Abstract
To facilitate the diagnosis of enterotoxigenic Escherichia coli (ETEC) infections in humans, we developed and evaluated real-time fluorescence PCR assays for the Roche LightCycler (LC) against the enterotoxin genes commonly present in strains associated with human illness. Separate LC-PCR assays with identical cycling conditions were designed for the type I heat-labile enterotoxin (LT I) and the type I heat-stable enterotoxin (ST I) genes, using the LC hybridization probe format. A duplex assay for ST I with two sets of amplification primers and three hybridization probes was required to detect the major nucleotide sequence variants of ST I, ST Ia and ST Ib. LC-PCR findings from the testing of 161 E. coli isolates of human origin (138 ETEC and 23 non-ETEC) were compared with those obtained by block cycler PCR analysis. The sensitivities and specificities of the LC-PCR assays were each 100% for the LT I and ST I genes. The LC-PCR and block cycler PCR assays were also compared for their abilities to detect LT I and ST I genes in spiked stool specimens with different methods of sample preparation. Findings from these experiments revealed that the limits of detection for the LC-PCR assays were the same or substantially lower than those observed for the block cycler PCR assay. Melting curve analysis of the amplified LT I and ST I genes revealed sequence variation within each gene, which for the ST I genes correlated with the presence of ST Ia and ST Ib. The rapidity, sensitivity, and specificity of the LC-PCR assays make them attractive alternatives to block cycler PCR assays for the detection and characterization of ETEC.
Enterotoxigenic Escherichia coli (ETEC) is a major cause of sporadic diarrheal disease in humans, affecting mainly children in developing countries (1, 3, 21) and travelers from industrialized countries visiting tropical or subtropical areas (9). ETEC strains have also caused waterborne outbreaks on cruise ships (6), food-borne outbreaks at schools (16) and restaurants (5, 11), and outbreaks among persons serving in the military (12, 20, 27). ETEC strains from humans cause mild or severe watery diarrhea by producing a heat-labile enterotoxin (LT I) (similar in structure to cholera toxin), heat-stable enterotoxins (ST Ia and/or ST Ib), or both (17).
Initially, detection of ETEC strains was accomplished with animal assays and cell culture techniques that required specific antibodies to reliably demonstrate the presence of the target enterotoxins. Later, enzyme-linked immunosorbent assays and membrane-based DNA hybridization assays, which improved speed and ease of use, became popular. Further enhancements in methods for the detection of ETEC gave rise to PCR assays (18, 24, 26, 28). These assays were attractive alternatives to the earlier methods because of their sensitivity and specificity, as well as their speed and use of readily available reagents. Recent advances in PCR amplification and fluorescence-based detection technologies have brought significant improvements to conventional block cycler PCR assays, thereby enabling the simultaneous, sequence-specific detection of multiple products in real time. The development of thermal cyclers with the ability to rapidly amplify genes and the capacity to continuously monitor the accumulation of specific PCR products labeled with fluorescent probes obviates the need for labor-intensive agarose gel procedures and provides a real-time, reliable identification of target genes (2, 7, 14, 15, 22, 23).
In the present study, we describe the development and evaluation of a set of real-time PCR assays for the LightCycler (LC) (Roche Diagnostics, Mannheim, Germany), using primers and hybridization probes that target the LT I, ST Ia (also referred to as STp), and ST Ib (also referred to as STh) genes. We used the hybridization probe format for these assays because it permitted a melting curve analysis to be performed after the amplification phase to achieve additional characterization of the target genes. The availability of rapid and specific assays for the detection and characterization of ETEC should facilitate the detection and control of outbreaks caused by ETEC.
MATERIALS AND METHODS
Bacterial strains.
The E. coli strains included in this study were selected from the culture collections of our institutions and are described in Table 1. We studied 161 E. coli isolates, consisting of 138 ETEC and 23 non-ETEC strains, and 168 strains from the following bacterial genera (the number of strains is in parentheses): Treponema (1), Borrelia (3), Leptospira (1), Helicobacter (2), Legionella (5), Neisseria (2), Branhamella (1), Xanthomonas (1), Moraxella (2), Kingella (1), Acinetobacter (1), Sphingomonas (1), Pseudomonas (4), Brucella (2), Bordetella (4), Shigella (4, including Shigella dysenteriae type 1), Salmonella (2), Citrobacter (2), Klebisella (2), Enterobacter (2), Serratia (2), Proteus (2), Morganella (1), Providencia (1), Yersinia (2), Vibrio (51, including 50 cholera toxin-positive V. cholerae O1 and O139 strains), Aeromonas (1), Pasteurella (1), Haemophilus (3), Actinobacillus (1), Bacteroides (1), Campylobacter (3), Fusobacterium (1), Eikenella (1), Prevotella (1), Porphyromonas (1), Rickettsia (1), Ehrlichia (1), Coxiella (1), Bartonella (2), Chlamydia (3), Mycoplasma (2), Ureaplasma (1), Micrococcus (1), Staphylococcus (5), Streptococcus (5), Leuconostoc (1), Pediococcus (1), Peptostreptococcus (3), Bacillus (2), Clostridium (3), Lactobacillus (2), Listeria (1), Corynebacterium (5), Gardnerella (1), Propionibacterium (1), Actinomyces (2), Mycobacterium (4), Nocardia (2), Tsukamorella (1), and Rhodococcus (1). The ETEC strains were isolated from outbreaks investigated by the Foodborne and Diarrheal Diseases Branch, Centers for Disease Control and Prevention, Atlanta, Ga., during the period 1975 to 2002. The O:H serotypes and presence of the genes for LT I, ST Ia, and ST Ib for the E. coli isolates are summarized in Table 1.
TABLE 1.
