Abstract
A DNA microarray for detection of Campylobacter spp. was recently developed and applied to detect Campylobacter spp. directly from chicken feces. Sixty-five pooled chicken cloacal swab samples from 650 individual broiler chickens were included in the study. The results of Campylobacter sp. detection obtained with DNA microarrays were compared to those obtained by conventional culture and gel electrophoresis. By conventional culture, 60% of the samples were positive for either Campylobacter jejuni or Campylobacter coli. By PCR and capillary electrophoresis, 95% of the samples were positive for Campylobacter spp., whereas with DNA microarrays all samples were positive for Campylobacter spp. By application of DNA microarray analysis, the isolates in 4 samples (6%) could not be identified to the species level, whereas by PCR-capillary electrophoresis, the isolates in 12 samples (19%) remained unidentified. Interestingly, PCR-capillary electrophoresis analysis revealed that two (3%) of the samples were positive for both C. jejuni and C. coli, while DNA microarray analysis revealed that nine (14%) of the samples were positive for both species. Of 65 samples, 2 samples were identified to contain C. coli by conventional culture but were positive for C. jejuni by both PCR-capillary electrophoresis and DNA microarray analysis. The discrepancy between the methods is discussed.
Campylobacter is the most common cause of intestinal disorders in humans in many industrial countries. An incidence of 86 campylobacteriosis cases per 100,000 inhabitants in 2001 (3) makes Campylobacter infection the most common food-borne pathogen in Denmark.
Poultry and poultry products are considered important sources of human campylobacteriosis and play important roles in disease transmission (5, 6, 8, 11). In Denmark, a systematic sampling procedure for continued evaluation of the prevalence rates and epidemiology of Campylobacter in poultry has been in place since 1998 (3). At the retail level, 30 to 40% of poultry at slaughter have been reported to be contaminated with Campylobacter (31). Isolation and identification of Campylobacter by the conventional culture method are laborious due to the slow growth rate, the lack of phenotypic differences in the bacteria, and often, the failure to identify Campylobacter to the species level by the available culture methods. There is a need for the development of a sensitive, rapid method for Campylobacter detection and identification to the species level.
Several PCR assays have successfully been applied to the detection of Campylobacter spp. in water (15, 26), some dairy products (9, 12, 29, 32), and chicken litter (13). The PCR method allows detection not only of viable bacteria but also of noncultivable forms of Campylobacter (12, 32). A multiplex PCR assay suitable for mass screening to detect Campylobacter directly from chicken feces has been developed (1). Agarose gel electrophoresis is often used for the PCR assays. Gel electrophoresis has a number of drawbacks, however, such as a poor ability to differentiate PCR products of approximately the same size in a multiplex PCR, and the ethidium bromide used to stain the DNA is a carcinogen.
Recently, a DNA microarray suitable for detection of Campylobacter at the species level was developed (14). In this study, the method was applied to detect Campylobacter directly from chicken fecal samples. Six hundred fifty cloacal swab specimens from broiler chickens were collected at slaughter and were tested in pools of 10 by conventional culture methods, PCR-capillary electrophoresis analysis, and DNA microarray analysis. The results were compared and are discussed here.
MATERIALS AND METHODS
Bacterial reference strains.
In this study Campylobacter jejuni CCUG 11284 (Culture Collection of the University of Gothenburg [CCUG], Gothenburg, Sweden) and Campylobacter coli CCUG 11283 were used for isolation of chromosomal DNA and were used as positive controls in the multiplex PCR.
Cloacal swab samples.
Cloacal swab samples were collected at random from 650 individual broiler chickens representing 65 broiler flocks at slaughter. The swabs were transferred to the laboratory in screw-cap centrifuge tubes with 15 ml of brain heart infusion transport medium (brain heart infusion broth [37 g/liter; Difco-BD, Brøndby, Denmark], 5% sterile defibrinated calf blood, and 0.5% agar [Oxoid, Greve, Denmark] [pH 7.4]). On arrival at the laboratory, the swabs were immediately subjected to laboratory processing. Ten swabs from each flock were transferred to 3 ml of sterile water and left at room temperature for 10 to 20 min to release the bacteria. The suspensions of feces and bacteria were used directly for Campylobacter detection.
Isolation and identification of Campylobacter spp. by conventional culture methods.
Ten microliters of the bacterial and fecal suspension was spread on the surface of a charcoal cefoperazone deoxycholate agar plate (CM 739 [Oxoid] with cefoperazone selective supplement SR 155E). The plate was incubated under microaerophilic conditions (6% O2, 6% CO2, 4% H2, and 84% N2) at 42°C for 48 h. A single colony suspected of being Campylobacter was examined morphologically by phase-contrast microscopy and was further purified on blood agar plates (Blood Agar Base No. 2 [Oxoid] supplemented with 5% sterile defibrinated calf blood). All the isolates were characterized to the species level by their catalase reactions, abilities to hydrolyze hippurate and indoxylacetate, and susceptibilities to nalidixic acid and cephalothin, by standard procedures (22, 23). The isolates were subsequently stored at −80°C in brain heart infusion broth with 15% glycerol until further investigations.
DNA techniques.
