Abstract
We developed a diagnostic array of oligonucleotide probes targeting species-specific variable regions of the genes encoding topoisomerases GyrB and ParE of respiratory bacterial pathogens. Suitable broad-range primer sequences were designed based on alignment of gyrB/parE sequences from nine different bacterial species. These species included Corynebacterium diphtheriae, Fusobacterium necrophorum, Haemophilus influenzae, Legionella pneumophila, Moraxella catarrhalis, Mycoplasma pneumoniae, Staphylococcus aureus, Streptococcus pneumoniae, and Streptococcus pyogenes. Specific probe sequences were selected by comparative analysis against the European Bioinformatics Database, as well as gyrB/parE sequences generated for this study. To verify specificity, at least six initial oligonucleotide probe sequences per bacterial species were tested by hybridization on a solid glass support using culture collection strains as templates. Finally, three oligonucleotide probes per bacterial species were utilized to examine 65 middle ear fluid and 29 throat swab samples. The sensitivities of the developed assay compared to classic culture from middle ear fluid samples for H. influenzae, M. catarrhalis, and S. pneumoniae were 96 (93 for culture), 73 (93 for culture), and 100% (78% for culture), respectively. No cross-reactivity with bacterial species belonging to the normal oral flora was observed when the 29 throat swab samples were studied. The sensitivity of the assay to detect S. pyogenes from these samples was 93% (80% for culture). These results provide a proof of concept for the diagnostic use of microarray technology based on broad-range topoisomerase gene amplification, followed by hybridization and specific detection of bacterial species.
Modern high-density arrays of DNA probes (microarrays) have proved to be powerful tools to study complex genetic profiles of cellular life. However, only a few experimental assays based on this valuable tool have been introduced for microbiological diagnostics. We present here novel data demonstrating the application of microarray technology combined with broad-range PCR amplification of topoisomerase genes to the diagnosis of bacterial respiratory infections.
Broad-range bacterial PCR is based on the use of primers that recognize conserved sequences of bacterial genes encoding essential molecules. The resulting DNA amplicons contain variable regions that provide a basis for the analysis of phylogenetic relationships and identification of different bacterial species. Furthermore, some genetically hypervariable regions provide targets for bacterial-species-specific oligonucleotide sequences. These oligonucleotides that target hypervariable regions may be used in a diagnostic array of probes for detection of bacterial pathogens.
The use of broad-range ribosomal DNA (rDNA) PCR technologies for identification of bacterial and fungal culture isolates in the clinical laboratory is well established (3, 28), and there are commercial kits available for that purpose. However, the kits are not widely used for microbial identification in diagnostic microbiological laboratories due to the time and skill needed to perform the assays, as well as the necessity for costly equipment, e.g., DNA sequencers. The more challenging task of utilizing broad-range PCR analysis directly on clinical specimens has been performed mainly by academic institutions and research laboratories (10, 15, 19, 24, 25, 35). In the direct approach, the molecular finding is not dependent on viable microorganisms or on those species that cannot readily be cultured by classical methodologies. In fact, studies based on broad-range molecular DNA amplification have shown that the diversity of human microbial flora is much broader than has been observed by using classic culture-based methodologies (16). The use of broad-range PCR amplification directly from clinical specimens has proved advantageous compared to bacterial culture, particularly when applied to infections caused by microbial species with unusual growth requirements and during antimicrobial treatment of the patient (2, 14, 23).
Dependence on DNA sequencing for molecular identification of relevant pathogens has limited the value of the broad-range bacterial PCR methodology in clinical diagnostics. In particular, the need to perform extensive sequencing experiments of clone libraries to identify possible multiple clinically relevant bacterial species in clinical samples has limited the practical use of this approach.
Topoisomerases are proteins that help to relieve helical winding and tangling problems during DNA replication. These enzymes change the structure of DNA by transiently breaking one or two strands in such a way that the strands remain unaltered as their shape is changed. All cellular organisms have type I and type II topoisomerases. In most bacteria, there are two type II topoisomerases: gyrase and topoisomerase IV. However, in some bacteria, topoisomerase IV seems to be missing. Bacterial DNA gyrase consists of two subunits, GyrA (gyrA) and GyrB (gyrB). The same is true with topoisomerase IV, with the genes coding for the two subunits, called ParC and ParE. gyrA and parC, as well as gyrB and parE, are homologous genes (9). Topoisomerase genes contain conserved and variable regions, similar to those encoding rDNA. Based on our preliminary DNA database-mining work, we chose gyrB and parE as the target genes for our diagnostic array of probes.
Acute upper respiratory tract infections (sinusitis, pharyngitis, bronchitis, and otitis) are among the leading reasons for physician office visits, and these infections account for ∼75% of antibiotic courses prescribed in developed countries (5). However, many of the antibiotic courses are unnecessary or inappropriate, because a significant proportion of these infections are caused by viruses. Clinical differential diagnosis to distinguish between bacterial and viral infections is challenging due to the similarity of the symptoms (22). The overuse and improper use of antibiotics are thought to be the main reasons for the development of resistance among bacterial pathogens causing respiratory tract infections. Therefore, to avoid unnecessary use of antimicrobials, there is a need to develop reliable and rapid methods for laboratory diagnosis of bacterial pathogens.
The most common bacterial species causing upper respiratory infections are Streptococcus pneumoniae, Haemophilus influenzae, Streptococcus pyogenes, and Moraxella catarrhalis. Occasionally, Staphylococcus aureus, Mycoplasma pneumoniae, and group C streptococci, as well as anaerobic bacteria, e.g., Fusobacterium necrophorum, are detected (17, 34). Corynebacterium diphtheriae is a rare finding in vaccinated populations; however, diphtheria is endemic in some developing countries. Legionella pneumophila has also been detected in upper respiratory tract specimens (30). We describe below a low-density array of oligonucleotide probes to be used for detection and identification of nine of these bacterial species after PCR amplification of topoisomerase gene sequences.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains listed in Table 1 were used to test the sensitivity and specificity of oligonucleotide probes. The bacterial strains or bacterial genomic DNAs were obtained from the American Type Culture Collection or from the German Collection of Microorganisms and Cell Cultures. Additionally, seven S. pneumoniae and five S. pyogenes clinical isolates were included in the study.