Characteristics of 138 ETEC and 23 non-ETEC strains tested by block cycler PCR and LC-PCR assays
| Genotypea
|
No. of strains | Serotypeb | ||
|---|---|---|---|---|
| LT I | ST Ia | ST Ib | ||
| + | − | − | 27 | O6:31 (1), O8:H9 (1), O15:NM (1), O25:NM (2), O60:H9 (1), O63:NM (1), O64:NM (4), O78:H12 (1), O127:H8 (1), O152:H2 (1), O159:H4/21 (3), O162:H28 (1), O166:H15 (1), O167:H5/Hund (2), Ound:H7/H10/H15/H32/NM (5), Orou:H28 (1) |
| − | + | − | 33 | O27:H7/H20/Hund (14), O99:NM (1), O159:NM (2), O169:H41/NM (14), Orou:H21 (1), Ound:NM (1) |
| − | − | + | 31 | O6:H10 (1), O25:NM (15), O25:H42 (1), O27:H20 (1), O34:H10 (1), O49:NM (1), O63:H12 (1), O78:H12 (2), O128:H7/H21/H27 (4), O135:H18 (1), O136:H12 (1), O148:H28 (1), O153:H45 (1) |
| + | + | − | 12 | O17:NM (1), O24:NM (1), O64:Hund (1), O78:H11 (2), O99:H33 (1), O148:H28 (4), Ound:H10/NM (2), |
| + | − | + | 32 | O6:H16 (18), O8:H9 (1), O75:Hund (1), O78:H12/Hund (4), O148:H28 (5), O167:H5 (1), Orou:Hund (1), Ound:Hund (1) |
| + | + | + | 3 | O78:H11 (1), O148:H28 (2) |
| − | − | − | 23 | O2:H4 (1), O103:H2 (1), O104:H28 (2), O111:NM (1), O123:H21 (1), O149:H7 (1), O154:H19 (1), O157:H7 (8), Ound:Hund (2), Orou:H4/H7/H10/Hund/NM (6) |
The presence of the LT I, ST Ia, and ST Ib genes was determined by the block cycler PCR assays of Ølsvik and Strockbine (19) and Schultsz et al. (24).
Ound and Hund, O and H antigens, respectively, not determined with available typing sera; Orou, O antigen not determined because of incomplete or rough lipopolysaccharide; NM, nonmotile. Numbers in parentheses indicate the number of strains with that serotype.
Positive control strains included E. coli strains H10407 (serotype O78:H11), containing the genes for LT I, ST Ia, and ST Ib; E2539 (serotype O25:NM), containing the gene for LT I; TX1 (serotype O78:H12), containing the gene for ST Ib; and B4106 (serotype O27:H7), containing the gene for ST Ia. Negative control strains included the following diarrheagenic E. coli strains that lack the LT and ST enterotoxin genes: Shiga toxin-producing E. coli ATCC 43895 (serotype O157:H7), enteropathogenic E. coli B170 (serotype O111:NM), enteroinvasive E. coli EDL1284 (serotype O124:NM), and enteroaggregative E. coli 3591-78 (serotype O75:NM). Bacterial isolates were stored frozen at −70°C in tryptic soy broth containing 10% glycerol and were subcultured prior to testing on plates of either MacConkey agar (Merck, Darmstadt, Germany) or tryptic soy agar containing 5% sheep blood.
Serologic characterization.
Determination of O:H serotypes was performed by standard methods, with the exception that the O serology was performed with microtiter plates instead of tubes (8).
Preparation of template DNA.
Template DNA from pure bacterial cultures was prepared as previously described (23). Briefly, a 1-cm-long sweep of bacterial growth from the quadrant of a blood or MacConkey agar plate with the heaviest growth was suspended in 300 μl of distilled water or 300 μl of lysis buffer composed of 1% Triton X-100, 0.5% Tween 20, 10 mM Tris-HCl (pH 8.0), and 1 mM EDTA and heated at 99.9°C for 10 min. In the case of overnight enrichment broths, 500 μl of broth was used for template preparation. After boiling, the lysate was then centrifuged at 1,000 × g for 2 min to sediment the debris. Aliquots of the clear supernatant (2 to 5 μl) were used as the source of template DNA for the block cycler PCR and LC-PCR assays.
Template DNA from the spiked stool specimens was extracted directly from sampling swabs by using the MagnaPure LC automated DNA extraction system (Roche Diagnostics) with the total nucleic acid isolation kit (Roche Diagnostics). Before processing in the automated DNA extraction system, the stool swabs were immersed in 500 μl of physiological saline and heated at 95°C for 10 min to inactivate infectious material. For outbreak-associated stool specimens, which were submitted as 50% (wt/vol) suspensions of stool in Cary-Blair transport medium, the stool suspensions were diluted 1:3 with 0.01 M phosphate-buffered saline and heated at 95°C for 10 min. A 200-μl aliquot of the heated mixture was then used as the sample for the automated extraction of template DNA. Aliquots of the extracted DNA were used either directly for template DNA in the LC-PCR assay or after a second heating at 99.9°C for 10 min in the block cycler PCR assay.
Block cycler PCR assays.
Detection and subtyping of LT I and ST I genes were accomplished by using the block cycler PCR methods described by Ølsvik and Strockbine (19) and Schultsz et al. (24). The method of Ølsvik and Strockbine was used initially to test all strains for the presence of LT I and ST I genes. It expanded the assay described by Olive (18) for LT I by adding a set of primers for ST I. In this duplex assay, a 322-bp fragment for LT I was amplified with primers LT1 (5′-TCT CTA TGT GCA TAC GGA GC-3′) and LT2 (5′-CCA TAC TGA TTG CCG CAA T-3′), and a 147-bp fragment for ST I was amplified with primers ST1 (5′-TTA ATA GCA CCC GGT ACA AGC AGG-3′) and ST2 (5′-CTT GAC TCT TCA AAA GAG AAA ATT AC-3′). A 5 μl-aliquot of supernatant was used unpurified as template DNA in a 50-μl reaction mixture consisting of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.5 mM MgCl2, 0.01% gelatin, 200 μM deoxynucleoside triphosphates, 2 μl of LT primer mix (50 μM each), 2 μl of ST primer mix (100 μM each), and 1.25 U of Taq DNA polymerase. Amplification conditions were as follows: an initial denaturation step at 94°C for 30 s; 35 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 1 min, and extension at 72°C for 1 min; and a final extension step at 72°C for 10 min.
The 161 E. coli strains (ETEC and non-ETEC) and any stool isolates that were positive in the initial PCR assay were also tested in the PCR assay of Schultsz et al. (24), which included three sets of primers for the detection of LT I (696-bp amplification product), ST Ia (186-bp amplification product), and ST Ib (166-bp amplification product), to confirm their enterotoxin genotype and to subtype the ST I genes. A control containing water instead of template was included in each experiment to exclude the possibility of reagent contamination. Amplified fragments were separated by agarose gel electrophoresis and visualized after ethidium bromide staining with UV illumination.
Oligonucleotide primers and fluorescence-labeled probes for the LC-PCR assays.
DNA oligonucleotide primers and hybridization probes were synthesized by TIB Molbiol, Berlin, Germany, and by the Biotechnology Core Facility, Centers for Disease Control and Prevention, Atlanta, Ga. Hybridization probes were labeled with fluorescein, LC Red 640, or LC Red 705. The nucleotide sequences of the primers and hybridization probes and their corresponding locations within the designated GenBank sequences are shown in Table 2. Oligonucleotide sequences for the hybridization probes were selected by using Oligo version 6.0 (Molecular Biology Insights, Inc., Cascade, Colo.) to have comparable melting temperatures (Tms), G+C contents within the range of 35 to 60 mol%, and minimal secondary structure due to self-complementarity or palindromic sequences.