Chromosomal DNA from the Campylobacter reference strains was extracted from 24-h blood agar plate cultures by using the QIAamp kit (Qiagen, Hilden, Germany). The DNA was eluted in 200 μl of preheated (65°C) sterile water. The DNA concentrations were measured on a spectrophotometer (Ultrospec 2000; Pharmacia Biotech, Cambridge, United Kingdom), and the DNA was stored at −20°C.
Chromosomal DNA from broiler chicken fecal samples was extracted by a previously described method (18) by using a King Fischer magnetic particle processor (Labsystems, Vantaa, Finland).
Oligonucleotide sequences.
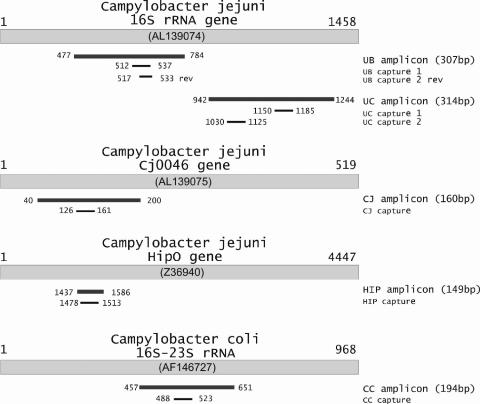
The sequences of the PCR primers, the oligonucleotide capture probes, and an oligonucleotide used as a positive hybridization control were described previously (14) and are summarized in Fig. 1 and Table 1. Briefly, two primers, UB-FW and UB-RW, were selected and used to amplify a 307-bp fragment from the bacterial 16S rRNA gene (1). Within the amplified region, two capture probes (universal bacterial [UB] capture probes 1 and 2 [UB capture 1 and 2, respectively]) were designed to target the bacterial 16S rRNA amplicons on the DNA microarray. The UB capture probes were located at nucleotides 512 to 537 (UB capture 1) and at nucleotides 517 to 533 (UB capture 2) on the complementary sequence from the start of the gene. This area was also used as a positive control for both multiplex PCR amplification and on the DNA microarray to confirm the presence of bacterial DNA (1). Two primers, UC-FW and UC-RW, were designed on the basis of the published sequences (17). These primers were used to specifically amplify a 314-bp fragment located at nucleotides 948 to 1244 of the 16S rRNA gene of the Campylobacter genus. Two 35-mer universal Campylobacter (UC) capture probes were designed in order to target the Campylobacter genus 16S rRNA amplicon on the DNA microarray. UC capture 1 probe was located at nucleotides 1090 to 1125, whereas UC capture 2 probe was located at nucleotides 1150 to 1185 of the 16S rRNA gene. The C. jejuni-specific PCR was performed on the basis of amplification of two C. jejuni-specific genes, the Cj0046 and the hippuricase (hipO) genes. Two primers, namely, primers CJ-FW and CJ-RW, which target the Cj0046 gene and which have been described previously (33), while two other primers, namely, primers HIP-FW and HIP-RW, which target the hipO gene, were selected for amplification of the hipO gene (14, 27a). A 35-mer capture probe, namely, the CJ capture probe, which is specific for C. jejuni, was located at nucleotides 126 to 161 of the C. jejuni Cj0046 gene (28); and the HIP capture probe targeted the sequences of the C. jejuni hippuricase gene at nucleotides 1478 to 1513. Finally, a 35-mer capture probe (the CC capture probe), located at nucleotides 488 to 523 of the C. coli 16S-23S rRNA intergenic spacer region, as described by O'Sullivan et al. (24), was used to detect a specific PCR amplicon derived from the C. coli 16S-23S rRNA intergenic spacer region at nucleotides 457 to 651. In order to identify suitable hybridization capture probes, both strands of the PCR products were labeled with cyanine 5 (Cy5)-labeled primers in the PCRs.
FIG. 1.
Schematic representation of the sequences used for specific PCR amplification. Sequence names and accession numbers in parentheses refer to the National Center for Biotechnology Information GenBank entry. The first and the last nucleotide positions of the sequences, the amplicons obtained, and the capture probe areas used are shown.
TABLE 1.