TABLE 1.
Bacterial strains used to test sensitivities and specificities of oligonucleotide probes
| Bacterium | Provider and code |
|---|---|
| Target organisms | |
| Corynebacterium diphtheriae | DSM 44123 |
| Fusobacterium necrophorum | DSM 20698 |
| Haemophilus influenzae | ATCC 51907Da |
| Legionella pneumophila | ATCC 33152D |
| Moraxella catarrhalis | DSM 9143 |
| Mycoplasma pneumoniae | ATCC 15531D |
| Staphylococcus aureus | DSM 20231 |
| Streptococcus pneumoniae | DSM 20566 |
| Streptococcus pyogenes | DSM 20565 |
| Normal-flora controls | |
| Haemophilus parainfluenzae | DSM 8978 |
| Haemophilus ducreyi | DSM 8925 |
| Moraxella caviae | ATCC 14659 |
| Moraxella cuniculi | ATCC 14688 |
| Pasteurella pneumotropica | ATCC 13669 |
| Streptococcus oralis | DSM 20627 |
| Streptococcus mitis | DSM 12643 |
D, genomic DNA.
Patient samples.
In total, 65 middle ear fluid samples (C100 to C164) and 29 throat swabs (C212 to C240) were analyzed. The middle ear fluid samples were collected between March 1996 and May 1997 during a randomized controlled trial, the procedures of which were approved by the Ethics Committee of Turku University Hospital (27). The participating young children had acute (duration, <48 h) otorrhea. The middle ear fluid samples were suctioned through a tympanostomy tube with a sterile glass suction tip. A swab was dipped into the sample for routine bacterial culture, and the rest of the middle ear fluid was frozen in virus isolation medium and stored at −70°C. The throat swabs were collected during the spring of 2003 and were taken for diagnostic bacterial culture. These anonymous and coded specimens were provided by Yhtyneet Laboratoriot, Helsinki, Finland. Microarray analysis of all clinical samples was performed blindly so that the analyst was not aware of the bacterial culture results at the time of analysis and interpretation of the results. Bacterial culture from patient samples was performed using standard methodologies.
Design of universal primers to amplify bacterial gyrB/parE.
gyrB genes of S. pyogenes (AE006540), S. pneumoniae (AE007387), and H. influenzae (U32738) were aligned with the BioEdit (8) program using the ClustalW alignment algorithm (31). Areas that were conserved in all these bacteria were selected for primer design. Subsequently, when the gyrB gene sequence of M. catarrhalis was obtained, the primers were modified according to this sequence information. The designed PCR primers gB1f (5′-CGTCCWGGKATGTAYATHGG-3′) and gB2r (5′-CCHACRCCRTGWAAWCCDCC-3′) amplify an ∼300-bp region of the bacterial gyrB and parE genes.
DNA extraction from bacterial cultures and patient samples.
DNA was extracted from bacterial cultures by using the QIAamp DNA Mini Extraction kit (Qiagen, Hilden, Germany). The throat swabs were transported to the laboratory in Transpocults (Orion Diagnostica, Espoo, Finland). The sampling stick was inserted into an Eppendorf tube containing 1 ml of 1× phosphate-buffered saline, followed by incubation in phosphate-buffered saline for 2 h at room temperature (RT). After incubation, the stick was removed, and the tubes were centrifuged for 10 min at 7,500 rpm at RT on a Heraeus Biofuge Pico instrument (Kendra, Hanau, Germany). After centrifugation, the supernatant was removed, and the obtained pellet was resuspended in 180 μl of ATC buffer provided by Qiagen. The DNA was then extracted by using the QIAamp DNA Mini Extraction kit (Qiagen). From middle ear fluid samples, DNA was extracted directly with the QIAamp DNA Mini kit.
Broad-range PCR for sequencing and cloning.
Broad-range bacterial gyrB/parE PCR was performed in a total volume of 50 μl. The reaction mixture contained 1× Hot StarTaq PCR buffer (Qiagen) with 2.8 mM MgCl2, 200 μM (each) deoxynucleoside triphosphate, 2.5 U of HotStarTaq DNA polymerase (Qiagen), 20 pmol of each primer (gB2r and gB1f), and 25 ng of template. Amplification was performed with a GenAmp PCR system 2700 thermocycler (Applied Biosystems, Foster City, Calif.) for 36 cycles (45 s at 95°C, 45 s at 52°C, and 45 s at 72°C). The cycles were preceded by a denaturation step at 95°C for 15 min and followed by an extension step at 72°C for 10 min.
Cloning and sequencing of gyrB/parE genes.
Partial gyrB/parE sequences were obtained from the EMBL public sequence database (http://www.ebi.ac.uk/), or they were determined in this study (M. catarrhalis, F. necrophorum, L. pneumophila, Streptococcus mitis, Streptococcus oralis, and Haemophilus parainfluenzae). The 300-bp gyrB/parE gene fragment of interest was confirmed either by direct sequencing or after being cloned from all bacterial species included in this study. Molecular cloning of amplified PCR products from all gram-positive bacteria was performed, since the universal primers presented in this study amplify both the gyrB and parE genes. Therefore, direct sequencing of the PCR-amplified products was not possible. To sequence the PCR products, they were first cloned into a TOPO TA cloning vector (Invitrogen, Carlsbad, Calif.). The vectors were then sequenced using an automated PRISM 3100 sequencer (Applied Biosystems) and BigDye Terminator version 3.0 cycle-sequencing chemistry. For each bacterial strain, at least five PCR fragments from different colonies were sequenced. The DNA sequences were analyzed with the Vector NTI suite version 7 program (InforMax, Bethesda, Md.).
