Abstract
The genome of retroviruses, including human immunodeficiency virus type 1 (HIV-1), consists of two identical RNA strands that are packaged as noncovalently linked dimers. The core packaging and dimerization signals are located in the downstream part of the untranslated leader of HIV-1 RNA—the Ψ and the dimerization initiation site (DIS) hairpins. The HIV-1 leader can adopt two alternative conformations that differ in the presentation of the DIS hairpin and consequently in their ability to dimerize in vitro. The branched multiple-hairpin (BMH) structure folds the poly(A) and DIS hairpins, but these domains are base paired in a long distance interaction (LDI) in the most stable LDI conformation. This LDI-BMH riboswitch regulates RNA dimerization in vitro. It was recently shown that the Ψ hairpin structure is also presented differently in the LDI and BMH structures. Several detailed in vivo studies have indicated that sequences throughout the leader RNA contribute to RNA packaging, but how these diverse mutations affect the packaging mechanism is not known. We reasoned that these effects may be due to a change in the LDI-BMH equilibrium, and we therefore reanalyzed the structural effects of a large set of leader RNA mutations that were presented in three previous studies (J. L. Clever, D. Mirandar, Jr., and T. G. Parslow, J. Virol. 76:12381-12387, 2002; C. Helga-Maria, M. L. Hammarskjold, and D. Rekosh, J. Virol. 73:4127-4135, 1999; R. S. Russell, J. Hu, V. Beriault, A. J. Mouland, M. Laughrea, L. Kleiman, M. A. Wainberg, and C. Liang, J. Virol. 77:84-96, 2003). This analysis revealed a strict correlation between the status of the LDI-BMH equilibrium and RNA packaging. Furthermore, a correlation is apparent between RNA dimerization and RNA packaging, and these processes may be coordinated by the same LDI-BMH riboswitch mechanism.
Retroviral particles package a dimeric RNA genome that is subsequently reverse transcribed into DNA by the virion-associated reverse transcriptase enzyme. The cis-acting RNA sequences and the trans-acting viral proteins that mediate RNA dimerization and RNA packaging have been studied extensively for several retroviruses, including human immunodeficiency virus type 1 (HIV-1) (9, 20, 35). It has proven particularly difficult to accurately map the RNA signals that execute these two processes, but most studies have indicated that these elements cluster within the untranslated leader region of the HIV-1 genome. In vitro studies have identified the dimerization initiation signal (DIS) as the primary dimerization signal, which acts through base pairing of a palindromic 6-nucleotide (nt) sequence motif in the exposed loop of the DIS hairpin (4, 27, 32, 40). Nevertheless, studies with mutant virions have indicated that the in vivo situation is much more complex, suggesting the possibility of multiple accessory dimerization signals (6, 21, 38). The Ψ hairpin has been implicated as the core element that mediates packaging, but there is ample evidence for the involvement of additional sequence elements (25, 28). It has also been suggested that the process of RNA dimerization may be coupled to RNA encapsidation; such a coupling seems to be an elegant mechanism for ensuring that only dimeric RNA genomes are packaged (18).
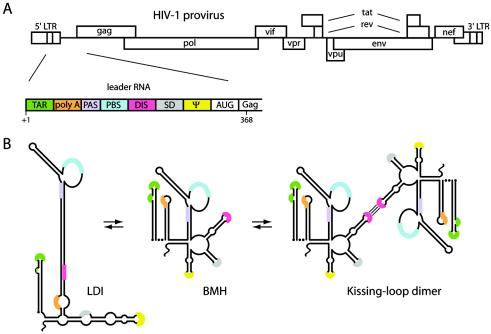
Huthoff and Berkhout demonstrated that the HIV-1 untranslated leader RNA can adopt two mutually exclusive conformations that differ in the presentation of the DIS hairpin motif in vitro; this riboswitch has been shown to regulate the process of RNA dimerization (Fig. 1) (23). More recently, Abbink and Berkhout implicated downstream leader sequences, including the Ψ hairpin, in this conformational change (1), suggesting the possibility that both RNA dimerization and RNA packaging are functionally coupled through the same riboswitch. This mechanism is not exclusive for HIV-1, as the HIV-2 leader also adopts alternative conformations to regulate dimerization in vitro (17, 26). Mutations throughout the leader RNA can affect the delicate equilibrium between the dimerization-incompetent long-distance interaction (LDI) structure, in which the DIS element is masked by an LDI with the upstream poly(A) domain, and the branched multiple-hairpin (BMH) structure, which exposes the DIS hairpin for “kissing-loop” dimerization (Fig. 1) (17, 23). We reasoned that the phenotype of leader mutants should be evaluated in the context of this LDI-BMH equilibrium. In vitro studies indeed confirmed the correlation between BMH-folding capacity and the ability to form dimers (24). Furthermore, the Mfold algorithm was found to provide a very useful tool for accurately predicting the impact of RNA mutations on the LDI-BMH equilibrium (23). To analyze the in vivo effects of leader mutations that affect the LDI-BMH riboswitch, we used the Mfold algorithm to analyze the folding properties of a large set of mutant virions that were previously analyzed for packaging and dimerization (9, 20, 35).