PCR primers, primer and capture probe sequences, sizes of PCR amplicons, and references used in this studya
| Primer name | Sequence | PCR amplicon size | Reference(s) | Tm (°C) |
|---|---|---|---|---|
| UB-FW | 5′-Cy5-GCTAACTCCGTGCCAGCAGCCGCGG-3′ | 307 bp | Bang et al. (1) | 86.9 |
| UB-RW | 5′-Cy5-GGGCGTGGACTACCAGGGTATC-3′ | 65.8 | ||
| UC-FW | 5′-Cy5-GCGAAGAACCTACCYGGRCTTGATA-3′ | 314 bp | Linton et al. (17); this study | 77.3 |
| UC-RW | 5′-Cy5-TCGCGRTATTGCGTCTCATTGTATATG-3′ | 75.9 | ||
| CC-FW | 5′-Cy5-GTTAAGAGTCACAAGCAAGT-3′ | 194 bp | O'Sullivan et al. (24); Keramas et al. (14) | 53.7 |
| CC-RW | 5′-Cy5-CTAAAAATATCTAAACTAAGTCG-3′ | 51.7 | ||
| CJ-FW | 5′-Cy5-CAAATAAAATTAGAGGTAGAATGT-3′ | 160 bp | Winters et al. (33); Keramas et al. (14) | 58.1 |
| CJ-RW | 5′-Cy5-CCATAAGCACTAGCTAGCTG-3′ | 57.3 | ||
| HIP-FW | 5′-Cy5-GTACTGCAAAATTAGTGGCG-3′ | 149 bp | Slater et al., (27a); Keramas et al. (14) | 61.1 |
| HIP-RW | 5′-Cy5-GCAAAGGCAAAGCATCCATA-3′ | 55.3 | ||
| POS CTRL | 5′-Cy5-CTAAGATTTTCTGCATAGCATTAAT-3′ | 25 nt | Keramas et al. (14) | 61.5 |
| UB CAP 1 | NH2-C6-AGTGATTCCGAGTAACGCTTGCACC | 25 nt | Keramas et al. (14) | 75.2 |
| UB CAP 2 | NH2-C6-CGGAGGGTGCAAGCGTTACTCGGAATCACTGGGC | 34 nt | This study | 88.3 |
| UC CAP 1 | NH2-C6-ACGTATTTAGTTGCTAACAGTTNGGCTGAGCACTC | 35 nt | Keramas et al. (14) | 81.0 |
| UC CAP 2 | NH2-C6-AGGAAGGTGTGGACGACGTCAAGTCATCATGGCC | 34 nt | This study | 89.2 |
| CJ CAP | NH2-C6-GATGAGCAAGGAGAAGGAGCTATAGGTTTAGGCGT | 35 nt | Keramas et al. (14) | 82.7 |
| HIP CAP | NH2-C6-ACGCCTGTTTTTCCTATTTCCTCATAAACCTCATA | 35 nt | Keramas et al. (14) | 78.6 |
| CC CAP | NH2-C6-TTGAGTTTTATCCTTTAACAAGTCCTGTAAAATTG | 35 nt | Keramas et al. (14) | 75.5 |
| POS and NEG CTRL CAP | NH2-C6-ATTAATGCTATGCAGAAAATCTTA | 24 nt | Keramas et al. (14) | 61.5 |
Abbreviations: Tm, melting temperature; nt, nucleotides; CAP, capture; POS, positive; NEG, negative; CTRL, control.
PCR conditions.
PCR was performed in a PTC-200 thermal cycler (MJ Research Inc., Waltham, Mass.) for 28 cycles, with each cycle consisting of 94°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 15 s. All PCR mixtures (25 μl) contained 0.2 mM (each) dATP, dCTP, dGTP, and dTTP; 2.5 mM MgCl2; 0.25 μl (200 nmol) of each primer (DNA Technology, Aarhus, Denmark) in a single-reaction PCR mixture; and 1× Taq DNA polymerase buffer containing 10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl (pH 8.3), and 0.5 μl (2.5 U) of Taq DNA polymerase (Roche, Hvidovre, Denmark). Multiplex PCR was performed by using individually adjusted concentrations of each primer to achieve the maximum PCR product; 150 nmol of universal bacterial forward and reverse primers, 300 nmol of universal Campylobacter forward and reverse primers, 500 nmol of C. jejuni-specific forward and reverse primers, 300 nmol of C. coli-specific forward and reverse primers, and 100 nmol of C. jejuni hippuricase-specific forward primer and 200 nmol of Campylobacter hippuricase-specific reverse primer.
DNA analysis.
All PCR products were analyzed by capillary electrophoresis in a 2100 Bioanalyzer (Agilent Technologies, Palo Alto, Calif.). DNA fragments up to 500 bp in length could be analyzed with DNA 500 chips.
DNA microarray preparation.
Prior to microarray printing, the capture probes were diluted in phosphate buffer (150 mM) to a final concentration of 30 μM. The capture probes were spotted in duplicate by using a Q-Array spotting robot (Genetix, New Milton, United Kingdom).
The printed microarrays were baked for 4 h at 80°C and blocked for 5 min in 60 mM sodium borohydrate (Sigma, Vallensbæk, Denmark) in phosphate-buffered saline (Sigma) with 25% ethanol (Merck, VWR, Roskilde, Denmark). The microarrays were then washed for 2 min in prewarmed 95°C distilled water and once for 1 min in 99.9% ethanol (Merck) at room temperature to remove unbound oligonucleotides. The microarrays were dried under a gentle stream of nitrogen and were stored until use. Each glass slide consisted of three individual microarrays framed with a 25-μl Geneframe (AB-0576; Abgene, Epsom, United Kingdom), which provided individual reaction chambers.
Hybridization process.
For the hybridizations, 12.5 μl of the PCR product was mixed with 12.5 μl of PerfectHyb Plus hybridization buffer (Sigma). Additionally 0.5 μl (corresponding to 0.025 pmol) of a Cy5-labeled oligonucleotide (which was used as a positive hybridization control in each test) was added prior to addition to the microarray. The hybridization mixture was denatured at 95°C for 2 min and then cooled to the hybridization temperature of 50°C and transferred to the microarray. Hybridization was performed for 2 h in a hybridization oven (Hybaid, Shaken-stack; Thermo Electron, Milford, Mass.). After hybridization, the Geneframes were removed and the microarrays were washed once with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.5% sodium dodecyl sulfate for 5 min, followed by a final wash with 0.1× SSC for 1 min with vigorous agitation in a microarray wash station (Telechem, Sunnyvale, Calif.). The microarrays were dried under a gentle stream of nitrogen before they were scanned. The microarrays were stored in a light-tight box to prevent photo bleaching.