Species-specific oligonucleotides.
For the design of probe oligonucleotides (Table 2), the sequences of the target group were aligned with nontarget sequences from closely related bacterial species. The alignments were performed with the BioEdit (8) program using the ClustalW alignment algorithm (31). Consensus sequences of the alignments were calculated, and less conserved areas were identified manually for further analysis. From these areas, oligonucleotide sequences with suitable lengths (20 to 25 nucleotides) were selected for comparison analysis. Using the FastA algorithm (20, 21), the oligonucleotide sequences were compared against the European Bioinformatics Database prokaryotic sequence database, which contained 2,251 relevant gyrB (284 parE) sequences representing ∼550 different bacterial species.
TABLE 2.
Sequences, melting temperatures, lengths, and positions of species-specific oligonucleotide probes used in this study
| Pathogen | Gene | Probe | Probe sequence 5′-3′ | Tma (°C) | Length (nt) | Nucleotides | Chip position |
|---|---|---|---|---|---|---|---|
| C. diphtheriae | gyrB | Cdiph 1 | TCTACCGGTGAACGCGGCCT | 62 | 20 | 109-128b | D4 |
| gyrB | Cdiph 2 | CTACCCACGTCGATGTGACC | 60 | 20 | 188-207b | C4 | |
| gyrB | Cdiph 3 | CGACCGTCCAAGTCGTTATGA | 58 | 21 | 287-307b | B4 | |
| F. necrophorum | gyrB | Fnec 1 | CAACATCAGCCAGGGGATTACAT | 58 | 23 | 110-132b | D1 |
| gyrB | Fnec 2 | GGAATTCCAGTAGACATACATCC | 56 | 23 | 250-272b | C1 | |
| gyrB | Fnec 3 | GGGAAGTGGTAGATAATTCTGTG | 56 | 23 | 143-165b | B1 | |
| H. influenzae | gyrB | Hinf 1 | ATGATAATTCTGTATCGGTGCAA | 53 | 23 | 194-216c | B5 |
| gyrB | Hinf 2 | AGCAGAAGTTATTATGACTGTGC | 54 | 23 | 270-292c | A5 | |
| gyrB | Hinf 3 | GATACCGATGACGGTACTGGTTTG | 60 | 24 | 85-108c | E4 | |
| L. pneumophila | gyrB | Lpneu 1 | GATGATAATTCCTACAAGGTATC | 53 | 23 | 313-335c | C3 |
| gyrB | Lpneu 2 | GTCATCATGACAGTCCTACATGC | 58 | 23 | 259-281c | B3 | |
| gyrB | Lpneu 3 | AAGAAGGCAAATCAGCAGCCGA | 58 | 22 | 254-275c | A3 | |
| M. catarrhalis | gyrB | Mcat 3 | TGTGGATATCCACCCTGAAGAAG | 58 | 23 | 237-259c | A4 |
| gyrB | Mcat 2 | GATGATAATTCATACAAAGTATC | 60 | 23 | 313-335c | E3 | |
| gyrB | Mcat 1 | ATACCGATGATGGTACAGGCTTG | 58 | 23 | 147-169c | D3 | |
| M. pneumoniae | parE | Mpneu 1 | GATAACGATTCTTATAAGATTGC | 51 | 23 | 324-346b | E5 |
| parE | Mpneu 2 | ATAACTGTTAGTGACAACGGTCG | 56 | 23 | 219-241b | D5 | |
| parE | Mpneu 3 | AGATTTCTACGATTGACACCGTC | 46 | 23 | 188-210b | C5 | |
| S. aureus | gyrB | Saur 1 | AGTGTGGGAAATTGTCGATAATA | 53 | 23 | 138-160b | B2 |
| gyrB | Saur 2 | GACGTCCAGCTGTCGAAGTTATT | 58 | 23 | 281-303b | A2 | |
| gyrB | Saur 3 | CGACTTCAGAGAGAGGTTTGCAC | 60 | 23 | 110-132b | E1 | |
| S. pneumoniae | gyrB | Spneu 1 | TTGAGACCGTCTTTACAGTCCTT | 56 | 23 | 293-315b | E2 |
| parE | Spneu 2 | CGACCGATGGCGCTGGTCTTCAT | 63 | 23 | 103-125b | D2 | |
| parE | Spneu 3 | GGTCAAGGTGGCTATAAGACATC | 58 | 23 | 324-346b | C2 | |
| S. pyogenes | parE | Spyo 1 | CAACAGATGCTACGGGATTGCAC | 60 | 23 | 103-125b | C6 |
| parE | Spyo 2 | GTCAGTGTGGCAGATAGCGGACG | 63 | 23 | 219-241b | B6 | |
| gyrB | Spyo 3 | GCTCGGTCAGTGTGGCAGAT | 60 | 20 | 215-234b | A6 |
Broad-range PCR for clinical analysis and template synthesis.
The primers gB1f and gB2r were used for target synthesis from study specimens. The reverse primer was modified to contain four phosphorothioates at its 5′ end. The forward primer was unmodified. PCR was performed in a total volume of 50 μl. The reaction mixture contained 1× HotStar Taq PCR buffer (Qiagen) with 2.8 mM MgCl2; 200 μM dATP, dGTP, and dTTP deoxynucleoside triphosphates; 140 μM dCTP and 25 nmol of Cy5-AP3-dCTP (Amersham Pharmacia Biotech, Little Chalfont, United Kingdom), 2.5 U of HotStart Taq DNA polymerase (Qiagen), 20 pmol of each primer, and 5 μl of DNA. The PCR conditions for template synthesis were the same as described above (see “Broad-range PCR for sequencing and cloning”). The PCR products were analyzed by electrophoresis in a 2% agarose gel containing ethidium bromide. The labeled PCR products were separated from unincorporated nucleotides and primers with the QIAquick PCR Purification kit (Qiagen). After the purification procedure, the double-stranded PCR product was treated with T7 gene 6 exonuclease (U.S. Biochemical Corp., Cleveland, Ohio) to produce a single-stranded template for the hybridization process. This reaction was performed in a total volume of 38.5 μl containing 30 μl of purified PCR product, 7 μl of T7 gene 6 exonuclease buffer, and 75 U of T7 gene 6 exonuclease at 37°C for 30 min. After hydrolysis, the enzyme was inactivated at 80°C for 15 min. A negative control (PCR mixture without DNA) was included in all PCR tests performed in this study.
