Abstract
After the initial development of a human immunodeficiency virus type 1 (HIV-1) subtype-screening tool by nested multiplex PCR, we further improve it through the redesign of subtype-specific primers based on subtype signature pattern (SSP) analyses and optimization of the PCR conditions. Extracted RNA from plasma samples was used in reverse transcription and the cDNA products were added to the first round PCR, in which universal primers in the gag region were used to detect HIV-1 M group isolates. In the second round of PCR, three pairs of subtype-specific primers, detecting subtypes B, C, and CRF01-AE, were added in one tube. Subtype determination was based on the different size of PCR products on the agarose gel electrophoresis. An additional set of primers detecting only the prevalent recombinant strains CRF07-BC and CRF08-BC was used to discriminate CRF07- and CRF08-BC from pure subtype C. Testing for all kinds of HIV subtype reference strains indicated that this assay was applicable. A panel of 252 HIV-positive samples and 30 HIV-negative samples was further used to evaluate and validate this assay. Compared to the assay of sequence-based phylogenetic analysis, the newly developed assay has an adequate designated subtype sensitivity, 93.2% (69 of 74) for subtype B, 95.1% (117 of 123) for subtype C, 94.0% (47 of 50) for CRF01-AE, and 95.0% (115 of 121) for CRF07-BC and CRF08-BC. Most importantly, the intersubtype specificity of the assay was found to be 100%. The assay specificity was also found to be 100% when used to test 30 HIV-negative samples. The average reproducibility was 96.0% for subtype B, 96.7% for subtype C, and 95.0% for CRF01-AE. We have developed a simple, rapid, and low cost assay for screening subtypes B, C, CRF01-AE, CRF07-BC, and CRF08-BC in China.
The unusual degree of genetic variability makes the human immunodeficiency virus type 1 (HIV-1) highly heterogeneous. Until now, strains of HIV-1 from diverse geographic regions have been divided into three groups—group M (major), O (outlier), and N (new or non-M, non-O). The group M isolates, comprising 9 “pure” subtypes (A to D, F to H, J, and K) and 15 circulating recombinant forms (CRF01 to CRF15), are primarily responsible for the global pandemic (7, 9, 18; Los Alamos National Laboratory [http://www.hiv.lanl.gov]).
In China, since the first case of HIV infection was identified in 1985, the number of HIV-infected people has increased alarmingly, especially in recent years. Although the recorded cumulative number of HIV infections and AIDS cases was only 40,560 at the end of 2002, it is estimated that the actual number of people with HIV/AIDS is more than 850,000 (NCAIDS, China CDC [http://www.chinaids.org.cn]). Considering the potential gravity of China's situation, all possible measures should be taken to prevent the further spread of HIV. Monitoring HIV genetic characteristics and subtyping prevalent isolates in China will provide essential data for HIV candidate vaccine design and HIV/AIDS prevention policy issues. The first national molecular epidemiology survey from 1996 to 2000 demonstrated that HIV-1 subtypes A, B, C, D, F, G, CRF01-AE, CRF07-BC, CRF08-BC, and HIV-2 have been found in China (5, 12, 13, 17, 19-22; Y. Shao, L. Su, X. Sun, H. Xing, P. Pan, H. Wolf, and J. Shen, XIIth Int. Conf. AIDS, abstr. 13132, 1998). HIV-1 subtype B, CRF07-BC, CRF08-BC, and CRF01-AE infections represented >90% of the circulating strains (13, 17, 19; Shao et al., XIIth Int. Conf. AIDS; unpublished data); other strain infections were scarce.
The primary assays for HIV subtyping are sequence-based phylogenetic analysis and the heteroduplex mobility assay (HMA). Sequence-based phylogenetic analysis is the “gold standard” for HIV subtyping, in which a part of the viral genome is sequenced and phylogenetic analysis is used in comparison with a set of reference sequences having known subtypes. This assay is reliable but labor-intensive and time-consuming. HMA, although much easier and more widely used, is also laborious (1, 3).
We have just completed the development of a simple, rapid, and low-cost subtype screening assay for subtypes B, C, and CRF01-AE that uses reverse transcription-PCR (RT-PCR) and a single nested multiplex PCR without the need for sequencing or HMA. After the initial assay development, improvements were made through the redesign of subtype-specific primers and optimization of PCR conditions. A panel of more than 200 clinical samples was used to evaluate the sensitivity and specificity of the assay.
MATERIALS AND METHODS
Sample collection.
Whole blood samples, anticoagulated with EDTA, were collected from several provinces in China. The plasma was separated within 8 h and stored at −80°C. All HIV-positive samples described here were identified by HIV enzyme immunoassay (Virostika HIV Uniform II plus O; Organon Teknika, The Netherlands) and Western blot assay (HIV blot 2.2, Genelab Technologies, Inc., Singapore). The 30 HIV-negative samples from volunteer blood donors were confirmed twice by the HIV enzyme immunoassay assay (the screening kit mentioned above and the HIV/1+2 antibody detection kit; Beijing Jinhao, Beijing, China).