Optical readout with scanner.
The microarrays were scanned with a laser (excitation, 633 nm; emission, 670 nm; ScanArray Lite; Packard Biosciences, Billerica, Mass.) and analyzed with OptiQuant software (Packard). For a result to be considered positive, the fluorescence value was required to be at least three times the average interassay coefficient of variation (CV) above the average background signal, according to the definition of Niessner (21). Briefly, the average CV for all capture probe duplicates in an array was measured and compared to the average background signal measured at different spots in the area surrounding the specific capture probes. Only signals with values at least three times higher than the average background values were considered positive.
RESULTS
Isolation and identification of Campylobacter spp. by conventional culture methods.
Of 65 samples tested, 39 (60%) samples were positive for Campylobacter, whereas 26 (40%) samples were negative for Campylobacter. Identification to the species level revealed that 35 (54%) isolates were identified as C. jejuni and 4 (6%) were identified as C. coli. The results of the detection of Campylobacter spp. by culture, PCR, and microarray analysis are presented in Table 2.
TABLE 2.
Results of culture, electrophoresis, and microarray analysisa
| Sample no. | Culture results
|
Electrophoresis results
|
Microarray results
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CJ | UB | UC | CC | CJ | HIP | UB | UC | CC | CJ | HIP | |
| 1 | − | + | + | + | − | + | + | + | + | − | + | + |
| 2 | − | − | + | − | − | − | − | + | +d | − | +c | +d |
| 3 | − | + | + | + | − | + | + | + | + | − | + | + |
| 4 | − | + | + | + | − | + | + | + | + | − | + | + |
| 5 | − | + | + | + | − | + | + | + | + | +c | + | + |
| 6 | − | + | + | + | − | − | − | + | + | +c | +d | +d |
| 7 | − | − | + | + | − | + | − | + | + | − | +b | +d |
| 8 | − | − | + | + | − | + | − | + | + | − | +b | +d |
| 9 | − | − | + | + | − | + | + | + | + | − | +b | + |
| 10 | − | − | + | + | − | + | + | + | + | − | +b | + |
| 11 | − | − | + | + | − | + | + | + | + | − | +b | + |
| 12 | − | − | + | + | − | + | + | + | + | +c | + | + |
| 13 | − | + | + | + | − | + | + | + | + | − | + | + |
| 14 | − | + | + | + | − | + | + | + | + | − | + | + |
| 15 | − | + | + | + | − | + | + | + | + | − | + | + |
| 16 | − | + | + | + | − | + | + | + | + | − | + | + |
| 17 | − | − | + | + | − | − | − | + | + | +d | − | − |
| 18 | − | − | + | + | − | − | − | + | + | − | − | − |
| 19 | − | − | + | + | − | − | − | + | + | +c | − | − |
| 20 | − | + | + | + | − | + | + | + | + | − | + | + |
| 21 | − | + | + | + | − | + | + | + | + | − | + | + |
| 22 | − | + | + | + | − | + | + | + | + | − | + | + |
| 23 | − | + | + | + | − | + | + | + | + | − | + | + |
| 24 | − | + | + | + | − | + | + | + | + | − | + | + |
| 25 | − | + | + | + | − | + | + | + | + | − | + | + |
| 26 | − | + | + | + | − | + | + | + | + | − | + | + |
| 27 | + | − | + | + | + | + | − | + | + | + | +b | +d |
| 28 | − | + | + | + | − | + | − | + | + | +c | + | +d |
| 29 | − | − | + | + | + | + | − | + | + | + | + | +d |
| 30 | − | − | + | + | − | + | + | + | + | − | + | + |
| 31 | − | − | + | + | − | + | + | + | + | − | + | + |
| 32 | + | − | + | + | − | − | − | + | + | +c | − | − |
| 33 | − | − | + | + | − | − | − | + | + | +d | − | − |
| 34 | − | − | + | + | − | − | − | + | + | +d | − | − |
| 35 | − | + | + | + | − | + | + | + | + | − | + | + |
| 36 | − | + | + | + | − | + | + | + | + | − | + | + |
| 37 | − | + | + | + | − | + | + | + | + | − | + | + |
| 38e | + | − | + | + | − | + | + | + | + | − | + | + |
| 39 | − | + | + | + | − | + | + | + | + | +c | + | + |
| 40 | − | + | + | − | − | − | − | + | +d | − | +d | +d |
| 41 | − | + | + | − | − | − | − | + | +d | − | +d | +d |
| 42 | − | + | + | + | − | + | + | + | + | − | + | + |
| 43 | − | − | + | + | − | − | − | + | + | +d | − | − |
| 44 | − | − | + | + | + | − | − | + | + | + | − | − |
| 45 | − | − | + | + | + | − | − | + | + | + | − | − |