Amplification of human beta-globin gene.
To control for the possible presence of polymerase inhibitors in DNA extracted from clinical samples, human beta-globin sequences were amplified. The primers beta 1F (5′-GGGTTTGAAGTCCAACTCCT-3′) and beta 1R (5′-TCCACATGCCCAGTTTCTAT-3′) were designed for this study by using the Primer3 server (26). The PCR mixture and conditions were the same as those used for gyrB/parE sequencing and cloning.
Preparation of glass slides and spotting.
5′-amino-modified oligonucleotides (Sigma-Genosys, Haverhill, United Kingdom) were dissolved in a 400 mM sodium carbonate buffer (pH 9.0) to a final concentration of 50 μM. Genorama microarray slides coated with enhanced aminosilane (Asper Biotech Ltd., Tartu, Estonia) were spotted by using an OmniGrid microarrayer (GeneMachines, San Carlos, Calif.) and TeleChem (Sunnyvale, Calif.) SMP3 pins. The average diameter of the spots was 100 μm. 5′-amino-modified forward PCR primer oligonucleotides were also included as control spots. Oligonucleotides specific for bacterial species were used as negative controls between species. After the spotting, the slides were exposed to vaporized NH3, followed by three washes in distilled water. The spotted arrays were stored at RT.
Hybridization and analysis of microarrays.
Hybridization of the fluorescent target on the microarray was performed in a total volume of 38 μl. The hybridization reaction mixture contained 200 to 300 ng of labeled template, 20× SSC (final concentration, 3.4×) (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and 20% sodium dodecyl sulfate (SDS) (for a final concentration 0.3%). Before hybridization, the hybridization mixture was denatured at 95°C for 3 min and chilled on ice, followed by pipetting on the microarray slide. The slides were covered with 24- by 60-mm glass coverslips and placed in a hybridization cassette (TeleChem) for overnight hybridization at 57°C. After hybridization, the slides were washed once with 0.1% SDS for 5 min at 57°C, once with 0.5× SSC-0.01% SDS for 5 min at RT, and once with 0.06× SSC for 5 min at RT. Finally, the microarray slides were dried with compressed air. Fluorescent images of the microarrays were generated by scanning the slides with the Agilest (Palo Alto, Calif.) DNA Microarray Scanner. The slide images were analyzed visually. A clinical sample was defined as positive for a given pathogen with the microarray analysis if two or three species-specific spots in the array gave positive signals.
Statistical analysis.
For statistical analysis, a sample was considered positive for a given pathogen if it was putatively positive either by the microarray DNA analysis or by classical culture or if it was putatively positive by both methodologies. A sample was considered negative if it was putatively negative for the given pathogen by both culture and microarray analysis. Thus, sensitivity for a given pathogen-specific assay was calculated as the number of putatively positive samples divided by the number of positive samples, and specificity was calculated as the number of putatively negative samples divided by the total number of negative samples. Therefore, the specificities for both assays were 100%, and only the sensitivities could be compared. Statistical analysis was performed for the clinically relevant pathogens H. influenzae, M. catharralis, and S. pneumoniae in the middle ear effusion sample group (n = 65) and for S. pyogenes in the throat swab group (n = 29).
Nucleotide sequence accession numbers.
The EMBL accession numbers for the sequences determined in this study are AJ617787, AJ617788, AJ617789, AJ617790, AJ617791, and AJ617792. These sequences represent the partial gyrB genes for DNA gyrase subunit B of M. catarrhalis, F. necrophorum, L. pneumophila, S. mitis, S. oralis, and H. parainfluenzae.
RESULTS
Universality and sensitivity of the primers.
The effectiveness of the bacterial broad-range gyrB/parE primers gB2r and gB1f was first assessed with DNA extracts from 16 different bacterial species (Table 1). All these bacterial strains produced PCR products of the expected size, ∼300 bp. No amplification was seen in bacterial DNA-negative amplification control samples or in those containing only human genomic DNA. The sensitivity of the broad-range primer set gB2r and gB1f was tested with a serial dilution of bacterial DNA extracts. The sensitivity of the assay was 2.5 fg of purified DNA per assay for H. influenzae, 250 fg for M. catarrhalis, 2.5 fg for S. pneumoniae, and 25 fg for S. pyogenes.
Experimental selection of species-specific oligonucleotides.
At least six specific oligonucleotide probes were initially designed for every nine bacterial species. Labeled amplicons (templates) obtained from DNA extracts of the bacterial isolates were hybridized on arrays of these oligonucleotides. Additionally, seven bacterial species representing the normal human flora were also included in these hybridization studies as template controls (Table 1). Up to 54% of the tentative pathogen-specific probes were rejected due to cross-hybridization with these controls or other templates. After two to four repetitive rounds of hybridization, three oligonucleotide probes for each bacterial species were selected for further experiments with clinical samples.
