 70°C for 90 s, and a final extension at 70°C for 3 min. Detection and analysis of rep-PCR products were implemented using the DiversiLab system, in which the amplified fragments of various sizes and intensities (Fig. 1A) were separated and detected using microfluidics chips (Fig. 1B) with the Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, Calif.), and analysis was performed with the DiversiLab software version 2.1.66 (Fig. 1C). The resulting DNA fingerprint patterns can be viewed as electropherograms (Fig. 1B). Reports included the dendrogram, isolate information, and a gel-like image of each sample (Fig. 1C).
FIG. 1.
Overview of automated rep-PCR and the DiversiLab system. (A) rep-PCR primers bind to many specific repetitive sequences interspersed throughout the genome and amplify various fragments of different lengths. (B) A microfluidics chip separates amplified fragments based on size and charge. The fluorescence intensity and migration time are used to generate electropherograms from each DNA sample. (C) A sample DiversiLab report showing data analysis of the samples, gel-like images, and the dendrogram.
DNA sequencing and analysis.
The species identification of all 69 isolates was confirmed by sequencing of ITS1-5.8S-ITS2 as described by White et al. (44). Briefly, the fragments with an average size of 580 bp containing ITS regions were amplified. The amplicons were purified using the High Pure PCR product purification kit (Roche Diagnostics Co., Indianapolis, Ind.), and bidirectional sequencing was carried out using ITS1 and ITS4 primers and the BigDye Terminator version 3.1 cycle sequencing kit. The products were purified by PERFORMA DTR gel filtration cartridges (EdgeBioSystems, Gaithersburg, Md.) and sequenced on an ABI 377 sequencer (Applied Biosystems). The results were analyzed using Sequencing Analysis version 3.3. The contiguous sequences were constructed using SeqMan (DNAStar, Inc., Madison, Wis.) and identified using BLAST on the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). An unweighted pair group method with arithmetic mean (UPGMA)-based tree was created based on total ITS sequence identity. Average sequence similarity within ITS1 and ITS2 regions of each sample was tabulated using a reference sequence selected for each of the Aspergillus species (GenBank accession numbers AB008416, AF176662, and AF138290 for A. flavus, A. fumigatus, and A. terreus, respectively).
Study design.
The study was designed to (i) compare the traditional method of Aspergillus species identification by morphology to the automated rep-PCR typing method, (ii) determine the reproducibility of the automated rep-PCR system, and (iii) establish the discriminatory power of pattern formation among the isolates within each Aspergillus species (A. fumigatus, A. flavus, and A. terreus). We proceeded in four steps. In the first step, 27 morphologically characterized Aspergillus isolates (10 A. fumigatus, 9 A. flavus, and 8 A. terreus isolates) whose species identification was known were analyzed by rep-PCR and clustered into categories. The concordance between the rep-PCR-based identification and the morphological identification of Aspergillus species was then compared. The second step was to assess the reproducibility of the automated platform by testing the 27 samples as mentioned above in triplicate and then testing all 69 isolates from both the hyphal and conidial stages of growth. The third step was to demonstrate the utility of the DiversiLab software by building a database using the 27 known samples and assigning a species identification to the 42 blinded (previously characterized by morphology) Aspergillus samples (28 A. fumigatus, 6 A. flavus, and 8 A. terreus isolates). Finally, all 69 Aspergillus isolates were identified based on ITS1-5.8S-ITS2 region sequence analysis to validate identification based on morphological as well as DNA fingerprint analysis.
RESULTS
Overview of the DiversiLab system.
The DiversiLab system consists of (i) rep-PCR reagent kits, (ii) an Agilent 2100 bioanalyzer that separates the amplified fragments generated by rep-PCR on a microfluidic chip and detects each fragment based on the fluorescence intensity and migration time, and (iii) web-based DiversiLab software version 2.1.66. For the analysis, a proximity matrix is automatically calculated by the DiversiLab software using the Pearson correlation to calculate pair-wise similarities between all samples in the report. The dendrograms are generated from the proximity matrix using UPGMA to create a hierarchical view of the relationships among the samples, in which samples with high similarity are joined by shallow branches, as indicated by the percent similarity scale. Scatter plots are generated from the proximity matrix using multidimensional scaling, which traditionally represents relative distances while leaving axes undefined. This creates a spatial, nonhierarchical view of the relationships among the samples, in which samples close together on the plot have high similarity, as indicated by gridlines on the scatter plot. The report (e.g., Fig. 1C) generated by the system contains a dendrogram (e.g., Fig. 2A, below) and a scatter plot (e.g., Fig. 2B) for sample comparison. Reports can also include electropherograms (e.g., Fig. 1B), gel-like images (e.g., Fig. 1C), and selectable demographic fields to further aid interpretation of the data. DNA fingerprints of an unknown microbial species or strain can be then compared against the stored rep-PCR fingerprint library for identification and typing.