FIG. 1.
Alternative foldings of the HIV-1 leader RNA. (A) Schematic of the 9.2-kb HIV-1 genome. The 5′ and 3′ long terminal repeats (LTR) and all nine open reading frames are indicated. The untranslated leader RNA consists of several regulatory domains. poly A, polyadenylation domain with the AAUAAA sequence; PAS, primer activation signal; SD, major splice donor; ψ, core packaging signal; AUG, Gag start codon. (B) The HIV-1 leader RNA can adopt two conformations. Detailed structure models of the LDI and BMH structures have been presented elsewhere. The poly(A) (orange) and DIS (pink) sequences are base paired to form the LDI structure. The same sequences form the poly(A) and DIS hairpins in the BMH structure. The riboswitch model for regulated dimerization argues that the ground-state LDI structure must first be rearranged into the BMH conformation to expose the DIS hairpin, which mediates subsequent RNA dimerization.
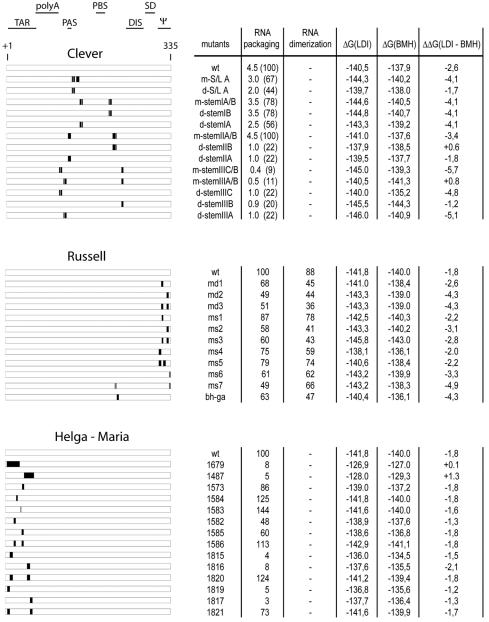
We reevaluated the results of previous studies of RNA packaging in mutant HIV-1 virions with respect to the LDI-BMH riboswitch concept. For this purpose, we focused on three studies that each provide a detailed quantitative analysis of a large set of RNA mutations throughout the HIV-1 leader (Fig. 2). Clever et al. (9) analyzed 13 mutations in the region surrounding the primer-binding site (PBS) element for their effect on RNA packaging. These changes include the opening of several base-paired stem segments in the PBS region, by mutating either the left or the right side of the stems, and double mutants in which base pairing is restored (Fig. 2). Packaging was measured as the ratio of full-length to spliced viral RNA within virion particles. Russell et al. (35) analyzed 14 HIV-1 mutants with changes in the leader and the 5′ part of the Gag open reading frame. They focused on the 11 leader mutations, and this set includes mutations in the loop and the stem of the Ψ hairpin and in the CU-rich stretch upstream of the DIS region (Fig. 2). The effect of these mutations on RNA packaging was expressed as the ratio of genomic to spliced RNA within virions. Russell et al. (35) also analyzed the dimerization status of the packaged RNA by native gel electrophoresis. Helga-Maria et al. (20) analyzed the effect of 14 mutations in the upstream trans-activation response element (TAR) hairpin on RNA packaging, measured as the ratio of virion-associated RNA to capsid (CA)-p24 protein. This set includes mutations that disrupt the TAR hairpin and double mutants in which the structure is restored (Fig. 2).