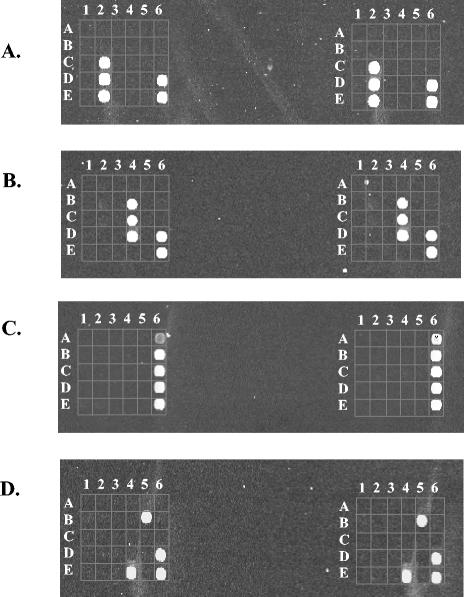
Representative results showing detection of S. pneumoniae and C. diphtheriae with three different species-specific oligonucleotide probes are shown in Fig. 1A and B, respectively. In Fig. 1A, hybridization was performed with a labeled amplicon from the S. pneumoniae culture collection strain (DSM 20566). All three S. pneumoniae-specific oligonucleotide probes (C2, D2, and E2), as well as the positive control oligonucleotide probes (D6 and E6), gave positive signals. The probe positions specific for the eight other bacterial species included in the figure were negative. This example also shows that specific recognition was obtained with oligonucleotide probes representing both the gyrB and parE genes. In Fig. 1B, hybridization was carried out with target amplified from the C. diphtheriae culture collection strain (DSM 44123). As in the previous example, positive signals were detected from all C. diphtheriae-specific oligonucleotide positions (B4, C4, and D4), as well as from the positive control positions (D6 and E6).
FIG. 1.
Hybridization results from bacterial cultures and clinical samples. Each hybridization result is presented in duplicate. The positive controls are located in positions D6 and E6. (A) S. pneumoniae hybridization. All three S. pneumoniae-specific oligonucleotides (C2, D2, and E2) gave positive signals. (B) C. diphtheriae hybridization. Positive signals were detected from all C. diphtheriae-specific oligonucleotides (B4, C4, and D4). (C) Hybridization results from a throat swab sample (patient C237). All three S. pyogenes-specific oligonucleotide probes gave a clear signal (A6, B6, and C6). (D) Hybridization results from a middle ear fluid sample (patient C108). Two out of three H. influenzae-specific oligonucleotides (B5 and E4) gave positive signals.
Hybridization results from clinical samples.
Broad-range bacterial gyrB/parE PCR was positive from all 29 throat swab samples, and only 4 middle ear fluid samples of the total 65 were negative. The possible presence of PCR inhibitors in these four samples was excluded by the successful amplification of a 400-bp fragment of the human beta-globin gene. All 90 positive broad-range bacterial gyrB/parE PCR samples were further characterized by hybridization experiments on the array of probes. The criteria for a positive diagnostic hybridization finding were that the (amino-modified forward primer) control probe was positive and that at least two different oligonucleotides specific for a certain bacterial species gave a positive signal. Water processed with the clinical samples was used as a negative PCR control. None of these negative controls were positive in the broad-range bacterial gyrB/parE PCR, as determined by agarose gel electrophoresis of the PCR products.
The four PCR-negative middle ear fluid samples were also negative when studied by bacterial culture. Of the remaining 90 samples, 10 were negative for clinically relevant bacterial species by bacterial culture and by hybridization on the oligonucleotide array. However, the broad-range bacterial gyrB/parE PCR had produced bacterial amplicons from those 10 samples, as indicated by positive signals from the hybridization control oligonucleotides included in each array of probes and as visualized by agarose electrophoresis of the PCR products. In these study specimens, this finding suggests the presence of DNA derived from probably clinically irrelevant nonpathogens (normal flora) not included in our diagnostic array panel. Original bacterial culture results from these samples were not available for further analysis.
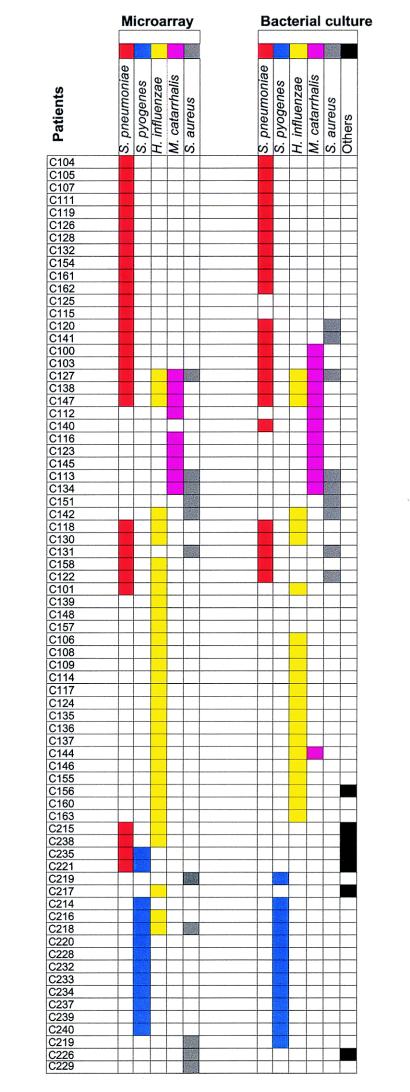
In seven cases, bacterial culture revealed pathogens not included in our diagnostic panel, and therefore, the hybridization results were negative. In all, five different pathogens of the nine included in the panel were identified by hybridization from the clinical samples. Those pathogens were S. pneumoniae, S. pyogenes, H. influenzae, M. catarrhalis, and S. aureus. A comparative summary of the hybridization and bacterial culture results is presented in Fig. 2. Results are shown only from those 73 samples from which one or more of the five pathogens listed above were detected by either hybridization, bacterial culture, or both methods.
FIG. 2.