| 46 | − | + | + | + | − | + | + | + | + | − | + | + |
| 47 | − | − | + | + | + | − | − | + | + | + | − | − |
| 48 | − | − | + | + | − | + | − | + | + | − | + | +d |
| 49 | − | − | + | + | − | − | − | + | + | − | +d | +d |
| 50e | + | − | + | + | − | + | + | + | + | − | + | + |
| 51 | − | − | + | + | − | + | + | + | + | − | + | + |
| 52 | − | − | + | + | + | − | − | + | + | +b | − | − |
| 53 | − | + | + | + | − | + | + | + | + | − | + | + |
| 54 | − | + | + | + | − | + | + | + | + | − | + | + |
| 55 | − | + | + | + | − | + | + | + | + | − | + | + |
| 56 | − | + | + | + | − | + | + | + | + | − | + | + |
| 57 | − | + | + | + | − | + | + | + | + | − | + | + |
| 58 | − | − | + | + | − | − | − | + | + | − | − | − |
| 59 | − | − | + | + | − | − | − | + | + | − | − | − |
| 60 | − | + | + | + | − | + | + | + | + | − | + | + |
| 61 | − | + | + | + | − | + | + | + | + | − | + | + |
| 62 | − | + | + | + | − | + | + | + | + | +c | + | + |
| 63 | − | + | + | + | − | + | + | + | + | +c | + | + |
| 64 | − | + | + | + | − | + | + | + | + | − | + | + |
| 65 | − | − | + | + | − | − | − | + | + | − | − | − |
| 66 | × | × | + | + | − | + | + | + | + | − | + | + |
| 67 | × | × | + | + | + | + | + | + | + | + | + | + |
| 67 | × | × | + | + | + | + | + | + | + | + | + | + |
| 68 | × | × | + | + | + | − | − | + | + | + | − | − |
| 69 | × | × | − | − | − | − | − | − | − | − | − | − |
| 70 | × | × | − | − | − | − | − | − | − | − | − | − |
Multiplex PCR was performed with DNA isolated from 65 pooled cloacal fecal samples, 3 samples containing chromosomal reference DNA, and 2 negative control samples. The samples tested by multiplex PCR were analyzed by both capillary electrophoresis and hybridization to microarrays. +, positive signal; −, no detectable signal; ×, not done. The abbreviations in the table match the abbreviations for the primer pairs and capture probes used: UB, universal bacterial; UC, universal Campylobacter; CJ, C. jejuni Cj0046 gene; HIP, C. jejuni hippuricase gene; CC, C. coli.
Positive by microarray analysis and electrophoresis but not by culture.
Positive by microarray analysis but not by electrophoresis or culture.
Positive by microarray analysis but not by electrophoresis.
The positive signal was altered between culture and PCR-based analysis.
Analysis of PCR products amplified from fecal samples detected by capillary electrophoresis chip.
Multiplex PCR was performed with DNA templates isolated from the 65 pooled samples as well as DNA isolated from the C. jejuni and C. coli reference strains and analyzed by DNA electrophoresis with a microchip-based Agilent 2100 bioanalyzer (Fig. 2). All 65 (100%) samples were positive for bacterial DNA by using universal bacterial primers, and 62 (95%) of the samples were positive for Campylobacter by using the universal Campylobacter primers. Of the 65 samples, 46 (71%) samples were positive for the C. jejuni Cj0047 gene, whereas only 40 (62%) were positive for the hipO gene. Six (9%) samples were positive with the C. coli-specific primers (Table 2 and Table 3).
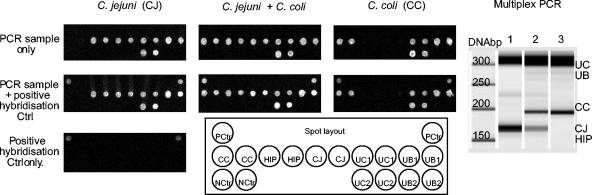
FIG. 2.
Microarray analysis. (Left panels) DNA microarrays were hybridized with amplicons generated by multiplex PCR. All capture probes specific for PCR amplicons or controls (ctrl) were printed in duplicate. Microarrays are shown hybridized with the product from the multiplex PCR (top three panels) or in combination with a positive hybridization control (middle three panels). The hybridization pattern of the positive hybridization control alone is shown in the lower panel (spot layout). A (negative) spotting buffer control (NCtr) was placed in the lower-left corner of the array. The spot diameters measure 150 μm. (Right panel) Gel-like image of capillary electrophoresis gel from analysis of multiplex PCR products with a 2100 BioAnalyzer and a DNA 500 chip (Agilent). The multiplex PCR was performed with DNA templates from C. jejuni (lane 1), C. coli (lane 3), or a mixture of DNA of both organisms combined (lane 2).
TABLE 3.