FIG. 2.
HIV-1 leader mutants. The RNA packaging and dimerization values for 38 HIV-1 leader RNA mutants were taken from previous studies. Black blocks indicate the positions of the mutations in the leader RNA; several regulatory domains within the leader are indicated at the top. The packaging efficiencies were measured as the ratio of genomic RNA to spliced RNA within virions in the studies of Clever et al. (9) and Russell et al. (35). In the study of Helga-Maria et al. (20), packaging was determined as the ratio of virion-associated RNA to CA-p24 protein. All RNA packaging values are relative to that of the corresponding wild-type (wt) construct, which was set at 100%. To obtain these values, we converted the data of Clever et al. into relative values (in parentheses). The dimerization values in the study of Russell et al. reflect the percentages of dimeric and monomeric RNA genomes from virions, as analyzed on a nondenaturing gel. The thermodynamic stabilities (ΔG, in kilocalories per mole) of the LDI and BMH structures were calculated by using the Mfold program and were used to calculate ΔΔG values for LDI-BMH.
We analyzed the impact of these 38 leader RNA mutations on the LDI-BMH equilibrium with the Mfold program, version 3.0 (http://mfold.burnet.edu.au) (29). Huthoff and Berkhout previously reported that structures predicted by the Mfold program are compatible with results from RNA structure probing studies (23). Furthermore, the Mfold program also accurately predicts the LDI-BMH folding characteristics of leader RNA mutants (23, 24). Settings were standard for all folding experiments, corresponding to conditions at 37°C and 1.0 M NaCl. We used a 500-nt sequence that includes the complete leader region (nt 1 to 335) of the respective viral isolates—HXB2 in the studies of Clever et al. (9) and Helga-Maria et al. (20) and BH10-HXB2 in the study of Russell et al. (35). All three wild-type isolates and 34 of the 38 mutants readily adopt the LDI structure as the energetically most favorable structure. To analyze the BMH structure of these transcripts, we forced the folding of the DIS hairpin by using the constraint option in the Mfold program. Only four mutants reversed the equilibrium and preferentially folded the BMH structure. The LDI conformation of these mutants was obtained in the Mfold program by prohibiting the folding of the DIS hairpin.
The thermodynamic stabilities (ΔG, in kilocalories per mole) of the LDI and BMH foldings are shown in Fig. 2. These energy values were used to calculate ΔΔG, which provides a measure of the status of the LDI-BMH equilibrium. A negative ΔΔG value for LDI-BMH indicates that the RNA structure preferentially folds the LDI structure, whereas a positive value reflects a preference for the BMH conformation. When both LDI and BMH foldings have equal ΔG values and thus a ΔΔG value of zero, the LDI and BMH conformations are present in an equimolar ratio. The ΔΔG values for the wild-type leader are −2.6, −1.8, and −1.8 kcal/mol in the studies of Clever et al. (9), Russell et al. (35), and Helga-Maria et al. (20), respectively. These data indicate that wild-type HIV-1 RNA is in the LDI conformation and that the BMH conformation is adopted by less than 10% of the molecules (16). The mutations in the study of Clever et al. result in either a further stabilization of the LDI structure or a shift toward the BMH conformation. All mutations introduced by Russell et al. are predicted to shift the equilibrium toward the LDI structure. Most TAR mutations in the study of Helga-Maria et al. shift the equilibrium toward the BMH conformation, according to the Mfold program (Fig. 2).