TABLE 2.
Oligonucleotide primers and LC hybridization probes used in the PCR assay
| Oligonucleotide | Sequencea | Target gene | Nucleotide position | GenBank accession no. | Reference |
|---|---|---|---|---|---|
| LTI-1 | GCG TTA CTA TCC TCT CTA TGT G | LT I | 874-895 | S60731 | Klaus Pfeffer, personal communication |
| LTI-2 | AGT TTT CCA TAC TGA TTG CCG C | LT I | 1213-1192 | S60731 | |
| LTI-HP-1 | AGG ATG AAG GAC ACA TTA AGA ATC ACA T-[FL] | LT I | 1102-1129 | S60731 | Present study |
| LTI-HP-2 | [Red 640]-TCT GAC CGA GAC CAA AAT TGA TAA ATT-[Ph] | LT I | 1131-1157 | S60731 | Present study |
| STIa-1 | CTG TAT TAT CTT TCC CCT CT | ST Ia | 317-336 | M25607 | Present study |
| STIa-2 | TTT AAT AAC ATG GAG CAC AG | ST Ia | 505-486 | M25607 | Present study |
| STIb-1 | CTG TAT TGT CTT TTT CAC CT | ST Ib | 79-98 | M34916 | Present study |
| STIb-2 | ATT AAT AGC ACC CGG TAC AA | ST Ib | 267-248 | M34916 | Present study |
| STI-HP-1a | AAA TCA GAA AAT ATG AAC AAC ACA T-[FL] | ST Ia | 430-454 | M25607 | Present study |
| STI-HP-1b | GTC CTG AAA GCA TGA ATA GTA GCA AT-[FL] | ST Ib | 193-218 | M34916 | Present study |
| STI-HP-2 | [Red 705]-ACT GCT GTG AAT TGT GTT GTA ATC CT-[Ph] | ST Ia | 458-482 | M25607 | Present study |
| ST Ib | 220-245 | M34916 |
[FL], fluorescein; [Red 640], LC-Red 640-N-hydroxy-succinimide ester; [Red 705], LC-Red 705-phosphoramidite; [Ph], 3′- phosphate.
LC-PCR assay and product detection.
All LC-PCR assays were performed with a fluorescence temperature cycler (LC; Roche Diagnostics). The LT amplification mixture consisted of 2 μl of 10× reaction mix (LC-FastStart master hybridization probes; Roche Diagnostics), 3 mM MgCl2, a 0.5 μM concentration of each oligonucleotide primer, a 0.2 μM concentration of each oligonucleotide probe, and 2 μl of template DNA in a final volume of 20 μl. The ST amplification mixture consisted of 2 μl of 10× reaction mix (LC-FastStart master hybridization probes; Roche Diagnostics), 3 mM MgCl2, 0.5 μM ST Ia and 0.125 μM ST Ih oligonucleotide primers, a 0.2 μM concentration of each oligonucleotide probe, and 2 μl of template DNA in a final volume of 20 μl. Samples were amplified as follows: an initial denaturation step at 95°C for 10 min to activate the FastStart Taq DNA polymerase, 40 cycles of denaturation at 95°C for 10 s and annealing at 50°C for 20 s, and an extension at 72°C for 30 s. The temperature transition rate was 20°C/s. The generation of target amplicons for each sample was monitored between the annealing and the elongation steps at 640 and 705 nm. At the end of the quantification phase of the reaction, samples positive for target genes were identified by the instrument at the cycle number where the fluorescence attributable to the target sequences exceeded that measured for background. Those scored as positive by the instrument were confirmed by visual inspection of the graphical plot (cycle number versus fluorescence value) generated by the instrument.
Following the amplification phase, a melting curve analysis was performed with a temperature transition rate of 0.2°C/s to determine the Tms for the sequences targeted by the hybridization probes. Tms were manually assigned from a plot generated by the instrument of the negative derivative of fluorescence versus temperature (−dF/dT) of the melting curve for amplification products measured at 640 and 705 nm. Positive controls were included in each LC run to monitor any variation of melting peak temperature determined in different runs. To avoid false-negative readings from possible variability in the target gene sequences, the final determination of the presence of the target genes in samples was performed at the completion of the melting curve analysis. Samples with amplicons having Tms above those obtained for the negative control samples (known negative strains and no-added-DNA template controls) were scored as positive for the respective target gene.
Sensitivity of LC-PCR assays for ETEC detection.
To assess the sensitivity of the PCR assays for the three target genes, E. coli H10407 strain ATCC 35401 was grown at 37°C in brain heart infusion broth (approximate optical density of 2.7 to 2.8 at 600 nm) and diluted 10-fold in phosphate-buffered saline to yield 101 to 1010 CFU/ml as estimated by a standard plating procedure. Crude DNA templates from a 300-μl aliquot of each dilution were prepared by heating the bacterial suspension at 99.9°C for 10 min to lyse the bacteria and centrifuging the lysate at 1,000 × g for 2 min to sediment debris. Aliquots of the clarified supernatant were tested in the LC-PCR assays and the block cycler PCR assays as described above to determine the minimum number of CFU per reaction that could be detected by each method.
To assess the sensitivity of the PCR assays for the three target genes in fecal specimens, aliquots of stool suspensions from four healthy volunteers (50% [wt/vol] in physiological saline) were inoculated with E. coli H10407 to yield final spiking levels of 100 to 107 CFU/g. Three swabs from each spiked stool suspension were prepared, and one swab from each spiking level was used to prepare DNA templates for PCR by direct DNA extraction, culture on a MacConkey agar plate (BBL, Cockeysville, Md.), or overnight enrichment in GN broth (BBL). Whole-cell templates from the overnight growth on the MacConkey plate and direct DNA extracts from the swabs were prepared as described above. The DNA templates were tested with the block cycler and real-time PCR assays to determine the limit of detection (LOD) for each assay. In this study, the LOD was defined as the lowest inoculation level at which three of the four spiked stool samples (75%) were positive.
Nucleotide sequence analysis of PCR products.
Amplification products were purified using the HighPure PCR product purification kit (Roche Diagnostics), and cycle sequencing reactions for the LT I, ST Ia, and ST Ib amplicons were performed as described in the PRISM Ready Reaction Dye Deoxy Terminator cycle sequencing kit protocol (Applied Biosystems, Weiterstadt, Gemany). Both strands of each amplicon originating from separate amplification reactions with the corresponding primers listed in Table 2 were sequenced in duplicate to rule out the possibility of Taq DNA polymerase-induced errors. The fluorescence-labeled reaction products were analyzed with an ABI PRISM 310 genetic analyzer (Applied Biosystems).