FIG. 2.
DiversiLab system-generated dendrogram and scatter plot of the 27 known Aspergillus isolates. The dendrogram (A) and scatter plot (B) show species clustering. A rep-PCR-based DNA fingerprinting library was simultaneously generated for comparison purposes. The horizontal bar at the bottom left of the dendrogram indicates the percent similarity coefficient within the species. The scatter plot shows a cluster of strains for each Aspergillus species, as indicated by the circles. Spacing between grid lines indicates increments of 5% similarity based on the proximity matrix results as represented using multidimensional scaling. All species assignments are based on morphological identification.
Creation of a rep-PCR-based DNA fingerprint library of Aspergillus species.
DNA was extracted from each of the 27 Aspergillus isolates that were identified as A. fumigatus (n = 10), A. flavus (n = 9), and A. terreus (n = 8) based on morphological characteristics. The rep-PCR-based DNA fingerprints of all 27 DNA samples were obtained using the DiversiLab system. The digitized images of the fingerprint profiles were stored in a database for further studies. Figure 2A shows a dendrogram containing gel-like images of all 27 known Aspergillus samples. The rep-PCR-based DNA fingerprint data of the different Aspergillus isolates were in concordance with the morphological identification, and all samples clustered into the three corresponding species categories. Since defining the criteria for strain-level identification is somewhat subjective (37) and rep-PCR fingerprinting has not traditionally been used for species identification, we used the following criteria: a ≤80% similarity indicated species discrimination, and 80 to 100% similarity indicated strain discrimination. Both the A. flavus and A. terreus species clusters showed further discrimination. All A. fumigatus isolates appeared to be the same strain both by the rep-PCR-based DNA fingerprint pattern visualized by the gel-like image and as indicated by a ≥98% similarity coefficient in the dendrogram. This was further evident in the tight clustering of all A. fumigatus strains on the scatter plot analysis (Fig. 2B).
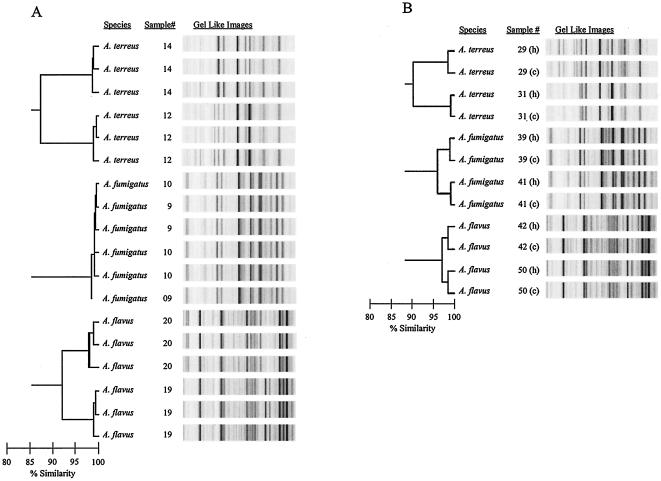
Reproducibility and stability of the rep-PCR DNA fingerprint patterns.
DNA from all 27 Aspergillus isolates was extracted three times and amplified using rep-PCR. Due to constraints regarding publication space and image resolution, only the DNA fingerprints of two isolates from each of the three Aspergillus species (isolates 14 and 12 of A. terreus, 10 and 9 of A. fumigatus, and 20 and 19 of A. flavus) are presented in Fig. 3A. The DNA fingerprint of each replicate of the rep-PCR was indistinguishable for a given isolate in the gel-like image as well as in terms of similarity coefficient (>98%) on the dendrogram. These results demonstrated that the rep-PCR profile of each of the 27 Aspergillus isolates in the library was reproducible with the DiversiLab system. In addition, the rep-PCR-based amplified products of the 27 Aspergillus isolates were processed using two different Agilent 2100 bioanalyzers. The rep-PCR profiles were found to be indistinguishable for a given isolate and, therefore, stable (data not shown).