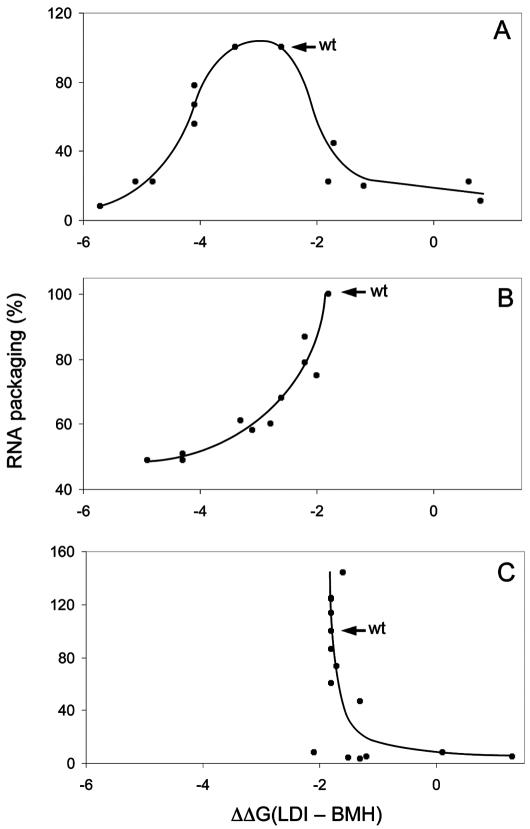
To study the effect of a shift in the LDI-BMH equilibrium on RNA packaging, we plotted the ΔΔG values of the mutants and the corresponding wild-type isolates against the RNA packaging efficiencies measured in the three studies. We first analyzed the data of Clever et al. (35) with mutations that shift the equilibrium toward either the LDI or the BMH structure (Fig. 3A). The results show a striking correlation between the ΔΔG values for the LDI-BMH equilibrium and RNA packaging. The wild type and the m-stem IIA/B mutant with ΔΔG values of approximately −3 kcal/mol are the most efficient in packaging. The packaging efficiency drops rapidly when the ΔΔG value is either increased or decreased. These results indicate that the wild-type LDI-BMH equilibrium is optimal for efficient packaging, but further stabilization of either the LDI or the BMH structure is incompatible with efficient packaging. A gradual loss of packaging efficiency is observed in both directions, and a profound packaging defect (<20% wild-type packaging) is apparent for mutants with ΔΔG values of less than −5 kcal/mol (more firmly LDI) and ΔΔG values of more than −1.5 kcal/mol (toward BMH).
FIG. 3.
HIV-1 RNA packaging is controlled by the LDI-BMH equilibrium. The relative RNA packaging efficiencies of the mutant sets shown in Fig. 2 are plotted against the ΔΔG values for LDI-BMH. (A, B, and C) Data sets from the studies of Clever et al. (9), Russell et al. (35), and Helga-Maria et al. (20), respectively. The wild-type (wt) constructs are indicated by arrows.
A similar pattern is apparent for mutants in the study of Russell et al. (35) (Fig. 3B). The wild type (ΔΔG value, −1.8 kcal/mol) is the most efficient in packaging, and a change in the equilibrium toward the LDI structure gradually decreases the packaging efficiency. The study of Helga-Maria et al. (20) provides data toward the other end of the spectrum because most of the mutants that they tested shift the equilibrium toward the BMH structure (Fig. 3C). Wild-type RNA is packaged efficiently, and some mutants with very similar ΔΔG values in fact demonstrate a slight increase in packaging efficiency. However, a further shift in the LDI-BMH equilibrium toward the BMH structure results in a dramatic packaging defect. These combined results indicate that a finely tuned equilibrium between the two leader RNA structures is important for RNA packaging.
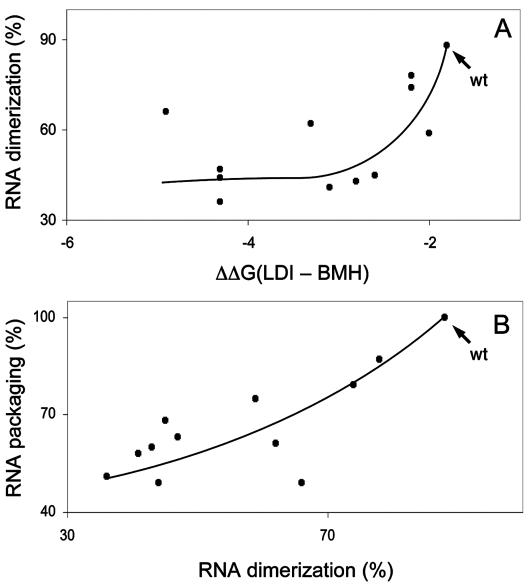
Russell et al. (35) also measured the amounts of RNA dimers by native gel electrophoresis of the virion-associated RNA. This analysis provides an opportunity to assess whether in vivo dimerization is also affected by changes in the LDI-BMH equilibrium. We therefore plotted the dimerization efficiencies against the ΔΔG values (Fig. 4A). The wild type demonstrates an optimal RNA dimerization level, which was gradually decreased when the LDI structure was further stabilized in the mutants. Interestingly, this pattern is very similar to that observed for RNA packaging (Fig. 3B), indicating that both processes are similarly affected by the status of the LDI-BMH equilibrium. To illustrate this finding, we plotted the RNA packaging values against the dimerization efficiencies (Fig. 4B). A linear correlation is apparent between RNA packaging and RNA dimerization, a finding which underscores the notion that these two processes are functionally and mechanistically coupled (35-37).