Nucleotide sequence accession numbers.
The nucleotide sequences of amplicons derived from representative strains harboring LT I and ST I variants were deposited in GenBank under the following accession numbers: AY342056 (strain F5176, LT I variant), AY342057 (strain F7682, ST Ia variant), AY342058 (strain C4046, ST Ib variant), and AY342059 (strain R554, ST Ib variant).
RESULTS
Strategy for duplex LC-PCR assay design.
We designed two separate real-time fluorescence PCR assays for the LC to detect the two main types of enterotoxin genes commonly present in ETEC strains causing human disease. One assay targets the LT I gene, while the other targets the ST Ia and ST Ib genes. For the LT I assay, primers spanning a 340-bp region of the B-subunit gene and a set of hybridization probes (anchor and sensor probes) internal to this region were designed against sequences that were conserved among known LT I genes (GenBank accession numbers M17874 and M17873) and that were divergent from the analogous region of the cholera toxin gene from Vibrio cholerae.
Due to considerable sequence variation between the ST Ia and ST Ib genes (homology of only 83.9% between the ST Ia [GenBank accession number M25607] and ST Ib [GenBank accession number M34916] sequences), we designed two sets of primers targeting conserved regions between members of each variant toxin type to give 189-bp amplicons for each gene. From multiple-sequence alignments of the ST I genes (25), we identified a 34-bp region internal to the primers with a high degree of homology between ST Ia and ST Ib. We selected this region to serve as the target sequence for a common anchor hybridization probe (STI-HP-2) for both toxins. We designed the sensor hybridization probes for ST Ia and ST Ib (STI-HP-1a and STI-HP-1b, respectively) to differ from each other in their theoretical Tms (4) in order to differentiate between the presence of ST Ia and ST Ib by melting curve analysis. The use of a common anchor probe reduced the complexity and cost of the ST I multiplex by employing seven oligonucleotides (four primers and three hybridization probes) instead of eight oligonucleotides.
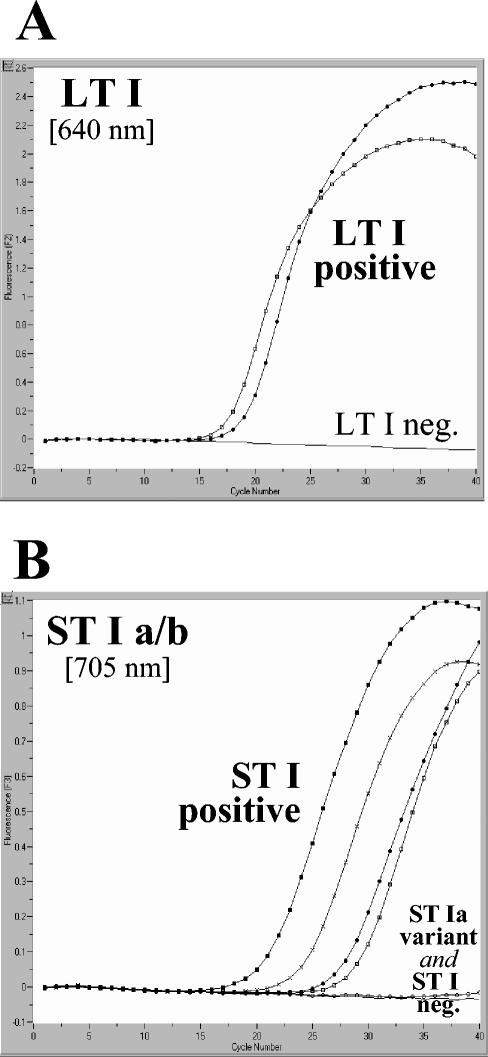
We designed the LT I and ST Ia/ST Ib LC-PCR assays to have identical amplification conditions so that both could be run simultaneously on the same instrument. Graphs depicting the accumulation of fluorescence throughout the LC-PCRs for LT I, ST Ia, and ST Ib are shown for a panel of representative strains in Fig. 1. The curve for strain H10407, which is positive for all three target genes, illustrates the clear discrimination between target genes attainable with the LC-PCR assays.
FIG. 1.
Amplification of LT and ST genes in separate LC-PCR assays. The graphs depict fluorescence at 640 and 705 nm versus cycle number for E. coli strains with various combinations of target genes. (A) LT genes. •, H10407 (LT I positive, ST Ia positive, ST Ib positive); □, F5176 (LT I positive, ST Ia negative, ST Ib negative); —, negative control. (B) ST genes. •, H10407 (LT I positive, ST Ia positive, ST Ib positive); ▪, TX1 (LT I negative, ST Ia negative, ST Ib positive); ×, C4046 (LT I negative, ST Ia negative, ST Ib positive); □, R554 (LT I negative, ST Ia negative, ST Ib positive); ○, F7682 (LT I negative, ST Ia positive, ST Ib negative); —, negative control.
Sensitivity and specificity of LC-PCR assays for detecting target genes in pure bacterial cultures.
To assess the ability of the LC-PCR assays to detect LT I, ST Ia, and ST Ib enterotoxin genes in a diverse collection of bacteria, we tested 161 E. coli isolates and 168 isolates of other bacterial species, including 51 strains of V. cholerae that are positive for the cholera toxin genes, in the LC PCR assays and compared these findings with those obtained by block cycler PCR assays. The correlation of the LC-PCR and block cycler assays is shown in Table 3. There was complete correlation between the LC-PCR results and those obtained by the block cycler PCR assays. Among the strains tested, the positive and negative predictive values of the LC-PCR assays for LT I, ST Ia, and ST Ib were each 100%. In scoring the results, it was important to complete the melting curve analysis step of the LC-PCR assay to successfully detect all the target genes. This was a factor for one strain in the collection (F7682 [LT I negative, ST Ia positive, ST Ib negative) (Fig. 1; see Fig. 3). The ST Ia gene present in this strain was a sequence variant of ST Ia that was not detected during the quantification step, in large part because its optimal binding temperature for the hybridization probes was below that used during the amplification assay. Because the melting curve analysis extends over a lower temperature range than that used during the amplification assay, this step increases the sensitivity of the assay for detecting genes with sequence variability in the region of the hybridization probes. In addition, the melting curve analysis also increases the sensitivity of the assay for detecting target genes that have not amplified sufficiently to be scored as positive after the quantification step. The benefit of this step in detecting samples with weak amplification is attributable, in part, to the large number of data points collected throughout the melting curve analysis, in contrast to the limited number of readings that such samples receive during the quantification step due to their shortened log-linear phase.