FIG. 3.
Reproducibility and stability of the rep-PCR-based fingerprints of Aspergillus DNA. (A) DNA from each of 27 Aspergillus isolates was extracted and amplified in triplicate. The dendrogram shows rep-PCR profiles in triplicate for two strains of each of the three Aspergillus species. (B) DNA from hyphae and conidia of each of the 42 blinded Aspergillus isolates was extracted and amplified in duplicate. The dendrogram shows rep-PCR profiles of DNA from hyphae (h) and conidia (c) of two strains of each of the three Aspergillus species. The horizontal bar at the bottom left of each of the dendrograms indicates the percent similarity coefficient within the strains.
Forty-two blinded Aspergillus isolates from both hyphae and conidia were used to extract DNA and to carry out the rep-PCR. Figure 3B shows the rep-PCR profile from six of these Aspergillus isolates (numbers 29, 31, 39, 41, 42, and 50). The DNA fingerprint in terms of the similarity coefficient (>98%) was indistinguishable for both sources of DNA (conidia and hypha) for all isolates, indicating that the rep-PCR profile was consistent in both developmental phases of Aspergillus growth (conidia and hyphae). Additionally, the similarities of the rep-PCR fingerprint patterns were 100% concordant with species clustering.
Assignment of presumptive identification of Aspergillus species.
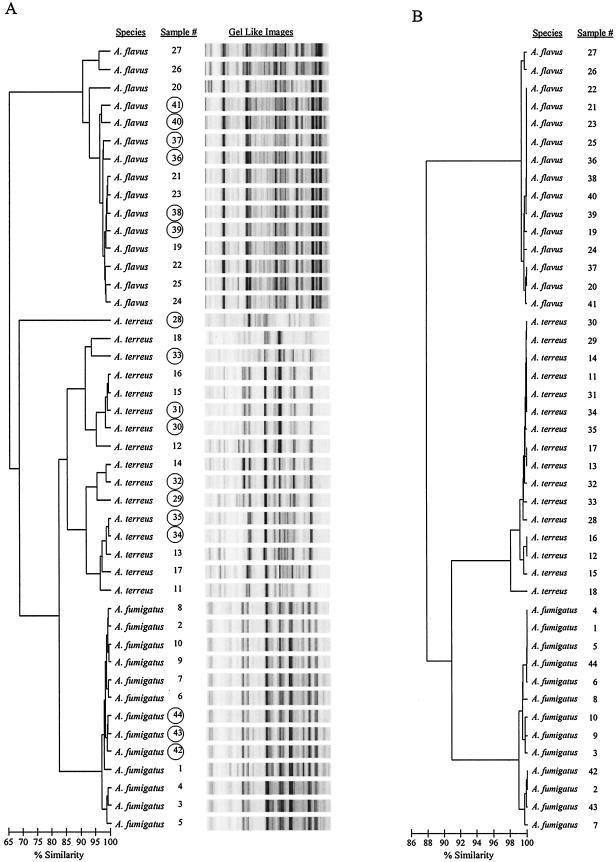
Forty-two blinded Aspergillus samples were analyzed using rep-PCR as described in Materials and Methods. Figure 4A shows rep-PCR profiles of 17 representative blinded Aspergillus isolates along with their presumptive identifications, generated automatically using the DiversiLab software and pair-wise groupings with the Aspergillus DNA fingerprint library previously created (Fig. 2A) from the 27 known isolates. The dendrogram in Fig. 4A shows a presumptive species identification of 42 Aspergillus samples in three major groups. When the 42 Aspergillus isolates were unblinded, the results showed 98% concordance with previous identifications based on morphological and cultural characteristics.
FIG. 4.
Comparison between rep-PCR-based DNA fingerprint and ITS1-5.8S-ITS2 region sequence analysis of 69 Aspergillus isolates. DNA from each of the 69 (27 known and 42 blinded) Aspergillus isolates was extracted, amplified by rep-PCR, and sequenced. (A) rep-PCR dendrogram shows the rep-PCR-based DNA profiles of 27 known and 17 of the 42 blinded (shown in circle) Aspergillus isolates. (B) ITS1-5.8S-ITS2 sequence-based phylogenetic tree of the same Aspergillus isolates. The horizontal bar at the bottom left of the dendrogram and the phylogenetic tree indicate the percent similarity coefficient within the strains.