Comparison of positive identification results for five pathogens. These results are based on the oligonucleotide array assay detection and conventional bacterial culture. In all, 73 patient samples consisting of 53 middle ear fluid (C100 to C163) and 20 throat swab (C214 to C240) samples were included in this comparison. Each bacterial species is indicated by a specific color.
Representative results of positive hybridizations from patient samples are presented in Fig. 1C and D. In Fig. 1C, hybridization was performed with labeled amplicons of DNA extracted from a throat swab specimen (patient C237). All three oligonucleotide probes specific for S. pyogenes (A6, B6, and C6) showed positive signals. Positive signals were also detected from the positive control probes (D6 and E6), whereas the other probes were negative, indicating absence of cross-reactivity with the normal oral bacterial flora. Detection of S. pyogenes was consistent with bacterial culture (Fig. 2). In the second example in Fig. 1D, DNA extracted from a middle ear effusion sample (patient C108) was studied. The only species-specific oligonucleotides showing positive signals were those specific for H. influenzae. This result was in accordance with bacterial culture (Fig. 2). However, it is noteworthy that only two out of three H. influenzae-specific oligonucleotides were positive (B5 and E4).
Our assay was also successful in identifying polybacterial infections. In 17 cases, more than one pathogenic bacterial species was present in a clinical sample, as determined by bacterial culture. In nine of these cases, the same bacterial species were detected with our array (Fig. 2).
As calculated from the analysis of 65 middle ear fluid samples, the sensitivities of the developed assay and those of classic culture for H. influenzae, M. catarrhalis, and S. pneumoniae were 96 (93 for culture), 73 (93 for culture), and 100% (78% for culture), respectively. No cross-reactivity with bacterial species belonging to the normal oral flora was observed when the 29 throat swab samples were studied. The sensitivity of the hybridization assay to detect S. pyogenes from these samples was 93% (80% by culture).
DISCUSSION
In this study, we present a novel low-density microarray method for the detection and identification of nine bacterial pathogens. For this method, universal primers amplifying a conserved region of the bacterial gyrB/parE gene were used, followed by characterization of the single-stranded PCR products by hybridization onto an oligonucleotide array.
After initial database mining, tentative oligonucleotides included in our panel were selected based on hybridization results obtained by using bacterial culture strains as templates. All probe oligonucleotides presented in our study performed well when PCR-amplified DNA extracts of 28 bacterial culture isolates representing 16 bacterial species were used. Subsequently, these selected species-specific oligonucleotides were tested with clinical samples from patients suffering from upper respiratory infections. Results from these patient samples showed differences in the degree of hybridization with different species-specific oligonucleotides. This may be explained by the formation of a highly complex mixture of bacterial PCR amplicons also representing those species not included in our diagnostic array of probes. Competition in reagent use during PCR amplification may cause variation in the probe detection sensitivities for specific pathogens, particularly if they are represented in low copy numbers in the original specimen compared to other, more abundant species that, e.g., belong to the normal human flora. Due to this probe performance variation, more than one pathogen-specific oligonucleotide probe sequence is required for reliable diagnosis when clinical samples are analyzed. In a previous study, the necessity of using multiple oligonucleotides for each pathogen was also reported (1). The authors found that two or more oligonucleotides may be required for reliable species identification of clinical isolates due to minor sequence variation within the target region. This variation cannot be ruled out in our study either.
Structural rDNA sequences (16S and 23S rDNAs) and corresponding rRNAs have been extensively used as targets for modern diagnostic assays. As the 16S rDNA gene is present in multiple copies in the genomes of most of the human bacterial pathogens and there is also a large amount of 16S rDNA sequence data available in public databases, it is not surprising that this gene region has been an obvious choice when diagnostic tests have been developed. One important drawback of using 16S rDNA genes in clinical diagnostics, however, is their conservative nature. Identification of bacteria at the species level may require resolving the whole gene, yet in some cases, phylogenetically closely related bacterial species cannot be differentiated from each other (7). Sequencing the whole 16S rDNA gene for identification purposes may often also be too labor-intensive and expensive for routine microbiological diagnostics. Another drawback is that PCR products from samples of patients with multibacterial infections, or from sites containing multiple bacterial species, cannot be directly analyzed by DNA sequencing. Extensive experiments, including molecular cloning followed by sequence analysis of multiple transformants, have been necessary to study polymicrobial infections and clinical specimens containing multiple bacterial species (16, 19, 29).
There are only a few previous studies in which the oligonucleotide microarray technology has been used for bacterial identification. Oligonucleotide microarrays based on 16S or 23S rDNA sequences have been used for detection of Campylobacter from fecal cloacal swabs (13), for detection of intestinal bacterial species from human fecal samples (33), and for detection of bacteremia-causing bacteria from positive blood culture broths (1). Other approaches have been developed in addition to these oligonucleotide-based methods targeting bacterial rRNA sequences. In one recent study, a DNA probe matrix directed toward rRNA targets was used for identification of bacteria and fungi from blood cultures (18), whereas Christensen et al. used terminal restriction fragment length polymorphism for identification of bacteria from blood cultures (4).
In the protein-coding genes, including gyrB, there is more variation than in the structural RNA-coding genes. Therefore, in this study we chose to use topoisomerase genes as our targets. The amount of variation is especially important if short nucleotide sequences, i.e., oligonucleotides, are used for the identification of closely related bacterial species. Fukushima et al. (6) showed that a microarray assay targeting gyrB can identify mycobacteria at the species level, whereas in a previous study, a DNA array based on 16S rDNA sequence data could not distinguish between closely related mycobacterial species (32). Kakinuma et al. (11) reported similar results by using microarrays focusing on the gyrB gene region for detection and identification of Escherichia coli, Shigella, and Salmonella. The usefulness of the gyrB region for identification of bacterial species has been reported by Yamamoto and Harayama (36) and Kasai et al. (12) as well.