TABLE 3.
Correlation of conventional PCR and LC-PCR results for 329 bacterial isolates
| Target gene | No. of isolates with the following conventional PCR/LCycler-PCR results:
|
Sensitivity (%) | Specificity (%) | |||
|---|---|---|---|---|---|---|
| +/+ | −/+ | +/− | −/− | |||
| LT I | 74 | 0 | 0 | 255 | 100 | 100 |
| ST Ia | 48 | 0 | 0 | 281 | 100 | 100 |
| ST Ib | 66 | 0 | 0 | 263 | 100 | 100 |
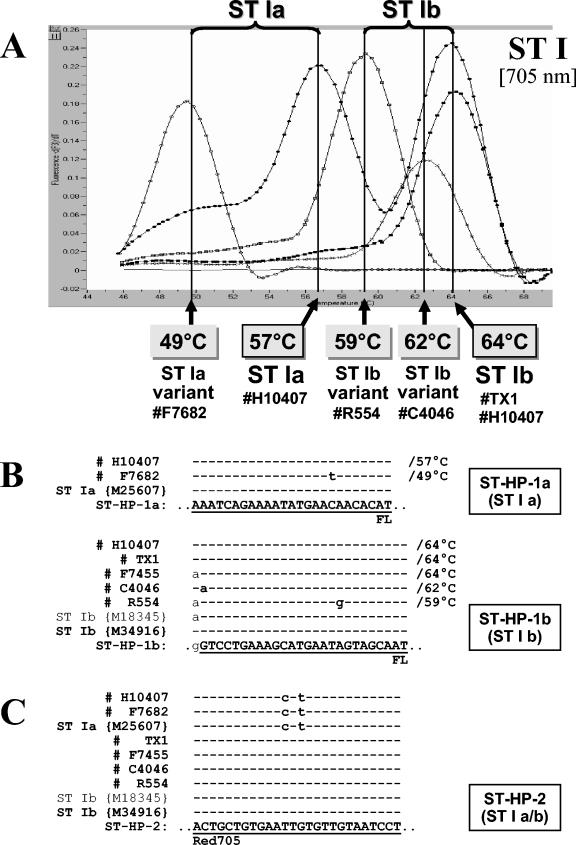
FIG. 3.
(A) Melting curve analysis performed on amplification products of four different ST-positive isolates, representative of the different ST genotypes observed in this study. Strain H10407 represents a typical ST Ia-positive and ST Ib-positive isolate, strain F7682 is an isolate harboring a variant ST Ia sequence, and strains C4046 and R554 are ST Ib-positive isolates harboring variant ST Ib sequences. (B and C) Sequence alignments with amplicons of the four different ST-genotypes; the sequences under GenBank accession numbers M25607 (ST Ia), M34916 (ST Ib), and M18345 (variant ST Ib); and ST Ia-specific sensor hybridization probe ST-HP-1a (B), ST Ib-specific sensor hybridization probe ST-HP-1b (B), or anchor hybridization probe ST-HP-2 (C). Sequence identity is indicated by dashes. The observed Tms are indicated.
Analytical sensitivity and specificity of the LC-PCR assays.
To determine the lower limit of detection for the LT I and ST I LC-PCR assays, we performed each of the assays on DNA preparations of reference strain H10407 at concentrations of 101 to 1010 CFU/ml in phosphate-buffered saline. Findings from the LC-PCR assays and those performed in parallel with the block cycler assay revealed a conservative lower limit of 102 organisms per reaction for the respective target genes in the LC-PCR assays as well as the block cycler PCR assay.
Because of the complexity of the hybridization probe detection format for the variants of ST I, we assessed the ability of the ST I LC-PCR assay to discriminate between the ST Ia and ST Ib genes by testing DNA preparations from E. coli strains that were positive for ST Ia, ST Ib, or both toxins in reaction mixtures that contained the four ST I primer oligonucleotides and either the ST Ia-specific or the ST Ib-specific pair of hybridization probes. Clearly positive or negative amplification curves were obtained, with no cross-reaction between the ST Ia-positive strains and the ST Ib-specific set of hybridization probes and vice versa. As expected, E. coli H10407, which is positive for both ST Ia and ST Ib, was positive in both sets of experiments.
Sensitivity of LC-PCR assays for detecting target sequences in fecal specimens.
The sensitivities of the LC-PCR and block cycler PCR assays for detecting ETEC in spiked fecal specimens from healthy donors were compared for the following types of template DNA samples: DNA extracted directly from the spiked fecal specimen, DNA released by boiling bacterial growth from the primary isolation plate (MacConkey agar), and DNA released by boiling of an aliquot of overnight enrichment broth (GN broth). Findings from these experiments are summarized in Table 4. The LODs for ETEC in fecal specimens DNA extracted directly from the stool specimen was used in the PCRs were 105 and 106 CFU/g for LT I and ST I, respectively, in the LC-PCR assays and >107 CFU/g for both target genes in the block cycler PCR assay. When DNA from boiled bacterial growth from the MacConkey primary isolation plate was used in the PCR, the LODs in the LC-PCR and block cycler assays for both target genes were the same, 103 CFU/g. Interestingly, when DNA from a boiled aliquot of overnight enrichment broth was used in the PCRs, the LOD for the LC-PCR assay was significantly improved, i.e., 1 CFU/g for LT I and 10 CFU/g for ST I. A similar improvement in sensitivity was observed for the block cycler PCR assay with the STI target (LOD, 10 CFU/g) but not with the LT I target (LOD, >107 CFU/g). The LODs for ST Ia and ST Ib in the LC-PCR assay were the same for all three experiments (data not shown).
TABLE 4.
Analytical sensitivity of the real-time and block cycler PCR assays for detection of ETEC LT I and ST I genes from DNA extracted directly from spiked stool samples (D), DNA extracted from spiked stool samples cultured in GN broth (GN), and DNA extracted from spiked stool samples cultured on MacConkey agar (MC)
| Inoculation level (CFU/g) | No. of positive samples/no. of samples analyzed
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Real-time PCR
|
Block cycler PCR
|
|||||||||||
| LT I
|
ST Ia
|
LT I
|
ST Ia
|
|||||||||
| D | GN | MC | D | GN | MC | D | GN | MC | D | GN | MC | |
| 107 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 0/4 | 0/4 | 4/4 | 1/4 | 4/4 | 4/4 |
| 106 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 4/4 | 0/4 | 0/4 | 4/4 | 0/4 | 4/4 | 4/4 |
| 105 | 4/4 | 4/4 | 4/4 | 0/4 | 4/4 | 4/4 | 0/4 | 0/4 | 4/4 | 0/4 | 4/4 | 4/4 |
| 104 | 0/4 | 4/4 | 4/4 | 0/4 | 4/4 | 4/4 | 0/4 | 0/4 | 3/4 | 0/4 | 4/4 | 3/4 |
| 103 | 0/4 | 4/4 | 4/4 | 0/4 | 4/4 | 4/4 | 0/4 | 0/4 | 3/4 | 0/4 | 4/4 | 3/4 |
| 102 | 0/4 | 4/4 | 0/4 | 0/4 | 4/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 4/4 | 0/4 |
| 101 | 0/4 | 4/4 | 0/4 | 0/4 | 4/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 4/4 | 0/4 |
| 100 | 0/4 | 4/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 |
ST Ia and ST Ib were detected with equal sensitivity.