Comparison of strategies for detection of Campylobacter spp., C. jejuni, and C. colia
| Primer or probe specificity | No. (%) of samples positive by:
|
||
|---|---|---|---|
| Culture | Electrophoresis | Microarray analysis | |
| Universal bacterial | NAb | 65 (100) | 65 (100) |
| Universal Campylobacter | 39 (60) | 62 (95) | 65 (100) |
| C. jejunic | 35 (54) | 46 (71) | 51 (79) |
| C. jejuni (hippuricase)c | 40 (62) | 51 (79) | |
| C. colic | 4 (6) | 6 (9) | 19 (29) |
| C. jejuni and C. coli | 0 | 2 (3) | 9 (14) |
| Campylobacter negative | 26 (40) | 3 (5) | 0 (0) |
| Undetermined Campylobacter sp. | 12 (19) | 4 (6) | |
Sixty-five pooled samples were analyzed and the results were scored.
NA, not applicable.
Including samples positive for both C. jejuni and C. coli.
Two (3%) samples were positive with both of the primers specific for C. jejuni and C. coli, and 12 (19%) samples were positive with the universal Campylobacter primer but could not be further resolved by gel electrophoresis analysis. Three (5%) samples were negative for Campylobacter spp. (Tables 2 and 3).
Microarray analysis of PCR products from fecal samples.
The multiplex PCR products were further tested by hybridization on DNA microarrays (Fig. 2). All 65 (100%) fecal PCR samples were positive with the universal bacterial amplicon, as shown by positive signals for hybridization to at least one of the two universal bacterial capture probes. In addition, all 65 samples also showed positive signals for hybridization to the genus-specific Campylobacter amplicon by hybridization to at least one of the two UC capture probes. Furthermore, 51 (79%) samples showed positive signals for hybridization to Campylobacter jejuni (the C. jejuni Cj0046 gene [CJ amplicon] and hippuricase gene [HIP amplicon]). Nineteen (29%) samples showed a positive hybridization signal for C. coli (CC amplicon). Nine samples (14%) were positive for both C. jejuni and C. coli, and four samples (6%) were positive for a member of the Campylobacter genus but were not further identified (Tables 2 and 3).
DISCUSSION
DNA-based methods for the detection of bacteria, particularly Campylobacter spp., have a number of advantages compared to conventional culture: (i) shorter assay times; (ii) the capability to identify different forms of Campylobacter; and (iii) the ability to target different genes. The method provides a possible means to detect Campylobacter spp. directly from fecal samples and to identify them to the species level. Numerous PCR-based methods for the detection of Campylobacter that target various genes of Campylobacter, such as the flagellin genes (4), the mapA gene (28), the guanosine triphosphatase gene (30), and bacterial ribosomal genes (9, 25-27), have been described. However, most PCR methods that have been described require additional steps to remove PCR inhibitors and a time-consuming preculturing step due to the low number of organisms present in the sample and the various PCR inhibitors in fecal or environmental samples (27). In most PCR assays, agarose gel electrophoresis is often used as the final detection method. Detection of the organism is dependent on the staining of DNA by intercalating dyes; thus, low-copy-number amplicons may be difficult to detect, especially when increased levels of background staining or contaminating DNA products are present (7). Use of a combination of a multiplex PCR with an extra hybridization step, for example, an enzyme-linked immunosorbent assay or membrane hybridization, has been shown to increase both the specificities and the sensitivities of the methods (16, 24).
Recently, a DNA microarray suitable for use for the mass screening of Campylobacter directly from chicken fecal samples was developed (14). In Denmark samples are collected from broiler chickens as individual fecal swabs and 10 swabs are pooled for testing, as recommended by the Danish National Surveillance for Campylobacter in Broiler Production program. The bacteria are then eluted from the swabs and grown by conventional culture. In this study, detection by DNA microarray analysis was evaluated by comparison to detection by the conventional culture method and multiplex PCR. By the conventional culture methods, 39 of the 65 samples tested were positive for Campylobacter spp., and the species identification revealed that 35 of the samples contained C. jejuni and 4 contained C. coli (Table 2).
The multiplex PCR included five different primer sets targeting four Campylobacter gene fragments (Fig. 1). The multiplex PCR was shown to be highly specific, since no PCR amplicons were detected when the method was applied to DNA from a set of bacterial reference strains, including different Campylobacter species, Campylobacter-related bacteria, and other enterobacteria (14). By use of the multiplex PCR in combination with capillary electrophoresis for PCR amplicon analysis, 23 more samples were found to be positive for members of the genus Campylobacter, and 2 of these samples were positive for both C. jejuni and C. coli (Table 3). Interestingly, by application of DNA microarray analysis, all 65 samples were positive for Campylobacter and 9 were positive for both C. jejuni and C. coli. The results obtained in the present study, in which higher numbers of samples were found to be positive by PCR-capillary chip electrophoresis and DNA microarray analysis, resemble the results of a recent study comparing conventional culture to DNA probe-based PCR assays (19). However, it has been shown that PCR-based methods can detect not only viable Campylobacter cells but also noncultivable and dead Campylobacter cells (2, 20), an advantage in, e.g., food testing, in which noncultivable forms present a potential risk to humans. Recently, we showed that the use of the DNA microarray increased the sensitivity by a factor of 100 in comparison to the sensitivity of capillary chip electrophoresis with the same PCR samples (14).