All cellular organisms have type I and type II topoisomerases, and in most bacteria there are two type II topoisomerases: gyrase and topoisomerase IV (parE). In some bacteria, the same conserved amino acid sequences can be found both in gyrase and topoisomerase IV proteins. This means that the universal primers that amplify gyrB may also amplify parE genes from certain bacterial groups. This seems to be the situation with most of the gram-positive bacteria. We did not find this amplification of two different genes to be a problem in our detection system. It is possible to design probes for both gyrB and parE genes, and this may even be a benefit, as it provides more options for target sequence selection. When both gene regions can be used for probe design, there are more possibilities to select those probes in which the length, melting temperature, and specificity are optimal.
Nucleic acid testing-based methodologies for detection of microbial pathogens can generally be divided into three steps: (i) nucleic acid purification, (ii) nucleic acid amplification, and (iii) detection of specific amplification products. In our present assay, all these steps have to be performed individually. Thus, it may not be suitable as an efficient tool for diagnosis of common respiratory illnesses. A combination of all these steps in one platform would make the assay more desirable for clinical use. The incorporation of gene markers for antibiotic susceptibility testing and virus-specific probes for differential diagnosis of respiratory viral and bacterial infections is an attractive prospect for further development. The microarray technology described here may form the basis for the development of user-friendly assays for bacterial- and infectious-disease diagnostics. This could be achieved by transforming the specific oligonucleotides from the classical Pat Brown glass platform (http://cmgm.stanford.edu/pbrown) utilized in our study onto more sophisticated and user-friendly low-density DNA array platforms. It is noteworthy that the primers developed for our study also amplify a broad range of other bacterial species not represented in our diagnostic array of probes. Therefore, future expansion of the diagnostic array to include more bacterial species is possible.
In conclusion, we have shown that the microarray technique based on the gyrB/parE genes as targets can be applied to the identification and detection of bacterial species from culture isolates and directly from clinical throat and middle ear fluid samples. Compared to the conventional broad-range bacterial PCR method, this technique offers many advantages, with the primary advantage being the possibility to detect and identify multiple pathogens directly from clinical samples without the need for extensive sequencing experiments.
Acknowledgments
This work was supported by the National Technology Agency (TEKES) and the Finnish National Fund for Research and Development (SITRA).
We thank Outi Monni from the Biomedicum Biochip Center for technical support related to array printing and scanning. We also thank Anni Virolainen-Julkunen and Ritva Heikkilä from Yhtyneet Laboratoriot for providing us with the throat swab samples. Finally, we thank Tiina Haarala, Marika Karjalainen, Merja Lindfors, Jonna Tallila, and Minna Veini for technical assistance.
REFERENCES
- 1.Anthony, R. M., T. J. Brown, and G. L. French. 2000. Rapid diagnosis of bacteremia by universal amplification of 23S ribosomal DNA followed by hybridization to an oligonucleotide array. J. Clin. Microbiol. 38:781-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosshard, P. P., A. Kronenberg, R. Zbinden, C. Ruef, E. C. Böttger, and M. Altwegg. 2003. Etiologic diagnosis of infective endocarditis by broad-range polymerase chain reaction: a 3-year experience. Clin. Infect. Dis. 37:167-172. [DOI] [PubMed] [Google Scholar]
- 3.Bosshard, P. P., S. Abels, R. Zbinden, E. C. Böttger, and M. Altwegg. 2003. Ribosomal DNA sequencing for identification of aerobic gram-positive rods in the clinical laboratory (an 18-month evaluation). J. Clin. Microbiol. 41:4134-4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen, J. E., J. A. Stencil, and K. D. Reed. 2003. Rapid identification of bacteria from positive blood cultures by terminal restriction fragment length polymorphism profile analysis of the 16S rRNA gene. J. Clin. Microbiol. 41:3790-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fendrick, A. M., S. Saint, I. Brook, M. R. Jacobs, S. Pelton, and S. Sethi. 2001. Diagnosis and treatment of upper respiratory tract infections in the primary care setting. Clin. Ther. 23:1683-1706. [DOI] [PubMed] [Google Scholar]
- 6.Fukushima, M., K. Kakinuma, H. Hayashi, H. Nagai, K. Ito, and R. Kawaguchi. 2003. Detection and identification of Mycobacterium species isolates by DNA microarray. J. Clin. Microbiol. 41:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukushima, M., K. Kakinuma, and R. Kawagu. 2002. Phylogenetic analysis of Salmonella, Shigella, and Escherichia coli strains on the basis of the gyrB gene sequence. J. Clin. Microbiol. 40:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 9.Huang, W. M. 1996. Bacterial diversity based on type II DNA topoisomerase genes. Annu. Rev. Genet. 30:79-107. [DOI] [PubMed] [Google Scholar]
- 10.Jalava, J., P. Kotilainen, S. Nikkari, M. Skurnik, E. Vanttinen, O. P. Lehtonen, E. Eerola, and P. Toivanen. 1995. Use of the polymerase chain reaction and DNA sequencing for detection of Bartonella quintana in the aortic valve of a patient with culture-negative infective endocarditis. Clin. Infect. Dis. 21:891-896. [DOI] [PubMed] [Google Scholar]
- 11.Kakinuma, K., M. Fukushima, and R. Kawaguchi. 2003. Detection and identification of Escherichia coli, Shigella, and Salmonella by microarrays using the gyrB gene. Biotechnol. Bioeng. 20:721-728. [DOI] [PubMed] [Google Scholar]
- 12.Kasai, H., K. E. Watanabe, A. Gasteiger, K. Bairoch, S. Isono, S. Yamamoto, and S. Harayama. 1998. Construction of the gyrB database for the identification and classification of bacteria. Genome Inform. Ser. Workshop Genome Inform. 9:13-21. [PubMed] [Google Scholar]
- 13.Keramas, G., D. D. Bang, M. Lund, M. Madsen, S. E. Rasmussen, H. Bunkenborg, P. Telleman, and C. B. V. Christensen. 2003. Development of a sensitive DNA microarray suitable for rapid detection of Campylobacter spp. Mol. Cell. Probes 17:187-196. [DOI] [PubMed] [Google Scholar]
- 14.Khulordava, I., G. Miller, D. Haas, H. Li, J. McKinsey, D. Vanderende, and Y.-W. Tang. 2003. Identification of the bacterial etiology of culture-negative endocarditis by amplification and sequencing of a small ribosomal RNA gene. Diagn. Microbiol. Infect. Dis. 46:9-11. [DOI] [PubMed] [Google Scholar]
- 15.Kotilainen, P., J. Jalava, O. Meurman, O.-P. Lehtonen, E. Rintala, O.-P. Seppälä, E. Eerola, and S. Nikkari. 1998. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J. Clin. Microbiol. 36:2205-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kroes, I., P. W. Lepp, and D. A. Relman. 1999. Bacterial diversity within the human subgingival crevice. Proc. Natl. Acad. Sci. USA 96:14547-14552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandell, G. L., J. E. Bennett, and R. Dolin (ed.).2000. Mandell, Douglas, and Benett's principles and practice of infectious diseases, 5th ed., vol. 1, p. 651-701. Churchill Livingstone, Ltd., Edinburgh, Scotland.