In addition to the spiked fecal specimens, six stool specimens from persons who were part of an outbreak caused by ST Ia-positive E. coli O27:H7 were tested for the presence of ETEC in the LC-PCR and block cycler PCR assays. The findings obtained with both methods by using sweeps of bacterial growth from the primary isolation plates for DNA template preparation agreed completely; three of six samples were positive for ST Ia, and three were negative. For the three positive stool samples, positive findings for ST Ia genes were obtained after approximately 40 min (cycles 23 to 27) in the LC-PCR, in contrast to over 4 h in the block cycler PCR assay.
Time requirements for LC-PCR and block cycler PCR assays.
We performed 40 amplification cycles for the LC-PCR assays, which typically took 70 min to accomplish. For the majority of samples (greater than 90% of isolates, enrichment broths, and growth from primary isolation plates), however, we could discriminate between PCR-positive and -negative samples after only 30 cycles, which took approximately 40 min to accomplish. LT I and ST I target genes in DNA extracted directly from stool specimens at 105 and 106 CFU/g, respectively, typically required 35 to 38 cycles to detect. Block cycler PCR assays with agarose gel detection of amplicons typically took 4.5 to 5 h to complete. If a sequence-based identification of amplicons from the block cycler was performed, the block cycler assay would require 7 to 18 h, depending on the format of the detection assay (enzyme immunoassay or membrane-based hybridization with DNA probes).
Differentiation of LT, ST Ia, and ST Ib variants by melting curve analysis.
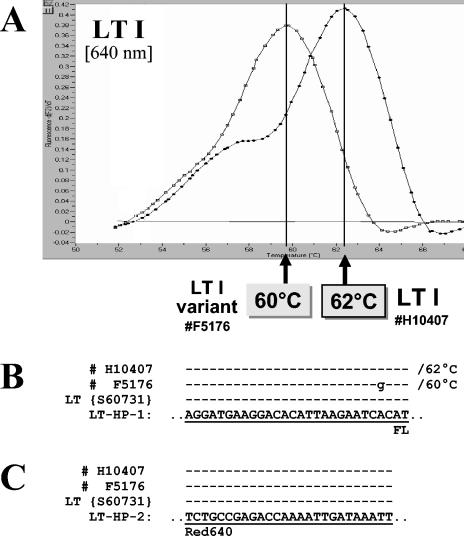
Sharp and clearly interpretable melting curves (plotted as the negative derivative of fluorescence versus temperature) were obtained with all 138 ETEC isolates tested. Findings from the melting curve analyses of these isolates and the representative isolates sequenced for each Tm are summarized in Table 5. In the course of this study, the interassay precision of the LC melting curve analysis was ±1°C. Melting curve profiles of representative strains and the sequences of their target genes at the annealing region of the hybridization probes are shown in Fig. 2 and 3. A predominant Tm was detected for each of the target genes. The Tms of the control strains for LT I, ST Ia, and ST Ib were 62, 57, and 64°C, respectively. These Tms reflect complete homology between the hybridization probes and their target sequences.
TABLE 5.
Distribution of Tms among 137 ETEC strains and associated sequence variation in the region of the hybridization probes
| Target gene (no. of strains positive for target gene) | Tm (°C) (±1°C) | No. of strains having designated Tm | No. of nucleotide differences between target gene and probesa | Representative ETEC strain(s) sequenced |
|---|---|---|---|---|
| LT I (74) | 62 | 67 | 0 | H10407 (O78:H11), E2539 (O25:NM) |
| 60 | 7 | 1 | F5176 (O167:H5) | |
| ST Ia (48) | 57 | 47 | 0 | H10407 (O78:H11) |
| 49 | 1 | 3 | F7682 (O136:H12) | |
| ST Ib (66) | 64 | 59 | 0 | H10407 (O78:H11), TX1 (O78:H12) |
| 62 | 6 | 1 | C4046 (O148:H28) | |
| 59 | 1 | 1 | R554 (O25:H42) |
Number of nucleotide differences between the target gene sequences of representative strains at each Tm and the corresponding hybridization probe sequences.
FIG. 2.
(A) Melting curve analysis performed on amplification products of two different LT-positive isolates. Strain H10407 represents a typical LT I-positive isolate, and strain F51761 is an ETEC isolate harboring a variant LT I sequence. (B and C) Sequence alignments with amplicons of the two different LT I-genotypes, the sequence under GenBank accession number S60731, and sensor hybridization probe LT-HP-1 (B) or anchor hybridization probe LT-HP-2 (C). Sequence identity is indicated by dashes. The observed Tms are indicated.
A small number of strains possessed genes with sequence variation in the region of the hybridization probes. We noted a reduced Tm of 60°C for seven LT I-positive strains, a Tm of 49°C for one ST Ia strain, and Tms of 62 and 59°C for six ST Ib strains and one ST Ib strain, respectively (Table 5). Sequencing data obtained for the amplicons of representative strains at each Tm showed one to three nucleotide mismatches per strain between the target sequence and that of the hybridization probes. In general, there was a correlation between the number of mismatches between the target and probe sequences and the degree to which the Tms of the variant strains were depressed relative to strains with target sequences completely homologous to the probes. In several cases the nucleotide changes resulted in amino acid changes; however, the effect of these changes on protein function is not known (10, 13).
DISCUSSION
We focused on the development of a reliable real-time PCR assay for the detection of LT I and ST I genes. Our set of real-time fluorescence PCR assays for LT I, ST Ia, and ST Ib covers the enterotoxin genes commonly associated with human disease. In the process of evaluating our assay, we by chance discovered a number of heretofore-unknown mutations within the amplification products of several ETEC strains tested. The sequences of the variant enterotoxin amplicons have been deposited in GenBank to help guide future assay development. Although the variants we detected were the exception and not the rule among the diverse collection of strains examined in this study, the presence of sequence variation in the toxins illustrates the advantage of the hybridization probe format and melting curve analysis for providing strain characterization over traditional block cycler PCR or other real-time PCR formats that lack the capacity for melting curve analyses. Strains with unusual Tms will be easy to monitor and track if they are to cause an outbreak.