FIG. 4.
HIV-1 RNA dimerization and RNA packaging are coupled. (A) The RNA dimerization of the mutant set of Russell et al. (35) shown in Fig. 2 is plotted against ΔΔG values for LDI-BMH. (B) The RNA packaging of the mutant set of Russell et al. is plotted against the RNA dimerization. The wild-type (wt) constructs are marked by arrows.
It has been notoriously difficult to accurately map the HIV-1 RNA dimerization and RNA packaging signals. Although the DIS and Ψ signals are generally thought to form the core elements, several additional elements, including sequences in the upstream leader domain and the first part of the Gag open reading frame, have been reported to positively or negatively affect these functions (8, 9, 14, 20, 30). Two possible explanations come to mind. First, studies with mutant viruses may be complicated by the fact that several domains of the untranslated leader encode multiple overlapping functions (5). For instance, both upstream TAR and poly(A) hairpins contribute to packaging (12, 30). However, these elements also play a role in transcriptional activation and RNA processing, respectively, and a subsequent analysis indicated that mutation of the poly(A) hairpin reduced the amount of intracellular HIV-1 RNA to the same extent, such that no net packaging defect is apparent (13). This example underscores the intricacies of dealing with the multifunctional HIV-1 leader RNA. Because of the superimposed demands of multiple viral replication steps, it is also difficult to study RNA dimerization and RNA packaging functions separately.
Second, it is possible that the effect of leader RNA mutations should be interpreted in the context of the overall leader RNA structure. It was proposed that the complete leader RNA is required to fold a specific tertiary RNA structure that is recognized in the processes of RNA dimerization and packaging (4). Evidence for a specific conformation of the complete leader was subsequently reported (7), but the analysis indicated that it was not a tertiary folding but an alternative secondary structure with long-distance base pairing between the poly(A) and DIS domains (23). This ground-state LDI structure can refold into the more widely known BMH structure that exposes the poly(A) and DIS hairpins. This new riboswitch concept explains the in vitro dimerization properties of diverse HIV-1 RNA mutants. Mutants with stable LDI folding show a dimerization defect, and mutants with spontaneous folding of the BMH structure dimerize much more efficiently than the wild-type control (22, 23).
We now provide evidence that the impact of diverse leader RNA mutations on the LDI-BMH equilibrium can also explain the dimerization and packaging properties of 38 mutant RNA genomes that were previously tested in vivo (9, 20, 35). Mutants with mutations that do not grossly affect the LDI-BMH equilibrium package RNA efficiently, but a change in the equilibrium toward the LDI or BMH structure leads to a loss of RNA packaging; the dimerization properties of the mutant set of Russell et al. (35) follow this trend. Thus, freezing of either leader RNA conformation causes a packaging defect, indicating that both structures are required in the multistep processes of RNA packaging and RNA dimerization. In vivo, the viral RNA will be coated by cellular and viral proteins that modulate the properties of the RNA molecule. It is therefore striking that in vivo packaging and dimerization properties can be explained by inspection of the LDI-BMH equilibrium. The data indicate that the leader RNA molecule plays an active role in these processes and that viral and/or cellular proteins apparently have no major influence on the LDI-BMH equilibrium.