Two samples (Table 2, samples 38 and 50) were found to be positive for C. coli by conventional culture but were positive for C. jejuni only when they were analyzed by PCR followed by capillary electrophoresis and microarray analysis. This could be interpreted in one of two ways: either the samples contained both C. coli and C. jejuni, of which only C. coli was detected by the conventional culture method, or the samples contained a hippuricase hydrolysis-negative C. jejuni strain, as described by Hani and Chan (10).
The application of the DNA microarray to the detection of Campylobacter showed that use of the method provides the possibility to decrease the assay time as well as decrease the sample volume, reagents, and materials needed. By the present approach, detection of Campylobacter directly from fecal swabs was performed in less than 3 h, with a hands-on time of only 20 min. The readout from the custom-made microarrays provided information on both the genus and the species detected. The simplicity and speed of the PCR-based assays make them highly applicable in the analysis of foods for the detection and identification of Campylobacter species and well suited for use for routine analysis and incorporation into an automated mass screening system.
Acknowledgments
We thank Mette Hansen, Elizabeth B. Sørensen, Annie Brandstrup, and Tuan Ming Nguyen (Danish Veterinary Institute, Aarhus, Denmark) for technical assistance.
This work was financially supported by the Danish Research Council (SUE project no. 2027-00-0008 and DABIC project no. 2014-00-0003), the Danish Broiler Meat Association, and the Danish Ministry of Food, Agriculture and Fisheries.
REFERENCES
- 1.Bang, D. D., K. Pedersen, and M. Madsen. 2001. Development of a PCR assay suitable for Campylobacter spp. mass screening programs in broiler production. J. Rapid Methods Aut. Microbiol. 9:97-114. [Google Scholar]
- 2.Beumer, R. R., J. de Vries, and F. M. Rombouts. 1992. Campylobacter jejuni non-culturable coccoid cells. Int. J. Food Microbiol. 15:153-163. [DOI] [PubMed] [Google Scholar]
- 3.Brøndsted, T., T. Hald, and F. Bager. 2001. Annual report on zoonoses in Denmark. The Danish Zoonosis Centre, Copenhagen, Denmark.
- 4.Comi, G., C. Pipan, G. Botta, L. Cocolin, C. Cantoni, and M. Manzano. 1996. A combined polymerase chain reaction and restriction endonuclease enzyme assay for discriminating between Campylobacter coli and Campylobacter jejuni. FEMS Immunol. Med. Microbiol. 16:45-49. [DOI] [PubMed] [Google Scholar]
- 5.Deming, M. S., R. V. Tauxe, P. A. Blake, S. E. Dixon, B. S. Fowler, T. S. Jones, E. A. Lockamy, C. M. Patton, and R. O. Sikes. 1987. Campylobacter enteritis at a university: transmission from eating chicken and from cats. Am. J. Epidemiol. 126:526-534. [DOI] [PubMed] [Google Scholar]
- 6.Evans, S. J. 1992. Introduction and spread of thermophilic campylobacters in broiler flocks. Vet. Rec. 131:574-576. [PubMed] [Google Scholar]
- 7.Fredricks, D. N., and D. A. Relman. 1999. Application of polymerase chain reaction to the diagnosis of infectious diseases. Clin. Infect. Dis. 29:475-486. [DOI] [PubMed] [Google Scholar]
- 8.Genigeorgis, C., and M. C. P. Hassuney. 1986. Campylobacter jejuni infection on poultry farms and its effect on poultry meat contamination during slaughter. J. Food Prot. 49:895-903. [DOI] [PubMed] [Google Scholar]
- 9.Giesendorf, B. A., W. G. Quint, M. H. Henkens, H. Stegeman, F. A. Huf, and H. G. Niesters. 1992. Rapid and sensitive detection of Campylobacter spp. in chicken products by using the polymerase chain reaction. Appl. Environ. Microbiol. 58:3804-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hani, E. K., and V. L. Chan. 1995. Expression and characterization of Campylobacter jejuni benzoylglycine amidohydrolase (hippuricase) gene in Escherichia coli. J. Bacteriol. 177:2396-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris, N. V., N. S. Weiss, and C. M. Nolan. 1986. The role of poultry and meats in the etiology of Campylobacter jejuni/coli enteritis. Am. J. Public Health 76:407-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hazeleger, W., C. Arkesteijn, B. Toorop, and R. Beumer. 1994. Detection of the coccoid form of Campylobacter jejuni in chicken products with the use of the polymerase chain reaction. Int. J. Food Microbiol. 24:273-281. [DOI] [PubMed] [Google Scholar]
- 13.Itoh, R., S. Saitoh, and J. Yatsuyanagi. 1995. Specific detection of Campylobacter jejuni by means of polymerase chain reaction in chicken litter. J. Vet. Med. Sci. 57:125-127. [DOI] [PubMed] [Google Scholar]
- 14.Keramas, G., D. D. Bang, M. Lund, M. Madsen, S. E. Rasmussen, H. Bunkenborg, P. Telleman, and C. B. V. Christensen. 2003. Development of a sensitive DNA microarray suitable for rapid detection of Campylobacter spp. Mol. Cell Probes 17:187-196. [DOI] [PubMed] [Google Scholar]