Sequence analysis of the ITS1-5.8S-ITS2 region.
The ITS regions are located between the 18S and 28S rRNA genes. The rRNA gene for 5.8S RNA separates the two ITS regions, and the sequence variation of ITS regions has led to their use in phylogenetic studies of many organisms (17, 38). Sequencing of the ITS1-5.8S-ITS2 region from each of the 69 Aspergillus isolates was performed as described in Materials and Methods. Identification of the samples was based on the ITS1-5.8S-ITS2 region sequence variations, using the highest percentage match of these sequences through BLAST searches of the GenBank database. A UPGMA tree was created based on the total ITS sequence identity (Fig. 4B). Alignment of a contiguous sequence of the ITS1-5.8S-ITS2 region of each Aspergillus isolate showed both single nucleotide differences and short lengths of sequence variation (data not shown). The average sequence similarity within the ITS1 and ITS2 regions was tabulated using an appropriate reference sequence for each of the Aspergillus species (Table 1). We observed that the rep-PCR-based identification of each Aspergillus isolate was in 100% agreement with the identification based on the ITS region sequence analysis of the respective isolates. The sequence-based phylogenetic tree analysis (Fig. 4B) was concordant with the dendrogram results in Fig. 4A. Thus, identification of each Aspergillus isolate according to its rep-PCR profile was validated by the ITS region sequence analysis (Table 1). One sample (isolate 28) was not easily classified into a specific group; however, the nearest neighbor group was A. terreus (Fig. 4A). Upon further investigation of the sample by ITS1-5.8S-ITS2 region sequence analysis, the isolate was determined to be A. terreus (Fig. 4B).
TABLE 1.
Sequence similarities and variations within ITS1 and ITS2 regions
| Species of samples | Avg sequence similarity with reference straina
|
|||||
|---|---|---|---|---|---|---|
|
A. flavus
|
A. fumigatus
|
A. terreus
|
||||
| ITS1 | ITS2 | ITS1 | ITS2 | ITS1 | ITS2 | |
| A. flavus | 0.995 | 0.999 | 0.794 | 0.839 | 0.815 | 0.851 |
| A. fumigatus | 0.796 | 0.856 | 0.990 | 0.988 | 0.847 | 0.903 |
| A. terreus | 0.811 | 0.852 | 0.849 | 0.872 | 0.993 | 0.997 |
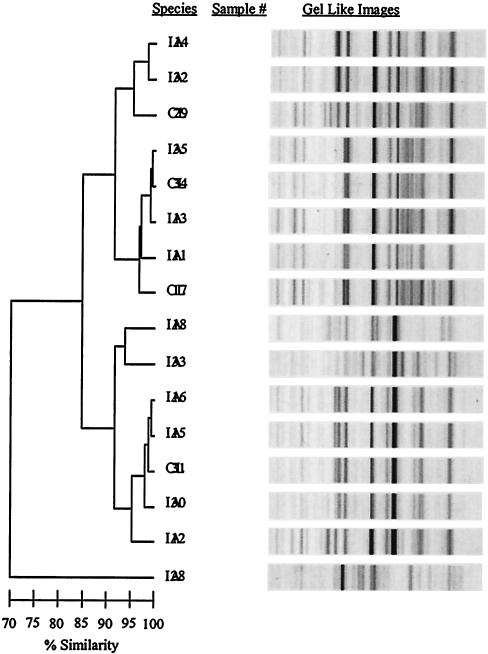
DiversiLab system-based strain discrimination of Aspergillus species.
The 69 Aspergillus clinical isolates were either colonizing strains (CL; n = 20) or strains associated with IA (proven or probable, n = 49) based on the National Institute of Allergy and Infectious Diseases Mycoses Study Group-European Oncology Research Cooperative criteria (1). Clonality was not observed within the CL strains or the IA strains for any of the three Aspergillus species. Figure 5 shows the data from all strains of A. terreus (n = 16), which included CL strains (n = 4) and strains involved in IA (n = 12). Although clonality was not seen within the CL strains or the strains causing IA, strain-level differentiation was evident in both the A. flavus (Fig. 2A and 4A) and A. terreus (Fig. 2A, 4A, and 5) strains.
FIG. 5.
rep-PCR-based strain discrimination of A. terreus. DNA from each of 16 strains of A. terreus was extracted and amplified via rep-PCR. The horizontal bar at the bottom left of the dendrogram indicates the percent similarity coefficient within the strains. CL, colonizing strain; IA, invasive strains.