It is possible that this riboswitch regulates early functions in the cell, e.g., translation of HIV-1 RNA (1), and that a Gag/nucleocapsid-mediated switch to the BMH structure occurs at a later stage during the RNA dimerization and RNA packaging processes. Furthermore, late functions within the virion, such as reverse transcription, could also be regulated by the same riboswitch (3).
It may seem strange that TAR mutations affect RNA packaging because the TAR hairpin is present in both LDI and BMH conformations (Fig. 1). However, it was suggested previously that opening of the TAR stem can induce the refolding of downstream sequences through base pairing with TAR sequences (19, 20, 22). Thus, mutation of TAR sequences may indirectly influence leader RNA structure and function.
Exposure of the DIS hairpin in the BMH conformation readily explains the ability of several mutants to dimerize efficiently in vitro. Unlike the DIS hairpin, the Ψ hairpin is present in both LDI and BMH structures, but its structural contexts differ significantly (1). The Ψ hairpin stem is extended in the LDI structure and includes the upstream Shine-Dalgarno (SD) signal and the downstream Gag start codon (Fig. 1). In the BMH structure, the lower part of this extended Ψ hairpin is opened to allow the formation of the upstream SD hairpin, whereas the downstream Gag sequences engage in base pairing with the upstream U5 sequences to form the U5-AUG duplex (1). The DIS, SD, and Ψ regions are high-affinity binding sites for Gag molecules (2, 10, 15, 39). Because the structures of these regions differ significantly in the LDI and BMH foldings, the riboswitch could provide a means for modulating RNA packaging (11). This notion would be consistent with our finding that the LDI-BMH equilibrium is highly important for efficient packaging.
Our current analysis demonstrates that mutant HIV-1 RNA molecules should be interpreted in terms of their impact on the LDI-BMH riboswitch. In leader RNA-mutated viruses, it is quite possible that compensatory changes occur in the Gag protein (33, 34). Different HIV-1 isolates may also differ slightly in the ΔΔG values of their leader RNAs; e.g., we measured values of −2.6 and −1.8 kcal/mol for the HXB2 and HXB2-BH10 isolates, respectively. However, these viruses are able to package viral genomic RNA efficiently. This finding is not surprising, considering that the leader RNA region is not the sole determinant for RNA packaging, which also involves Gag proteins. Presumably, each HIV-1 isolate possesses mutually adapted leader RNA sequences and Gag proteins that act in concert to efficiently encapsidate viral genomic RNA.
Using the detailed quantitative results of the study of Russell et al. (35), we also found a correlation between the dimerization and packaging properties of mutant RNAs in virions (Fig. 4B). This finding further supports the idea that these two processes are coupled, and the LDI-BMH riboswitch provides a mechanistic explanation. Further detailed RNA structural knowledge for this riboswitch may allow the design of nucleic acid or other compounds that affect this RNA equilibrium, for instance, by preferential binding to one of the conformations. Ooms et al. recently identified certain antisense DNA oligonucleotides that bind differentially to the two leader RNA conformations (31). Based on the results presented in this study, it seems possible that such compounds will interfere with several critical replication steps and thus represent interesting compounds for the generation of a new class of antiviral agents.
Acknowledgments
We thank Wim van Est for artwork.
RNA studies in the laboratory of Ben Berkhout are supported by NWO-CW and ZonMw (VICI Program).