Assay design.
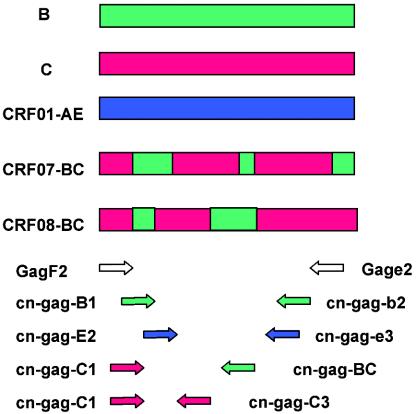
The basic design of the new assay is to use RT-PCR and nested PCR to subtype HIV strains, in which outer primers were universal for HIV-1 M group strains, and the inner primers were subtype-specific primers that were only reactive to one designated subtype of HIV-1. Accurate primer design plays a key role in this assay, especially in the generation of subtype-specific primers. To identify one subtype from others successfully, the sequences of these subtype-specific primers should be highly specific in intersubtype and highly conserved in intrasubtype comparisons. This pattern was actually a subtype signature pattern (SSP). Signature pattern analysis was first applied in a study of HIV transmission in dental practice in 1990 (8, 11). In the present study, SSPs were defined as unique nucleic acid sites that are distinctly representative of a subtype or CRF relative to a background of the rest of the HIV-1 subtypes or CRFs. The design of subtype-specific primers with SSPs ensures both high sensitivity and high specificity of the assay. Several primer sets were carefully designed and tested for the present study. The best set of primers was selected for subtypes B, C, and CRF01-AE (Table 1 and Fig. 1). In addition, recombinant strains CRF07-BC and CRF08-BC, prevalent CRFs in China, have subtype C backbones with small portions of subtype B (10, 13, 19) (Fig. 1). The subtype C-specific primer set cn-gag-C1/cn-gag-c3 was reactive to pure subtype C and CRF07- and CRF08-BC. To further distinguish CRF07- and CRF08-BC from pure subtype C, the additional primer set cn-gag-C1 and cn-gag-BC was designed to detect only CRF07- and CRF08-BC. However, this set of primers cannot differentiate between CRF07- and CRF08-BC.
TABLE 1.
All primers used in this study
| Primera | Sequenceb | Direction | HXB2 gag position | Usage |
|---|---|---|---|---|
| Gag F2 | 5′-ATG GGT GCG AGA GCG TCA RTA TTA A-3′ | Forward | 1∼25 | Outer primer set for HIV-1 M group strains |
| Gag e2 | 5′-TCC AAC AGC CCT TTT TCC TAG G-3′ | Reverse | 1243∼1222 | |
| cn-gag-B1 | 5′-GGA GCT AGA ACG ATT CGC AG-3′ | Forward | 117∼136 | Inner primer set for HIV-1 subtype B |
| cn-gag-b2 | 5′-TCA TCA TTT CTT CTA GTG TAG CTG CT-3′ | Reverse | 1042∼1017 | |
| cn-gag-C1† | 5′-GGG AAA GAA ACA CTA TAT GCT AAA ACA CC-3′ | Forward | 72∼100 | cn-gag-C1/c3 or cn-gag-C1/c2, inner primer set for HIV-1 subtype C |
| cn-gag-c3† | 5′-TAA GGC TTC TTT GGT GTC TCG T-3′ | Reverse | 303∼282 | |
| cn-gag-c2† | 5′-GTA CAC AAT AGA GAG TTG CTA CTG TGT TG-3′ | Reverse | 265∼237 | |
| cn-gag-BC | 5′-CTT GTC TTA TGT CCA GAA TGC TGG T-3′ | Reverse | 862∼838 | cn-gag-C1/BC, inner primer set for HIV-1 CRF07- and CRF08-BC |
| cn-gag-E2† | 5′-TAC AAT AGC AAC CCT CTG GTG CG-3′ | Forward | 240∼262 | Inner primer set for HIV-1 CRF01-AE |
| cn-gag-e3† | 5′-CTG GAT TCG CAT TTT GGA CTA GC-3′ | Reverse | 985∼963 | |
| 306 | 5′-GGG AAA AAA TTC GGT TAA GGC C-3′ | Forward | 47∼68 | Inner primer set for HIV-1 M group strains |
| cn-gag | 5′-TAG TTC CTG CTA TRT CAC TTC C-3′ | Reverse | 718∼697 |
The primers marked with a dagger (†) are subtype C- and CRF01-AE-specific primers. Since these primers do not match with subtype B reference strain HXB2, the position listed is only the equivalent position in HXB2.
R, A or G.
FIG. 1.
Schematic diagram of subtype-specific nested multiplex PCR. The upper part of the figure is the gag gene region diagram of subtypes B, C, CRF01-AE, CRF07-BC, and CRF08-BC (10). The lower part of the diagram shows the primer sets in nested multiplex PCR. Primer pair GagF2/Gage2 are universal primers for HIV-1 M group strains. Primer sets cn-gag-B1/cn-gag-b2, cn-gagC1/cn-gag-c3, cn-gag-E2/cn-gag-e3 are subtype-specific primers for subtypes B, C, and CRF01-AE, respectively. Primer set cn-gag-C1/cn-gag-BC is specific for CRF07- and CRF08-BC.