After evaluating the separate LT I and ST I LC-PCR assays, we attempted to combine them into a single multiplex reaction with six primer and five hybridization probe oligonucleotides in one capillary. The presence of LT I-specific genes was detected at 640 nm (channel F2), and the presence of ST I-specific genes was detected at 705 nm (channel F3). A melting curve analysis was performed to resolve the ST Ia and ST Ib variants. Although conditions that permitted the simultaneous detection and differentiation of the three target genes in a single reaction were identified (final concentrations of 5 mM MgCl2 and 0.2 μM primers LT-1 and LT-2), the sensitivity of the assay was observed to be affected by slight deviations in primer oligonucleotide concentrations that can routinely occur among different batches of commercially synthesized oligonucleotides. For this reason, we do not recommend combining the present assays in a single capillary for routine diagnostic testing.
Since the rational design of probes for microarray systems is based on a comprehensive database of nucleotide sequences, a potential use for the enterotoxin LC-PCR assays is to screen strains to identify sequence variants of the enterotoxin genes. In this study, the melting curve analysis proved to be a very efficient method for identifying a small number of strains with variant enterotoxin genes for subsequent sequencing to develop DNA array-based assays.
Findings from the experiment with spiked stool specimens revealed significant variation in the LODs for the LC-PCR and block cycler PCR assays with the three different methods of template DNA preparation and highlight the importance of this single step for the performance of the PCR assays. The LODs for both target genes in the LC-PCR and for the ST genes in the block cycler PCR assays were lowest with DNA templates prepared from the overnight GN enrichment broths. The failure of the block cycler PCR assay to detect the LT genes from the GN broth was unexpected and illustrates the unpredictable nature of PCR assays. Experiments to determine if a component in the GN broth may be responsible for this selective inhibition are planned.
The LODs for the LC-PCR and block cycler PCR assays were similar to each other when overnight growth from the MacConkey plate was used to prepare the DNA templates. Interestingly, these values were 100- to more than 1,000-fold higher that those from the overnight GN broth culture. The reason(s) for this is not clear. Possible factors that may have affected the sensitivity of these assays include inhibitors carried over from the agar, slower growth characteristics of the target bacterium relative to nontarget bacteria on MacConkey agar compared with GN broth, and denser cell suspensions prepared from the agar plates than from the GN broth, resulting in too much DNA being added to the PCR mixture.
The highest LODs were observed for the DNA template preparation extracted directly from stool; the LC-PCR assay was 100 to 1,000 times more sensitive than the block cycler PCR assay for detecting the ST I and LT I genes, respectively. In comparison with block cycler PCR assays, this difference offers a potential advantage for LC-PCR assays for rapidly diagnosing ETEC infections, by allowing detection of ETEC without enrichment in samples containing 105 to 106 CFU/g. The practical benefit of this difference is not clear, as the percentage of diarrheal specimens with ETEC in this range is not known.
The combination of simplex LT I-specific and duplex ST I-specific LC-PCR assays described in this report offers rapid cycling (1 h or less) with real-time, sequence-specific detection of amplicons. The fluorescence-labeled hybridization probes not only provide confidence in the identification of target genes but also reduce the risk of product contamination of the laboratory because the amplification reaction, detection of PCR products, and melting curve analyses are conducted in a single closed capillary tube. Although our current protocol for processing stool specimens for ETEC requires an overnight enrichment step in GN broth to achieve optimal sensitivity, the rapid amplification and reliable detection of the prototype LT I and ST I genes, and variants thereof, in samples should facilitate the diagnosis of individuals infected with ETEC.
While the diagnosis and management of individual patients infected with ETEC do not require an isolate, the isolation of ETEC from patient samples is critical for public health purposes. The association of the ETEC target genes with a particular bacterium requires testing of isolates, and isolates are still needed for molecular subtyping (DNA fingerprinting) and serologic characterization for surveillance purposes. For these reasons, clinical laboratories are strongly encouraged to consult with their public health authorities for guidance on forwarding positive samples for isolation of ETEC. The speed with which bacterial colonies can be screened for target genes and these genes can be characterized with the LC-PCR assays should facilitate the detection and investigation of ETEC outbreaks. Although LC-PCR assays are currently more expensive to perform than block cycler PCR assays, their speed, reduced hands-on time, and greater information and the reliability of the results make them attractive alternatives to conventional block cycler PCR assays for the detection and characterization of ETEC.
Acknowledgments
We thank Norbert Lehn and Stefan Lukas for their active support and Klaus Pfeffer (Institute of Medical Microbiology, University of Düsseldorf, Düsseldorf, Germany) for consultation in the design of the LT I primers. We gratefully acknowledge the excellent technical assistance of Markus Bollwein and Michaela Hien during the study.
This work has been supported in part by the Arab Fund for Economic and Social Development, Kuwait. M.T.Y. was a recipient of the Arab Fund's distinguished Scholar Award.
REFERENCES
- 1.Albert, M. J., S. M. Faruque, A. S. G. Faruque, P. K. Neogi, M. Ansaruzzaman, N. A. Bhuiyan, K. Alam, and M. S. Akbar. 1995. Controlled study of Escherichia coli diarrheal infections in Bangladeshi children. J. Clin. Microbiol. 33:973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bellin, T., M. Pulz, A. Matuseek, H.-G., Hempen, and F. Gunzer. 2001. Rapid detection of enterohemorrhagic Escherichia coli by real-time PCR with fluorescent hybridization probes. J. Clin. Microbiol. 39:370-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, R.E. 1986. The epidemiology of cholera and enterotoxigenic Escherichia coli diarrheal disease, p. 23-32. In J. Holmgren, A. Lindberg, and R. Mollby (ed.), Development of vaccine and drug against diarrhea. Eleventh Nobel Conference, Stockholm. Studentlitteratur, Lund, Sweden.
- 4.Breslauer, K. J., R. Frank, H. Blocker, and L. A. Marky. 1986. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 83:3746-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 1994. Foodborne outbreaks of enterotoxigenic Escherichia coli—Rhode Island and New Hampshire. Morb. Mortal. Wkly. Rep. 43:81-89. [PubMed] [Google Scholar]
- 6.Daniels, N. A., J. Neimann, A. Karpati, U. D. Parashar, K. D. Greene, J. G. Wells, A. Srivastava, R. V. Tauxe, E. D. Mintz, and R. Quick. 2000. Traveler's at the sea: three outbreaks of waterborne enterotoxigenic Escherichia coli on cruise ships. J. Infect. Dis. 181:1491-1495. [DOI] [PubMed] [Google Scholar]
- 7.Edwards, K. J., L. A. Metherell, M. Yates, and N. A. Saunders. 2001. Detection of rpoB mutations in Mycobacterium tuberculosis by biprobe analysis. J. Clin. Microbiol. 39:3350-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ewing, W. H. 1986. The genus Escherichia, p. 93-134. In Edwards and Ewing's identification of Enterobacteriaceae, 4th ed. Elsevier Science Publishing Co., Inc., New York, N.Y.