- 18.Marlowe, E. M., J. J. Hogan, J. F. Hindler, I. Andruszkiewicz, P. Gordon, and D. A. Bruckner. 2003. Application of an rRNA probe matrix for rapid identification of bacteria and fungi from routine blood cultures. J. Clin. Microbiol. 41:5127-5133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikkari, S., F. A. Lopez, P. W. Lepp, P. R. Cieslak, S. Ladd-Wilson, D. Passaro, R. Danila, and D. A. Relman. 2002. Broad range bacterial detection and the analysis of unexplained death and critical illness. Emerg. Infect. Dis. 8:188-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 21.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence analysis. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Putto, A., O. Ruuskanen, and O. Meurman. 1986. Fever in respiratory virus infections. Am. J. Dis. Child. 140:1159-1163. [DOI] [PubMed] [Google Scholar]
- 23.Rantakokko-Jalava, K., S. Nikkari, J. Jalava, E. Eerola, M. Skurnik, O. Meurman, O. Ruuskanen, A. Alanen, E. Kotilainen, P. Toivanen, and P. Kotilainen. 2000. Direct amplification of rRNA genes in diagnosis of bacterial infections. J. Clin. Microbiol. 38:32-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Relman, D. A., J. S. Loutit, T. M. Schmidt, S. Falkow, and L. S. Tomkins. 1990. The agent of bacillary angiomatosis—an approach to the identification of uncultured pathogens. N. Engl. J. Med. 323:1573-1580. [DOI] [PubMed] [Google Scholar]
- 25.Relman, D. A., T. M. Schmidt, R. P. MacDermott, and S. Falkow. 1992. Identification of the uncultured bacillus of Whipple's disease. N. Engl. J. Med. 327:293-301. [DOI] [PubMed] [Google Scholar]
- 26.Rozen, S., and H. J. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers, p. 365-386. In S. Krawetz and S. Misener (ed.), Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 27.Ruohola, A., T. Heikkinen, J. Jero, T. Puhakka, T. Juven, M. Narkio-Makela, H. Saxen, and O. Ruuskanen. 1999. Oral prednisolone is an effective adjuvant therapy for acute otitis media with discharge through tympanostomy tubes. J. Pediatr. 134:459-463. [DOI] [PubMed] [Google Scholar]
- 28.Tang, Y. W., N. M. Ellis, M. K. Hopkins, D. H. Smith, D. E. Dodge, and D. H. Persing. 1998. Comparison of phenotypic and genotypic techniques for identification of unusual aerobic pathogenic gram-negative bacilli. J. Clin. Microbiol. 36:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanner, M. A., D. Shoskes, A. Shahed, and N. R. Pace. 1999. Prevalence of corynebacterial 16S rRNA sequences in patients with bacterial and “nonbacterial” prostatitis. J. Clin. Microbiol. 37:1863-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Templeton, K. E., S. A. Scheltinga, P. Sillekens, J. W. Crielaard, A. P. van Dam, H. Goossens, and E. C. Claas. 2003. Development and clinical evaluation of an internally controlled, single-tube multiplex real-time PCR assay for detection of Legionella pneumophila and other Legionella species. J. Clin. Microbiol. 41:4016-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson, J. D., D. G., Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Troesch, A., H. Nguyen, C. G. Miyda, S. Desvarenne, T. R. Gingeras, P. M. Kaplan, P. Cros, and C. Mabilat. 1999. Mycobacterium species identification and rifampin resistance testing with high-density DNA probe arrays. J. Clin. Microbiol. 37:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, R.-F., M. L. Beggs, L. H. Robertson, and C. E. Cerniglia. 2002. Design and evaluation of oligonucleotide-microarray method for the detection of human intestinal bacteria in fecal samples. FEMS Microbiol. Lett. 213:175-182. [DOI] [PubMed] [Google Scholar]
- 34.Waris, M. E., P. Toikka, T. Saarinen, S. Nikkari, O. Meurman, R. Vainionpaa, J. Metsola, and O. Ruuskanen. 1998. Diagnosis of Mycoplasma pneumoniae pneumonia in children. J. Clin. Microbiol. 36:3155-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilbrink, B., I. M. van der Heijden, L. M. Schouls, J. D. van Embden, J. M. Hazes, F. C. Breedveld, and P. P. Tak. 1998. Detection of bacterial DNA in joint samples from patients with undifferentiated arthritis and reactive arthritis, using polymerase chain reaction with universal 16S ribosomal RNA primers. Arthritis Rheum. 41:535-543. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto, S., and S. Harayama. 1995. PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl. Environ. Microbiol. 61:1104-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]