Assay procedure. (i) RNA extraction and RT reaction.
RNA was extracted from plasma samples with a QIAamp viral RNA minikit (Qiagen, Hilden, Germany). RT was performed at 37°C for 1 h with 20 μl of an RT mixture consisting of 0.5 mM deoxynucleoside triphosphate, 1.5 μM reverse primer Gage2, 20 U of RNase inhibitor, 100 U of Moloney murine leukemia virus (New England Biolabs, Hitchin, United Kingdom), and 5 to 10 μl of RNA template. Then, the reaction was heated to 95°C for 5 min and placed on ice for 5 to 10 min.
(ii) Nested multiplex PCR.
In the first round PCR, all 20 μl of RT product was added to a 80-μl reaction mixture that ultimately contained 35 mM KCl (pH 9.0), 0.1% Triton X-100, 1.5 mM MgCl2, 0.4 μM forward primer GagF2, and 5 U of Taq polymerase (Promega, Shanghai, People’s Republic of China) without using the buffer for Taq polymerase provided by the company. Then, 5 μl of the first-round products was added to the second-round 50-μl mixtures containing 1× buffer, 1.5 mM MgCl2, 2.5 U of Taq polymerase, and 0.4 μM concentrations of the primer sets. In the second round, three primer sets detecting subtypes B (cn-gag-B1/cn-gag-b2), C (cn-gag-C1/cn-gag-c3), and CRF01-AE (cn-gag-E2/cn-gag-e3) were added to one tube. Because three sets of primers were added in one tube, it was called a nested multiplex PCR. Hot-start and touchdown techniques were used to decrease the frequency of nonspecific reactions (15). The reaction conditions are listed in Table 2. The PCR products were separated by electrophoresis on a 2% agarose gel (1× TAE, 100 V, 40 min) with ethidium bromide staining. Subtype determination was made based on PCR product size (subtype B, 900 bp; subtype C, 230 bp; subtype CRF01-AE, 740 bp). When the bands were not found in the expected subtype positions, the results were considered negative. Primer set cn-gag-C1/cn-gag-c2 can also be used as alternative subtype C-specific primers, but it was not as effective as cn-gag-C1/cn-gag-c3.
TABLE 2.
Primer sets for PCR and reaction conditions
| Primer set(s)a | Reaction conditions | No. of cycles |
|---|---|---|
| GagF2/Gage2 | 94°C for 5 min, 52°C for 1 min, 72 °C for 2 min | 1 |
| 94°C for 30 s, 52°C for 30 s, 72°C for 1 min 30 s | 35 | |
| 72°C for 10 min | 1 | |
| 306/cn-gag | 94°C for 2 min, 50°C for 1 min, 72°C for 2 min | 1 |
| 94°C for 30 s, 52°C for 30 s, 72°C for 1 min | 35 | |
| 72°C for 10 min | 1 | |
| cn-gag-B1/cn-gag-b2, cn-gag-C1/cn-gag-c3, cn-gag-E2/cn-gag-e3, and cn-gag-C1/cn-gag-BCb | 94°C for 2 min, 58°C for 1 min, 72°C for 2 min | 1 |
| 94°C for 30 s, 58°C for 30 s, 72°C for 1 min | 3 | |
| 94°C for 30 s, 57°C for 30 s, 72°C for 1 min | 3 | |
| 94°C for 30 s, 56°C for 30 s, 72°C for 1 min | 3 | |
| 94°C for 30 s, 55°C for 30 s, 72°C for 1 min | 3 | |
| 94°C for 30 s, 54°C for 30 s, 72°C for 1 min | 25 | |
| 72°C for 10 min | 1 |
A slash mark means that the first primer is a forward primer and the second primer is a reverse primer.
Touchdown PCR is described for these four primer sets.
(iii) Additional PCR for distinguishing CRF07- and CRF08-BC from pure subtype C.
If it was positive for subtype C, additional PCR with primer set cn-gag-C1/cn-gag-BC was applied to determine whether it was pure subtype C or CRF07- and CRF08-BC.
Evaluation of the new assay by using reference strains.
To evaluate the newly developed subtyping tool, we used it to subtype reference isolates. The following HIV-1 and HIV-2 reference strains were subtyped: M group subtype A, 92UG029 and 92RW008; M group subtype B, RL42 (B′) and 93BR029(B/F); M group subtype C, 92BR025, 93IN905, and 97CN001 (CRF07-BC); M group subtype D, 94UG114; CRF01-AE, CMU10, and 93TH023; M group subtype F, 93BR020; M group subtype G, JV1083; N group, YBF30; O group, MVP5180; and HIV-2 reference strain subtype A, UC2. In addition, M group subtype H and Euro-American B full-length cloning plasmids, p90CF056.1 and pNL4-3, were also analyzed. These reference strains and plasmids were generously provided by the AIDS Research and Reference Reagent Program of the Nationals Institute of Health. Virus RNA was extracted, and RT-PCR and nested multiplex PCR were performed as described above.