DISCUSSION
IA represents a formidable challenge in institutions that care for a large number of highly immunosuppressed patients (23). In view of the lack of effective antifungal prophylaxis and the inadequate activity of modern antifungal drugs, every effort should be made to prevent infections (23). Although the medical and economic benefits of a highly integrated, comprehensive infection control program that includes routine determination of clonality of Aspergillus species causing IA have not been proven, there is emerging evidence that such an approach might be cost-effective for other nosocomial pathogens (18).
Despite the growing number of molecular typing methods, currently there is no simple method that is optimal for species and strain identification of Aspergillus isolates. New technologies are being developed for the DNA fingerprinting of fungal strains, and they have recently been reviewed (15). Among them, sequence-based technology, such as multilocus sequence typing (5, 30), and hybridization-based diversity array technology (20) may provide rapid and accurate genotyping of fungi. Management and analysis of high-throughput data in microarray hybridization may not be suitable for low-volume laboratories. The costs associated with automated sequencing in the multilocus sequence typing approach (16, 32), in particular the start-up costs, which can be more than three times the cost of the DiversiLab system, may keep sequencing technologies out of the reach of most clinical laboratories.
rep-PCR was initially developed for the subspecies and strain differentiation of important bacterial nosocomial pathogens, such as Staphylococcus aureus (11), Streptococcus pneumoniae (41), and enteric gram-negative rods (40). This approach takes advantage of the fact that there are repetitive elements interspersed throughout the bacterial genome that, when amplified by PCR, give highly discriminatory profiles within bacterial species (10, 42, 43) and some eukaryotic organisms, such as yeast (26, 27). We hypothesized that this might hold true for the pathogenic molds, which are also known to harbor repetitive elements (35).
Traditionally, rep-PCR products are separated by agarose gel electrophoresis followed by gel staining and interpretation. Though rep-PCR is the most cost-effective typing method and has considerably better discriminatory power than most methods (32), one study indicated that manual rep-PCR gives inconsistent interlaboratory results and may be cumbersome in clinical laboratories (13). Here, we have shown that automated rep-PCR offers several advantages in molecular typing of fungal species. The DiversiLab Aspergillus kit was easy to use, and the automated detection and analysis by the DiversiLab system provided readily interpretable reports. The efficiency, high discriminatory capability, and reproducibility of the rep-PCR method enables database building and intralaboratory comparison, which in turn makes the method optimal for high-throughput applications. Specifically, the system uses a microfluidics platform for DNA fragment separation to achieve increased reproducibility and throughput and decreased labor costs. The small footprint of the automated system allows for portability, while the web-based interface enables remote access and data sharing for assessment of population profiles among medical institutions. This enables standardized comparisons of strains isolated in different laboratories. DiversiLab system-based results were generated in less than 4 h from isolate to identification in these studies. In view of the declining hours required for laboratory mycology education (31), additional cost savings could exist both in labor for sample processing and interpretation (less skilled) and reagents (quality-controlled kits) by developing a rep-PCR-based fungal identification system.
We observed that the ITS1 region showed more interspecies variation than the ITS2 region (Table 1). This supports the earlier findings by Henry et al. (19). Though ITS sequence-based identification of Aspergillus species was accurate, the time, cost, and labor in carrying out such an analysis were much higher than those required for implementing the rep-PCR-based analysis (i.e., 1.5 days versus 4 h, respectively). We demonstrated that the rep-PCR-based presumptive identifications of Aspergillus species were in agreement with those obtained from the ITS region sequence analysis (Fig. 4). In addition, rep-PCR produced a high level of strain discrimination among A. terreus and A. flavus, as both species clustered further into subspecies (Fig. 4A). The sequence-based phylogenetic tree of Aspergillus species was derived solely from the variations in the ITS region sequences (Table 1), possibly only a few base pair differences. This may explain the placement of isolate 28, relatively indistinguishable, in the A. terreus cluster of the sequence-based phylogenetic tree (Fig. 4B). Alternatively, the rep-PCR DNA profiles represent a molecular pattern surveying the entire genomic DNA of a given Aspergillus isolate, providing a larger template in which to detect genomic variations, which is demonstrated by isolate 28 in the rep-PCR dendrogram (Fig. 4A). This demonstrates the potential of the DiversiLab system in strain differentiation of Aspergillus species.