- 15.Kirk, R., and M. T. Rowe. 1994. A PCR assay for the detection of Campylobacter jejuni and Campylobacter coli in water. Lett. Appl. Microbiol. 19:301-303. [DOI] [PubMed] [Google Scholar]
- 16.Lawson, A. J., J. M. Logan, G. L. Neill, M. Desai, and J. Stanley. 1999. Large-scale survey of Campylobacter species in human gastroenteritis by PCR and PCR-enzyme-linked immunosorbent assay. J. Clin. Microbiol. 37:3860-3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linton, D., A. J. Lawson, R. J. Owen, and J. Stanley. 1997. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lund, M., A. Wedderkopp, M. Waino, S. Nordentoft, D. D. Bang, K. Pedersen, and M. Madsen. 2003. Evaluation of PCR for detection of Campylobacter in a national broiler surveillance programme in Denmark. J. Appl. Microbiol. 94:929-935. [DOI] [PubMed] [Google Scholar]
- 19.Maher, M., C. Finnegan, E. Collins, B. Ward, C. Carroll, and M. Cormican. 2003. Evaluation of culture methods and a DNA probe-based PCR assay for detection of Campylobacter species in clinical specimens of feces. J. Clin. Microbiol. 41:2980-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medema, G. J., F. M. Schets, A. W. van de Giessen, and A. H. Havelaar. 1992. Lack of colonization of 1 day old chicks by viable, non-culturable Campylobacter jejuni. J. Appl. Bacteriol. 72:512-516. [DOI] [PubMed] [Google Scholar]
- 21.Niessner, R. 1999. Enzyme and immunoassays. In Ullmann's encyclopedia of industrial chemistry. Wiley-VCH Verlag GmbH & Co., Weinheim, Germany.
- 22.On, S. L., and B. Holmes. 1991. Reproducibility of tolerance tests that are useful in the identification of campylobacteria. J. Clin. Microbiol. 29:1785-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.On, S. L., and B. Holmes. 1992. Assessment of enzyme detection tests useful in identification of campylobacteria. J. Clin. Microbiol. 30:746-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Sullivan, N. A., R. Fallon, C. Carroll, T. Smith, and M. Maher. 2000. Detection and differentiation of Campylobacter jejuni and Campylobacter coli in broiler chicken samples using a PCR/DNA probe membrane based colorimetric detection assay. Mol. Cell Probes 14:7-16. [DOI] [PubMed] [Google Scholar]
- 25.Owen, R. J., A. Fayos, J. Hernandez, and A. Lastovica. 1993. PCR-based restriction fragment length polymorphism analysis of DNA sequence diversity of flagellin genes of Campylobacter jejuni and allied species. Mol. Cell Probes 7:471-480. [DOI] [PubMed] [Google Scholar]
- 26.Oyofo, B. A., S. A. Thornton, D. H. Burr, T. J. Trust, O. R. Pavlovskis, and P. Guerry. 1992. Specific detection of Campylobacter jejuni and Campylobacter coli by using polymerase chain reaction. J. Clin. Microbiol. 30:2613-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen, H. N., J. E. Olsen, K. Jorgensen, and O. F. Rasmussen. 1996. Detection of Campylobacter jejuni and Campylobacter coli in chicken faecal samples by PCR. Lett. Appl. Microbiol. 23:363-366. [DOI] [PubMed] [Google Scholar]
- 27a.Slater, E. R., and R. J. Owen. 1997. Restriction fragment length polymorphism analysis shows that the hippicurase gene of Campylobacter jejuni is highly conserved. Lett. Appl. Microbiol. 25:274-278. [DOI] [PubMed] [Google Scholar]
- 28.Stucki, U., J. Frey, J. Nicolet, and A. P. Burnens. 1995. Identification of Campylobacter jejuni on the basis of a species-specific gene that encodes a membrane protein. J. Clin. Microbiol. 33:855-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uyttendaele, M., R. Schukkink, B. van Gemen, and J. Debevere. 1995. Detection of Campylobacter jejuni added to foods by using a combined selective enrichment and nucleic acid sequence-based amplification (NASBA). Appl. Environ. Microbiol. 61:1341-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Doorn, L. J., B. A. Giesendorf, R. Bax, B. A. van der Zeijst, P. Vandamme, and W. G. Quint. 1997. Molecular discrimination between Campylobacter jejuni, Campylobacter coli, Campylobacter lari and Campylobacter upsaliensis by polymerase chain reaction based on a novel putative GTPase gene. Mol. Cell Probes 11:177-185. [DOI] [PubMed] [Google Scholar]
- 31.Wedderkopp, A., E. Rattenborg, and M. Madsen. 2000. National surveillance of Campylobacter in broilers at slaughter in Denmark in 1998. Avian Dis. 44:993-999. [PubMed] [Google Scholar]
- 32.Wegmuller, B., J. Luthy, and U. Candrian. 1993. Direct polymerase chain reaction detection of Campylobacter jejuni and Campylobacter coli in raw milk and dairy products. Appl. Environ. Microbiol. 59:2161-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winters, D. K., A. E. Leary, and M. F. Slavik. 1997. Rapid PCR with nested primers for direct detection of Campylobacter jejuni in chicken washes. Mol. Cell Probes 11:267-271. [DOI] [PubMed] [Google Scholar]