Evaluation and validation of the assay by sequence-based phylogenetic analysis.
We used a panel of more than 200 HIV-positive and 30 HIV-negative clinical samples to evaluate the assay's sensitivity and specificity. To check whether the subtyping results by nested multiplex PCR were accurate, all HIV-positive samples were analyzed by sequencing and phylogenetic analysis. By PCR with one set of inner universal primers, 306/cn-gag, we obtained the products for sequencing. The first-round PCR products obtained by using GagF2/Gage2 were used as the template in this round of PCR, saving both time and DNA sample. The products were separated by agarose gel electrophoresis and purified by using a Qiagen gel extraction kit. DNA sequencing was performed by using fluorescent dye terminators (Prism BigDye terminator cycle sequencing ready reaction kit; Applied Biosystems) and an automated DNA sequencer (Applied Biosystems model 377). To exclude the possibility of nonspecific amplification, some of the nested multiplex PCR products amplified by subtype-specific primers were also purified and sequenced.
DNA sequences were aligned with reference sequences of HIV-1 M group subtypes A to D, F to H, J, and K and the N and O groups by using CLUSTAL X. Subtype reference sequences were downloaded from Los Alamos National Laboratory (http://www.hiv.lanl.gov). Genetic distances were calculated by using the Kimura two-parameter approach, and a phylogenetic tree was constructed by the neighbor-joining method, both included in the MEGA package (Molecular Evolutionary Genetics Analysis; version 2.1. [http://www.megasoftware.net]). The topology reliability of the phylogenetic tree was estimated by performing bootstrap analysis with 100 replicates. Bootstrap values of ≥70% were considered statistically significant.
We also randomly selected 31 samples to repeat the test five times to assess the reproducibility of the assay.
RESULTS
Subtype-specific primer design and SSP analysis.
Subtype-specific primer design is the key problem, as well as the basis for this assay. To search for these primers is actually to search for accurate oligonucleotide sequences with SSPs. Keeping the SSPs at the 3′ end of the primer was found to be important to increase the assay specificity. In SSP analysis, we used subtype consensus sequences downloaded from the Los Alamos National Laboratory, because they, being derived from a lot of same subtype sequences, were more representative than single or several ones. In the sequence alignments of all HIV-1 subtypes and CRFs (Fig. 2), we could easily find the SSPs. Based on the sequence analysis, subtype-specific primers with SSPs for subtype B and C used in the present study were very specific and mismatched with other subtypes at the 3′-terminal bases (Fig. 2). Nevertheless, the sequences of subtype A, CRF01-AE, CRF02-AG, CRF03-AB, and CRF04-cpx are so similar that it is hard to find strict SSPs for CRF01-AE. From the sequence analysis, the CRF01-AE-specific primers used in the present study may discriminate CRF01-AE from subtype A and CRF03-AB due to a mismatch with them; however, they may not work on CRF02-AG and CRF04-cpx because the forward primer matches well with CRF02-AG and CRF04-cpx, and only the fourth base at the 3′ end of the reverse primer mismatches with CRF02-AG and CRF04-cpx (Fig. 2). Fortunately, none case of CRF02-AG and CRF04-cpx infections were reported in China. Other SSPs for HIV-1 subtypes B, C, CRF01-AE, CRF07-BC, and CRF08-BC in gag region are listed in Table 3.
FIG. 2.
Nucleotide sequence alignments of HIV-1 subtypes and CRFs. The name of the subtype or CRF labeled with “CON_X” means consensus of subtype X or CRF X, e.g., “CON_B” stands for consensus of subtype B strains. Some subtypes or CRFs are only represented by one sequence, e.g., 05.VI1310, meaning CRF05-VI1310. The sequences are not continuous, numbered according to the HXB2. The positions for subtype-specific primers are underlined.
TABLE 3.
SSP analysis in gag region
| Subtype | Equivalent HXB2 gag position of SSPa |
|---|---|
| B | 91T, 100A, 136G, 150G, 173G, 264G, 321G, 645G, 838A, 839C, 1019C, 1153A, 1379A |
| C | 58A, 82C, 84C, 89T, 99C, 237C, 283C, 284G, 501G, 624G, 1074C, 1417G, 1497T |
| CRF01-AE | 117A, 121T, 258G, 261C, 488G, 792G, 964C, 966A, 1383A, 1385A |
| CRF07-BC | 58A, 82C, 84C, 89T, 99C, 237C, 283C, 284G, 332T, 838A, 839C, 1129C, 1293C, 1379A |
| CRF08-BC | 58A, 82C, 84C, 89T, 99C, 237C, 283C, 284G, 332T, 645G, 838A, 839C, 1074C, 1129C, 1293G, 1497T |
SSP positions used to design subtype-specific primers are underlined.