We additionally demonstrated that rep-PCR is useful in strain differentiation of A. flavus (Fig. 2A and 4A) and A. terreus (Fig. 2A, 4A, and 5). Our results clearly demonstrate the opportunistic nature of aspergilli, since a large number of isolates from diverse genetic backgrounds were capable of causing IA and little or no clonality among the invasive or the CL Aspergillus strains was seen. Therefore, preventive methods, especially environmental containment of molds, are important (23, 36). Further optimization of the rep-PCR protocol would enhance the discrimination between the A. fumigatus strains, which are responsible for the majority of IA cases (12, 28), similar to results obtained with modifications to rep-PCR protocols for bacteria (21).
The weaknesses of our study were the limited number of tested strains that came from a single geographical location, the lack of standard sampling of isolates, and the lack of environmental isolates. Another issue is the lack of consensus on the acceptable levels of discriminatory power and what defines genetic relatedness or unrelatedness at the species level. Therefore, one cannot easily define the threshold of sequence hypervariability that discriminates moderately related pathogens. Building larger databases and careful prospective multi-institutional infection control studies should help in addressing these issues.
In conclusion, our study demonstrates the potential utility of automated rep-PCR and the DiversiLab system in identifying Aspergillus isolates at the species and strain levels. The rep-PCR was highly reproducible. Further studies with larger sample sets of expanded geographical representation are needed to validate our preliminary findings. Such studies should compare and/or combine rep-PCR with other genotypic methods, in both epidemic and nonepidemic situations, not only in terms of facility, reproducibility, and discriminatory power but also in terms of cost and turnaround time. Finally, the performance of automated rep-PCR for other emerging and medically important opportunistic molds, such as the Fusarium species and the Zygomycetes, should be explored.
REFERENCES
- 1.Ascioglu, S., J. H. Rex, B. de Pauw, J. E. Bennett, J. Bille, F. Crokaert, D. W. Denning, J. P. Donnelly, J. E. Edwards, Z. Erjavec, D. Fiere, O. Lortholary, J. Maertens, J. F. Meis, T. F. Patterson, J. Ritter, D. Selleslag, P. M. Shah, D. A. Stevens, and T. J. Walsh. 2002. Defining opportunistic invasive fungal infections in immunocompromised patients with cancer and hematopoietic stem cell transplants: an international consensus. Clin. Infect. Dis. 34:7-14. [DOI] [PubMed] [Google Scholar]
- 2.Bart-Delabesse, E., C. Cordonnier, and S. Bretagne. 1999. Usefulness of genotyping with microsatellite markers to investigate hospital-acquired invasive aspergillosis. J. Hosp. Infect. 42:321-327. [DOI] [PubMed] [Google Scholar]
- 3.Bart-Delabesse, E., J. Sarfati, J. P. Debeaupuis, W. van Leeuwen, A. van Belkum, S. Bretagne, and J. P. Latge. 2001. Comparison of restriction fragment length polymorphism, microsatellite length polymorphism, and random amplification of polymorphic DNA analyses for fingerprinting Aspergillus fumigatus isolates. J. Clin. Microbiol. 39:2683-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertout, S., F. Renaud, T. De Meeus, M. A. Piens, B. Lebeau, M. A. Viviani, M. Mallie, J. M. Bastide, et al. 2000. Multilocus enzyme electrophoresis analysis of Aspergillus fumigatus strains isolated from the first clinical sample from patients with invasive aspergillosis. J. Med. Microbiol. 49:375-381. [DOI] [PubMed] [Google Scholar]
- 5.Bougnoux, M. E., S. Morand, and C. d'Enfert. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J. Clin. Microbiol. 40:1290-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buffington, J., R. Reporter, B. A. Lasker, M. M. McNeil, J. M. Lanson, L. A. Ross, L. Mascola, and W. R. Jarvis. 1994. Investigation of an epidemic of invasive aspergillosis: utility of molecular typing with the use of random amplified polymorphic DNA probes. Pediatr. Infect. Dis. J. 13:386-393. [PubMed] [Google Scholar]
- 7.Chazalet, V., J. P. Debeaupuis, J. Sarfati, J. Lortholary, P. Ribaud, P. Shah, M. Cornet, H. Vu Thien, E. Gluckman, G. Brucker, and J. P. Latge. 1998. Molecular typing of environmental and patient isolates of Aspergillus fumigatus from various hospital settings. J. Clin. Microbiol. 36:1494-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deak, T., J. Chen, and L. R. Beuchat. 2000. Molecular characterization of Yarrowia lipolytica and Candida zeylanoides isolated from poultry. Appl. Environ. Microbiol. 66:4340-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Debeaupuis, J. P., J. Sarfati, V. Chazalet, and J. P. Latge. 1997. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect. Immun. 65:3080-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bruijn, F. J., J. R. Lupski, and G. M. Weinstock. 1998. Bacterial genomes: physical structure and analysis. Chapman & Hall, New York, N.Y.