- 9.Gorbach, S. L., B. H. Kean, D. G. Evans, D. G. Evans, Jr., and D. Bessudo. 1975. Traveller's diarrhea and toxigenic Escherichia coli. N. Engl. J. Med. 292:933-936. [DOI] [PubMed] [Google Scholar]
- 10.Guzman-Verduzio, L. M., and Y. M. Kupersztoch. 1989. Rectification of two Escherichia coli heat-stable enterotoxin allele sequences and lack of biological effect of the change of the carboxy-terminal tyrosine for histidine. Infect. Immun. 57:645-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedberg, C. W., S. J. Savarino, J. M. Besser, C. J. Paulus, V. M. Thelen, L. J. Myers, D. N. Cameron, T. J. Barrett, J. B. Kaper, M. T. Osterholm, et al. 1997. An outbreak of foodborne illness caused by Escherichia coli O39:NM, an agent not fitting into the existing scheme for classifying diarrheagenic E. coli. J. Infect. Dis. 176:1625-1628. [DOI] [PubMed] [Google Scholar]
- 12.Huerta, M., I. Grotto, M. Gdalevich, D. Mimouni, B. Gavrieli, M. Yavzori, D. Cohen, and O. Shiplberg. 2000. A waterborne outbreak of gastroenteritis in the Golan Heights due to enterotoxigenic Escherichia coli. Infection 28:267-271. [DOI] [PubMed] [Google Scholar]
- 13.Inoue, T., T. Tsuji, M. Koto, S. Imamura, and A. Miyama. 1993. Amino acid sequence of heat-labile enterotoxin from chicken enterotoxigenic Escherichia coli is identical to that of human strain H 10407. FEMS Microbiol. Lett. 108:157-161. [DOI] [PubMed] [Google Scholar]
- 14.Logan, J. M., K. J. Edwards, N. A. Saunders, and J. Stanley. 2001. Rapid identification of Campylobacter spp. by melting peak analysis of biprobes in real-time PCR. J. Clin. Microbiol. 39:2227-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maguire, A. L., D. F. J. Brown, J. J. Gray, and U. Desselberger. 2001. Rapid screening technique for class 1 integrons in Enterobacteriaceae and nonfermenting gram-negative bacteria and its use in molecular epidemiology. Antimicrob. Agents Chemother. 45:1022-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitsuda, T., T. Muto, M. Yamada, N. Kobayashi, M. Toba, Y. Aihara, A. Ito, and S. Yokota. 1998. Epidemiological study of food-borne outbreak of enterotoxigenic Escherichia coli O25:NM by pulsed-field gel electrophoresis and randomly amplified polymorphic DNA analysis. J. Clin. Microbiol. 36:652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moseley, S. L., J. W. Hardy, M. I. Huq, P. Echeverria, and S. Falkow. 1983. Isolation and nucleotide sequence determination of a gene encoding a heat-stable enterotoxin of Escherichia coli. Infect. Immun. 39:1167-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Olive, M. 1989. Detection of enterotoxigenic Escherichia coli after polymerase chain reaction amplification with a thermostable DNA polymerase. J. Clin. Microbiol. 27:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ølsvik O., and N. A. Strockbine. 1993. PCR detection of heat-stable, heat-labile, and Shiga-like toxin genes in Escherichia coli, p. 271-276. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology. American Society for Microbiology, Washington, D.C.
- 20.Oyofo, B. A., L. F. Peruski, T. F. Ismail, S.H. el-Etr, A. M. Churilla, M. O. Wasfy, B. P. Petrucelli, and M. E. Gabriel. 1997. Enteropathogens associated with diarrhea among military personnel during Operation Bright Star 96, in Alexandria, Egypt. Milit. Med. 162:396-400. [PubMed] [Google Scholar]
- 21.Paniagua, M., F. Espinoza, M. Ringman, E. Reizenstein, A. M. Svennerholm, and H. Hallander. 1997. Analysis of incidence of infection with enterotoxigenic Escherichia coli in a prospective cohort study of infant diarrhea in Nicaragua. J. Clin. Microbiol. 35:1404-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reischl, U., H. J. Linde, M. Metz, B. Leppmeier, and N. Lehn. 2000. Rapid identification of methicillin-resistant Staphylococcus aureus and simultaneous species confirmation using real-time fluorescence. J. Clin. Microbiol. 38:2429-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reischl, U., M. T. Youssef, J. Kilwinski, N. Lehn, W. L. Zhang, H. Karch, and N. A. Strockbine. 2002. Real-time fluorescence PCR assays for the detection and characterization of Shiga toxin, intimin, and enterohemolysin genes from Shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 40:2555-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schultsz, C., G. J. Pool, R. van Ketel, B. de Wever, P. Speelman, and J. Dankert. 1994. Detection of enterotoxigenic Escherichia coli in stool samples by using nonradioactively labeled oligonucleotide DNA probes and PCR. J. Clin. Microbiol. 32:2393-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekizaki, T., H. Akashi, and N. Terakado. 1985. Nucleotide sequences of the genes for Escherichia coli heat-stable enterotoxin I of bovine, avian, and porcine origins. Am. J. Vet. Res. 46:909-912. [PubMed] [Google Scholar]
- 26.Tsen, H.-Y., and L.-Z. Jian. 1998. Development and use of a multiplex PCR system for the rapid screening of heat-labile toxin I, heat-stable toxin II and Shiga-like toxin I and II genes of Escherichia coli. J. Appl. Microbiol. 84:585-592. [DOI] [PubMed] [Google Scholar]
- 27.Wilshaw, G. A., T. Cheasty, B. Rowe, H. R. Smith, D. N. Faithful-Davies, and T. G. J. Brooks. 1995. Isolation of enterotoxigenic Escherichia coli from British troops in Saudi Arabia. Epidemiol. Infect. 115:455-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yavzori, M., N. Porath, O. Ochana, R. Dagan, R. Orni-Wasserlauf, and D. Cohen. 1998. Detection of enterotoxigenic Escherichia coli in stool specimens by polymerase chain reaction. Diagn. Microbiol. Infect. Dis. 31:503-509. [DOI] [PubMed] [Google Scholar]