Nested multiplex PCR of reference strains.
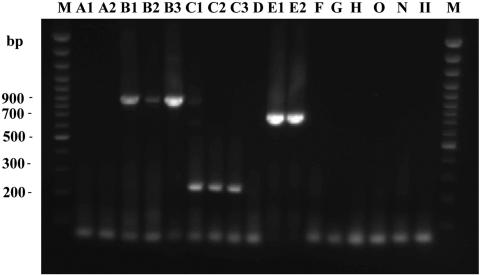
The nested multiplex PCR results of the HIV-1 group M subtype A, D, F, G, and H reference strains and the group N strain, group O strain and HIV-2 strain were all negative (Fig. 3). These samples were all reactive to universal primer sets, excluding the possibility of poor template quality or inappropriate template concentration. Subtype B (RL42, 93BR029, and pNL4-3), subtype C (92BR025 and 93IN905), CRF07-BC (97CN001), and CRF01-AE (CMU10 and 93TH023) samples were all positive for expected subtype PCR product sizes (subtype B, 900 bp; subtype C, 230 bp; subtype CRF01-AE, 740 bp). Of these, 93BR029 was identified as recombinant B/F, with a subtype B fragment in the gag region and a subtype F fragment in the env region. This assay is based on the gag region, so it was positive. Although the inner primer set matched the template well, mismatch by the outer primer set was probably responsible for the weakness of the 93BR029 band. Furthermore, the assay successfully discriminated between subtypes A and CRF01-AE.
FIG. 3.
Nested multiplex PCR of HIV reference isolates. Lanes: M, DNA Marker; A1, 92UG029; A2, 92RW008; B1, RL42; B2, 93BR029; B3, pNL4-3; C1, 92BR025; C2, 93IN905; C3, 97CN001; D, 94UG114; E1, CMU10; E2, 93TH023; F, 93BR020; G, JV1083; H, p90CF056.1; O, MVP5180; N, YBF30; II, HIV-2 UC2.
Subtype determination by nested multiplex PCR.
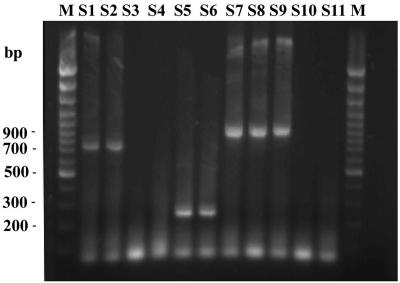
The agarose gel electrophoresis results of a representative sample amplified by nested multiplex PCR are shown in Fig. 4. In all, 69 subtype B, 117 subtype C, and 47 CRF01-AE samples were identified. Of 117 subtype C samples, 115 were found to be positive to CRF07- and CRF08-BC-specific primers. All 30 HIV-negative samples were negative in the nested multiplex PCR.
FIG. 4.
Agarose-gel electrophoresis of samples analyzed by nested multiplex PCR. Lanes: M, DNA Marker; S1, CQ2010 (CRF01-AE); S2, FJ9909 (CRF01-AE); S3, SC9712 (D); S4, JS11 (D); S5, SC0227 (C); S6, JS0121 (C); S7, HEN0217 (B′); S8, JS0127 (B′); S9, EC9005 (B); S10, SD20 (A); S11, JS17 (A).
Sequence-based phylogenetic analyses.
To validate the newly developed assay, we sequenced all of the samples and obtained a total of 252 HIV-1 sequences. Phylogenetic analyses of the sequences demonstrated that 74 samples belonged to subtype B (Euro-American B, 11; Thailand B, 63), 123 belonged to subtype C, 50 belonged to CRF01-AE, 3 belonged to subtype A, and 2 belonged to subtype D (the phylogenetic tree was not shown). The sequences amplified by subtype-specific primers are the same as the sequences from the universal primer set, excluding nonspecific amplification of similar-sized products. In subtype C samples, there are two samples closer to C.ETH2220 than CRF07-BC.97CN001 in genetic distance (not shown). The other 121 samples are more closely related to CRF07-BC.97CN001 and CRF08-BC.97CNGX6F, the representative strains of the CRF07- and CRF08-BC. According to the genetic distances, these subtype C samples may actually be considered CRF07- and CRF08-BC. This was confirmed by online subtyping as well (NCBI [http://www.ncbi.nlm.nih.gov/retroviruses/subtype/subtype.html]). We conclude that the 121 subtype C samples belong to CRF07- and CRF08-BC rather than pure subtype C strains in the gag region.
Assay validation by the phylogenetic analyses.
We compared the subtype results of the nested multiplex PCR with those of phylogenetic analyses (Table 4). The subtype determination by nested multiplex PCR corresponded well to the phylogenetic analyses. The subtype B-specific primer set can amplify both Thailand B (93.7% [59 of 63]) and Euro-American B (90.9% [10 of 11]) samples. The designated subtype sensitivities of this new assay were adequate: subtype B, 93.2% (69 of 74); subtype C, 95.1% (117 of 123); and CRF01-AE, 94.0% (47 of 50). In addition, the primer set for CRF07- and CRF08-BC detected 95.0% (115 of 121) of the samples correctly. Furthermore and most importantly, the intersubtype specificity of the assay was 100%, which means false-positive results were rare. Since the 30 HIV-negative samples were also found to be negative by this assay, the specificity of this assay for the presence of HIV is also 100%.