- 11.Del Vecchio, V. G., J. M. Petroziello, M. J. Gress, F. K. McCleskey, G. P. Melcher, H. K. Crouch, and J. R. Lupski. 1995. Molecular genotyping of methicillin-resistant Staphylococcus aureus via fluorophore-enhanced repetitive-sequence PCR. J. Clin. Microbiol. 33:2141-2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denning, D. W. 1998. Invasive aspergillosis. Clin. Infect. Dis. 26:781-803. [DOI] [PubMed] [Google Scholar]
- 13.Deplano, A., A. Schuermans, J. Van Eldere, W. Witte, H. Meugnier, J. Etienne, H. Grundmann, D. Jonas, G. T. Noordhoek, J. Dijkstra, A. van Belkum, W. van Leeuwen, P. T. Tassios, N. J. Legakis, A. van der Zee, A. Bergmans, D. S. Blanc, F. C. Tenover, B. C. Cookson, G. O'Neil, M. J. Struelens, and The European Study Group on Epidemiological Markers of the ESCMID. 2000. Multicenter evaluation of epidemiological typing of methicillin-resistant Staphylococcus aureus strains by repetitive-element PCR analysis. J. Clin. Microbiol. 38:3527-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Einsele, H., H. Hebart, G. Roller, J. Loffler, I. Rothenhofer, C. A. Muller, R. A. Bowden, J. van Burik, D. Engelhard, L. Kanz, and U. Schumacher. 1997. Detection and identification of fungal pathogens in blood by using molecular probes. J. Clin. Microbiol. 35:1353-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gil-Lamaignere, C., E. Roilides, J. Hacker, and F. M. Muller. 2003. Molecular typing for fungi—a critical review of the possibilities and limitations of currently and future methods. Clin. Microbiol. Infect. 9:172-185. [DOI] [PubMed] [Google Scholar]
- 16.Goulding, J. N., J. V. Hookey, J. Stanley, W. Olver, K. R. Neal, D. A. Ala'Aldeen, and C. Arnold. 2000. Fluorescent amplified-fragment length polymorphism genotyping of Neisseria meningitidis identifies clones associated with invasive disease. J. Clin. Microbiol. 38:4580-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guarro, J., J. Gene, and A. M. Stchigel. 1999. Developments in fungal taxonomy. Clin. Microbiol. Rev. 12:454-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hacek, D. M., T. Suriano, G. A. Noskin, J. Kruszynski, B. Reisberg, and L. R. Peterson. 1999. Medical and economic benefit of a comprehensive infection control program that includes routine determination of microbial clonality. Am. J. Clin. Pathol. 111:647-654. [DOI] [PubMed] [Google Scholar]
- 19.Henry, T., P. C. Iwen, and S. H. Hinrichs. 2000. Identification of Aspergillus species using internal transcribed spacer regions 1 and 2. J. Clin. Microbiol. 38:1510-1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaccoud, D., K. Peng, D. Feinstein, and A. Kilian. 2001. Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res. 29:E25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, J. R., and C. Clabots. 2000. Improved repetitive-element PCR fingerprinting of Salmonella enterica with the use of extremely elevated annealing temperatures. Clin. Diagn. Lab. Immunol. 7:258-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klinch, M. A. 2002. Identification of common Aspergillus species. Cenraalburaue voor Schimmelcultures, Utrecht, The Netherlands.
- 23.Kontoyiannis, D. P., and G. P. Bodey. 2002. Invasive aspergillosis in 2002: an update. Eur. J. Clin. Microbiol. Infect. Dis. 21:161-172. [DOI] [PubMed] [Google Scholar]
- 24.Larone, D. H. 1995. Medically important fungi: a guide to identification, 3rd ed. ASM Press, Washington, D.C.