TABLE 4.
Comparison of phylogenetic analyses and nested multiplex PCR
| Subtype by phylogenetic analysis | Subtype by nested multiplex PCRa
|
||||||
|---|---|---|---|---|---|---|---|
| B
|
C
|
CRF01-AE
|
Total no. | ||||
| No. positive | No. negative | No. positive | No. negative | No. positive | No. negative | ||
| Euro-American B | 10 | 1 | 0 | 0 | 0 | 0 | 11 |
| Thailand B | 59 | 4 | 0 | 0 | 0 | 0 | 63 |
| C | 0 | 0 | 117 | 6 | 0 | 0 | 123 |
| CRF01-AE | 0 | 0 | 0 | 0 | 47 | 3 | 50 |
| A | 0 | 0 | 0 | 0 | 0 | 0 | 3 |
| D | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| Total | 69 | 5 | 117 | 6 | 47 | 3 | 252 |
The percent sensitivity values for subtypes B, C, and CRF01-AE and for the total subtypes are 93.2, 95.1, 94.0, and 94.3%, respectively. The percent specificities were 100% for all for subtypes.
Assay reproducibility.
In 31 samples tested for assay reproducibility, 6 samples were Euro-American B, 9 were Thailand B, 12 were subtype C, and 4 were CRF01-AE (Table 5). The average reproducibility based on the Table 5 data is 96.0% for subtype B samples (72 of 75), 96.7% for subtype C (58 of 60), and 95.0% for CRF01-AE (19 of 20).
TABLE 5.
Reproducibility of the nested multiplex PCR
| Expt | Reproducibility resultsa for subtype:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Euro-American B
|
Thailand B (B′)
|
C
|
CRF01-AE
|
|||||
| % | No. positive/n | % | No. positive/n | % | No. positive/n | % | No. positive/n | |
| 1 | 100 | 6/6 | 100 | 9/9 | 91.7 | 11/12 | 100 | 4/4 |
| 2 | 100 | 6/6 | 88.9 | 8/9 | 100 | 12/12 | 100 | 4/4 |
| 3 | 100 | 6/6 | 88.9 | 8/9 | 100 | 12/12 | 75.0 | 3/4 |
| 4 | 83.3 | 5/6 | 100 | 9/9 | 100 | 12/12 | 100 | 4/4 |
| 5 | 100 | 6/6 | 100 | 9/9 | 91.7 | 11/12 | 100 | 4/4 |
| Avg (totals) | 96.7 | 29/30 | 95.6 | 43/45 | 96.7 | 58/60 | 95.0 | 19/20 |
n = the total number of strains.
DISCUSSION
The purpose of the present study was to develop a subtype screening tool for subtypes B, C, and CRF01-AE. The comparison between sequence-based phylogenetic analyses and the nested multiplex PCR clearly illustrates the high intersubtype specificity (100%), HIV specificity (100%) and designated subtype sensitivity of this new assay (subtype B, 93.2%; subtype C, 95.1%; CRF01-AE, 94.0%; and CRF07- and CRF08-BC, 95.0%) with satisfactory reproducibility (subtype B, 96.0%; subtype C, 96.7%; CRF01-AE, 95.0%). Detection of reference isolates also confirms the credibility of this new assay.
However, occasional nonspecific bands do appear. Weak bands appeared near the position of the subtype B-specific band in several subtype C samples. False-priming in several subtype C samples is likely responsible. After measures to reduce nonspecific reactions were taken, such as “hot-start” and “touchdown” techniques, the nonspecific bands were generally no longer visible. In addition, not only RNA samples from the plasma but also DNA samples from the peripheral blood mononuclear cells can be used in this assay. The subtype-specific primers in the second-round PCR could also be performed separately instead of being combined in one tube.
In the present study, the subtype determination was based on the size of the PCR products. Thus, the factors that influence the size of fragments, such as gene deletions or insertions, typically occurring with HIV, could interfere with the result interpretation (16). In our experiments, only one sample had two strong bands. One band was found near the position of subtype C, and the other was located at the position of subtype B. These two bands were each purified and DNA sequenced. The sequence analysis indicates that this sample belongs to subtype B. An ∼600-bp deletion was identified between the subtype B-specific primer pairs leading to bands representing both the deleted and nondeleted template forms. This pattern was unique to this sample. Because the target fragments we amplified are very short, gene deletion or insertion occurring exactly in this region is unusual. Insertions or deletions of the template DNA should only lead to three possible scenarios. (i) When DNA deletion or insertion happens out of the target region, it imposes no effect on the result. (ii) When deletions or insertions span upper, lower, or both primers, the result would be negative. (iii) When deletions or insertions occur between the upper and lower primers, the target PCR products would extend or shorten. Only the third condition will influence the result interpretation. Thus, if a strong band is found near but not on the subtype-specific position, the possibility of gene deletion or insertion should be considered. DNA sequencing is necessary for further subtype determination.
Globally, the prevalence of HIV infection is increasing rapidly. The variation among HIV strains will increase as additional mosaic strains are discovered. Characterizing HIV is important, but the currently available assays for HIV subtyping are laborious. Many scientists are developing new simple and rapid assays for HIV subtyping, including a solid phase plate assay (4) and a DNA enzyme immunoassay method for HIV subtyping (14). The sensitivity and specificity of these assays are satisfactory. Yagyu developed a PCR subtyping method only differentiating subtype B and CRF01-AE, but the specificity of that assay was found to be lacking (23). Recently, a real-time isothermal amplification assay has been developed to distinguish HIV-1 subtypes A, B, C, CRF01-AE, and CRF02-AG (2). Another real-time PCR with multiregion amplification and fluorescent, subtype-specific probes, the multiregion hybridization assay, is in fact a robust tool for HIV-1 subtyping, not only for one gene region genotyping but also for recombination detection (6). However, low sensitivity, local strain match, and high cost are still the barriers for its application in developing countries.
Our nested multiplex PCR assay is simpler, faster, and more practical for use in developing countries such as China. The specificity and sensitivity are the most important parameters for evaluation of any assay. Subtype-specific primers, based on the SSP analysis, were carefully designed. The use of two subtype-specific primers for this assay should be an improvement over other assays that use only one subtype-specific probe. The idea of the present study can also be applied in the other subtype-specific primer design. Because the selected samples in this panel were biased toward subtypes Thailand B, C, and CRF01-AE, additional subtype samples such as, for example, subtypes A, Euro-American B, CRF02-AG, and CRF04-cpx, are needed to further evaluate this assay if it is to be used outside of China.
In summary, we have developed a simpler and more rapid subtype screening assay for HIV-1 subtypes B, C, CRF01-AE’CRF07-BC, and CRF08-BC in China. This assay has potential applications in HIV laboratories located not only in China but also in other countries around the world.
Acknowledgments
We thank the AIDS Research and Reference Reagent Program, National Institutes of Health, for providing the reference strains. We thank the staff of the provincial Center for Disease Control and Prevention in China for supplying samples. We also thank Jonathan Li and Mimi Trager from the University of California, San Francisco, and Ray Chen from the National Institute of Allergy and Infectious Disease, National Institutes of Health, for proofreading.
This study was supported by China 973 National Key Project G1999054107 and National Outstanding Youth Grant 39925030.
REFERENCES
- 1.Agwale, S. M., K. E. Robbins, L. Odama, A. Saekhou, C. Zeh, A. Edubio, O. M. Njoku, N. Sani-Gwarzo, M. F. Gboun, F. Gao, M. Reitz, D. Hone, T. M. Folks, D. Pieniazek, C. Wambebe, and M. L. Kalish. 2001. Development of an env gp41-based heteroduplex mobility assay for rapid human immunodeficiency virus type 1 subtyping. J. Clin. Microbiol. 39:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Baar, M. P., E. C. Timmermans, M. Bakker, E. de Rooij, B. van Gemen, and J. Goudsmit. 2001. One-tube real-time isothermal amplication assay to identify and distinguish human immunodeficiency virus type 1 subtypes A, B, and C and circulating recombinant forms AE and AG. J. Clin. Microbiol. 39:1895-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delwart, E. L., E. G. Sharper, J. Louwagie, F. E. McCutchan, M. Grez, H. Rubsamen-Waigmann, and J. I. Mullins. 1993. Genetic relationships determined by a DNA heteroduplex mobility assay: analysis of HIV-1 env genes. Science 262:1257-1261. [DOI] [PubMed] [Google Scholar]
- 4.González-Villaseor, L. I., K. Wu, and L. Yang. 2000. A solid phase plate assay for HIV-1 genotyping. Mol. Cell. Probes 14:137-147. [DOI] [PubMed] [Google Scholar]
- 5.Graf, M., Y. Shao, Q. Zhao, T. Seidl, J. Kostler, H. Wolf, and R. Wagner. 1998. Cloning and characterization of a virtually full-length HIV type 1 genome from a subtype B′-Thai strain representing the most prevalent B-clade isolate in China. AIDS Res. Hum. Retrovir. 14:285-288. [DOI] [PubMed] [Google Scholar]