- 25.Lasker, B. A. 2002. Evaluation of performance of four genotypic methods for studying the genetic epidemiology of Aspergillus fumigatus isolates. J. Clin. Microbiol. 40:2886-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lasker, B. A., L. S. Page, T. J. Lott, and G. S. Kobayashi. 1992. Isolation, characterization, and sequencing of Candida albicans repetitive element 2. Gene 116:51-57. [DOI] [PubMed] [Google Scholar]
- 27.Lasker, B. A., L. S. Page, T. J. Lott, G. S. Kobayashi, and G. Medoff. 1991. Characterization of CARE-1: Candida albicans repetitive element-1. Gene 102:45-50. [DOI] [PubMed] [Google Scholar]
- 28.Latge, J. P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leenders, A., A. van Belkum, S. Janssen, S. de Marie, J. Kluytmans, J. Wielenga, B. Lowenberg, and H. Verbrugh. 1996. Molecular epidemiology of apparent outbreak of invasive aspergillosis in a hematology ward. J. Clin. Microbiol. 34:345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Negroni, R., D. Ellis, G. Bulmer, J. R. Graybill, and A. Restrepo. 1998. Teaching medical mycology in the year 2000. Med. Mycol. 36(Suppl. 1):106-108. [PubMed] [Google Scholar]
- 32.Olive, D. M., and P. Bean. 1999. Principles and applications of methods for DNA-based typing of microbial organisms. J. Clin. Microbiol. 37:1661-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radford, S. A., E. M. Johnson, J. P. Leeming, M. R. Millar, J. M. Cornish, A. B. Foot, and D. W. Warnock. 1998. Molecular epidemiological study of Aspergillus fumigatus in a bone marrow transplantation unit by PCR amplification of ribosomal intergenic spacer sequences. J. Clin. Microbiol. 36:1294-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Redkar, R. J., M. P. Dube, F. K. McCleskey, M. G. Rinaldi, and V. G. Del Vecchio. 1996. DNA fingerprinting of Candida rugosa via repetitive sequence-based PCR. J. Clin. Microbiol. 34:1677-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sampson, R. A. 1994. Current systematics of the genus Aspergillus, p. 261-267. In K. A. Powell, A. Renwick, and J. F. Peberdy (ed.), The genus Aspergillus: from taxonomy and genetics to industrial application. FEMS symposium no. 69. Plenum Press, New York, N.Y.
- 36.Sehulster, L., and R. Y. Chinn. 2003. Guidelines for environmental infection control in health-care facilities. Recommendations of CDC and the Healthcare Infection Control Practices Advisory Committee (HICPAC). Morb. Mortal. Wkly. Rep. Recomm. Rep. 52:1-42. [PubMed] [Google Scholar]
- 37.Soll, D. R. 2000. The ins and outs of DNA fingerprinting the infectious fungi. Clin. Microbiol. Rev. 13:332-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turenne, C. Y., S. E. Sanche, D. J. Hoban, J. A. Karlowsky, and A. M. Kabani. 1999. Rapid identification of fungi by using the ITS2 genetic region and an automated fluorescent capillary electrophoresis system. J. Clin. Microbiol. 37:1846-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Burik, J. A., D. Myerson, R. W. Schreckhise, and R. A. Bowden. 1998. Panfungal PCR assay for detection of fungal infection in human blood specimens. J. Clin. Microbiol. 36:1169-1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Versalovic, J., V. Kapur, T. Koeuth, G. H. Mazurek, T. S. Whittam, J. M. Musser, and J. R. Lupski. 1995. DNA fingerprinting of pathogenic bacteria by fluorophore-enhanced repetitive sequence-based polymerase chain reaction. Arch. Pathol. Lab. Med. 119:23-29. [PubMed] [Google Scholar]
- 41.Versalovic, J., V. Kapur, E. O. Mason, Jr., U. Shah, T. Koeuth, J. R. Lupski, and J. M. Musser. 1993. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J. Infect. Dis. 167:850-856. [DOI] [PubMed] [Google Scholar]
- 42.Versalovic, J., T. Koeuth, and J. R. Lupski. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823-6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Versalovic, J., and J. R. Lupski. 2002. Molecular detection and genotyping of pathogens: more accurate and rapid answers. Trends Microbiol. 10:S15-S21. [DOI] [PubMed] [Google Scholar]
- 44.White, T., T. Burns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, Calif.