- 6.Hoelscher, M., W. E. Dowling, E. Sanders-Buell, J. K. Carr, M. E. Harris, A. Thomschke, M. L. Robb, D. L. Birx, and F. E. McCutchan. 2002. Detection of HIV-1 subtypes, recombinants, and dural infections in east Africa by a multi-region hybridization assay. AIDS 16:2055-2064. [DOI] [PubMed] [Google Scholar]
- 7.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 8.Korber, B., G. Myers. 1992. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res. Hum. Retrovir. 8:1549-1560. [DOI] [PubMed] [Google Scholar]
- 9.Loussert-Ajaka, I., M. L. Chaic, B. Korber, F. Letournearu, E. Gomas, E. Allen, T. D. Ly, F. Brun-Vezinet, F. Simon, and S. Saragosti. 1995. Variability of human immunodeficiency virus type 1 group O strains isolated from Cameroonian patients living in France. J. Virol. 69:5640-5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McClutchan, F. E., J. K. Carr, D. Murphy, S. Piyasirisilp, F. Gao, B. Hahn, X.-F. Yu, C. Beyrer, and D. L. Birx. 2002. Precise mapping of recombination breakpoints suggests a common parent of two BC recombinant HIV type 1 strains circulating in China. AIDS Res. Hum. Retrovir. 18:1135-1140. [DOI] [PubMed] [Google Scholar]
- 11.Ou, C.-Y., C. A. Ciesielski, G. Myers, C. I. Bandea, C.-C. Luo, B. T. Korber, J. I. Mullins, G. Schochetman, R. L. Berkelman, A. N. Economou, J. J. Witte, L. J. Furman, G. A. Satten, K. A. Maclnnes, J. W. Curran, H. W. Jaffe, laboratory investigation group and epidemiology investigation group. 1992. Molecular epidemiology of HIV transmission in a dental practice. Science 256:1165-1171. [DOI] [PubMed] [Google Scholar]
- 12.Pan, P., C. Zeng, X. Fan, J. Yao, H. Xing, Y. Feng. Y. Shao. 1999. Identification and characterization of human immunodeficiency virus type 1 subtype F env gene in China. Chin. J. Virol. 15:97-101. [Google Scholar]
- 13.Piyasirisilp, S., F. E. McCutchan, J. K. Carr, E. Sanders-Buell, W. Liu, J. Chen, R. Wagner, H. Wolf, Y. Shao, S. Lai, C. Beyrer, and X. Yu. 2000. A recent outbreak of human immunodeficiency virus type 1 in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J. Virol. 74:11286-11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plantier, J. C., L. Vergne, F. Damond, S. MBoup, E. MPoudi-NGole, L. Buzelay, I. Farfara, D. Brand, M. Peeters, F. Brun-Vezinet, E. Delaporte, and F. Barin. 2002. Development and evaluation of a DNA enzyme immunoassay method for env genotyping of subtypes A through G of human immunodeficiency virus type 1 group M, with discrimination of the circulating recombinant forms CRF01-AE and CRF02-AG. J. Clin. Microbiol. 40:1010-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roux, K. H. 1998. Optimization of PCR, p. 33-41. In C. W. Dieffenbach and G. S. Dveksler (ed.), PCR primer: a laboratory manual. Science Publishing House, Beijing, Peoples Republic of China.
- 16.Sanchez, G., X. Xu, J. C. Chermann, and I. Hirsch. 1997. Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J. Virol. 71:2233-2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao, Y., F. Zhao, W. Yang, Y. Zhang, and X. Gong. 1999. The identification of recombinant HIV-1 strains in IDUs in southwest and northwest China. Chin. J. Exp. Clin. Virol. 13:109-112. [PubMed] [Google Scholar]
- 18.Sharp, P. M., D. L. Robertson, F. Gao, and B. H. Hahn. 1994. Origins and diversity of human immunodeficiency viruses. AIDS 8:S27-S42. [Google Scholar]
- 19.Su, L., M. Graf, Y. Zhang, H. von Briesen, H. Xing, J. Kostler, H. Melzl, H. Wolf, Y. Shao, and R. Wagner. 2000. Characterization of a virtually full-length human immunodeficiency virus type 1 genome of a prevalent intersubtype (C/B′) recombinant strain in China. J. Virol. 74:11367-11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su, L., H. Xing, H. Yang, X. Luo, and Y. Shao. 1997. Identification and characterization of HIV-1 subtype A in China. Chin. J. Virol. 13:265-268. [Google Scholar]
- 21.Xing, H., P. Pan, Y. Feng, X. Lin, J. Yu, X. Gong, Y. Dai, and Y. Shao. 2000. Viral sequence analysis of HIV-1 and HIV-2 dural infection firstly found in China. Chin. J. Virol. 16:111-115. [Google Scholar]
- 22.Xing, H., G. Qin, Y. Feng, G. Liu, P. Pan, X. Fan, and Y. Shao. 1999. Identification and characterization of HIV-1 subtype D strain in China. Chin. J. Exp. Clin. Virol. 13:157-162. [PubMed] [Google Scholar]
- 23.Yagyu, F., Y. Ikeda, K. Ariyoshi, W. Sugiura, S. Wongkhomthong, M. Masuda, and H. Ushijima. 2002. Differentiation of subtypes B and E of human immunodeficiency virus type 1 by polymerase chain reaction using novel env gene primers. J. Virol. Methods 101:11-20. [DOI] [PubMed] [Google Scholar]