REFERENCES
- 1.Abbink, T. E., and B. Berkhout. 2003. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 278:11601-11611. [DOI] [PubMed] [Google Scholar]
- 2.Amarasinghe, G. K., J. Zhou, M. Miskimon, K. J. Chancellor, J. A. McDonald, A. G. Matthews, R. R. Miller, M. D. Rouse, and M. F. Summers. 2001. Stem-loop SL4 of the HIV-1 psi RNA packaging signal exhibits weak affinity for the nucleocapsid protein. Structural studies and implications for genome recognition. J. Mol. Biol. 314:961-970. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout, B., M. Ooms, N. Beerens, H. Huthoff, E. Southern, and K. Verhoef. 2002. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J. Biol. Chem. 277:19967-19975. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout, B. 1996. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acids Res. Mol. Biol. 54:1-34. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout, B. 2000. Multiple biological roles associated with the repeat (R) region of the HIV-1 RNA genome. Adv. Pharmacol. 48:29-73. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout, B., and J. L. B. van Wamel. 1996. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 70:6723-6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkhout, B., and J. L. B. van Wamel. 2000. The leader of the HIV-1 RNA genome forms a compactly folded tertiary structure. RNA 6:282-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clever, J. L., D. A. Eckstein, and T. G. Parslow. 1999. Genetic dissociation of the encapsidation and reverse transcription functions in the 5′R region of human immunodeficiency virus type 1. J. Virol. 73:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clever, J. L., D. Mirandar, Jr., and T. G. Parslow. 2002. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 76:12381-12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Damgaard, C. K., H. Dyhr-Mikkelsen, and J. Kjems. 1998. Mapping the RNA binding sites for human immunodeficiency virus type-1 Gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 26:3667-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Darlix, J. L., M. L. Lastra, Y. Mely, and B. Roques. 2002. Nucleocapsid protein chaperoning of nucleic acids at the heart of HIV structure, assembly and cDNA synthesis, p. 69-88. In C. Kuiken, B. Foley, E. Freed, B. Hahn, P. Marx, F. McCutchan, J. Mellors, S. Wolinsky, and B. Korber (ed.), HIV sequence compendium 2002. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 12.Das, A. T., B. Klaver, and B. Berkhout. 1998. The 5′ and 3′ TAR elements of the human immunodeficiency virus exert effects at several points in the virus life cycle. J. Virol. 72:9217-9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das, A. T., B. Klaver, and B. Berkhout. 1999. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J. Virol. 73:81-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das, A. T., B. Klaver, B. I. F. Klasens, J. L. B. van Wamel, and B. Berkhout. 1997. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J. Virol. 71:2346-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Guzman, R. N., Z. Rong Wu, C. C. Stalling, L. Pappalardo, P. N. Borer, and M. F. Summers. 1998. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science 279:384-388. [DOI] [PubMed] [Google Scholar]
- 16.de Smit, M. H., and J. van Duin. 1990. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc. Natl. Acad. Sci. USA 87:7668-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirac, A. M., H. Huthoff, J. Kjems, and B. Berkhout. 2002. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic Acids Res. 30:2647-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harrich, D., C. W. Hooker, and E. Parry. 2000. The human immunodeficiency virus type 1 TAR RNA upper stem-loop plays distinct roles in reverse transcription and RNA packaging. J. Virol. 74:5639-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Helga-Maria, C., M. L. Hammarskjold, and D. Rekosh. 1999. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J. Virol. 73:4127-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill, M. K., M. Shehu-Xhilaga, S. M. Campbell, P. Poumbourios, S. M. Crowe, and J. Mak. 2003. The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J. Virol. 77:8329-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huthoff, H., and B. Berkhout. 2001. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res. 29:2594-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huthoff, H., and B. Berkhout. 2001. Two alternating structures for the HIV-1 leader RNA. RNA 7:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huthoff, H., and B. Berkhout. 2002. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry 41:10439-10445. [DOI] [PubMed] [Google Scholar]
- 25.Kim, H.-J., K. Lee, and J. J. O'Rear. 1994. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology 198:336-340. [DOI] [PubMed] [Google Scholar]
- 26.Lanchy, J. M., and J. S. Lodmell. 2002. Alternate usage of two dimerization initiation sites in HIV-2 viral RNA in vitro. J. Mol. Biol. 319:637-648. [DOI] [PubMed] [Google Scholar]
- 27.Laughrea, M., and L. Jette. 1994. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry 33:13464-13474. [DOI] [PubMed] [Google Scholar]
- 28.Luban, J., and S. P. Goff. 1994. Mutational analysis of cis-acting packaging signals in human immunodeficiency type 1 RNA. J. Virol. 68:3784-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 30.McBride, M. S., M. D. Schwartz, and A. T. Panganiban. 1997. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 71:4544-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ooms, M., K. Verhoef, E. Southern, H. Huthoff, and B. Berkhout. 2004. Probing alternative foldings of the HIV-1 leader RNA by antisense oligonucleotide scanning arrays. Nucleic Acids Res. 32:819-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paillart, J.-C., E. Skripkin, B. Ehresmann, C. Ehresmann, and R. Marquet. 1996. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 93:5572-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rong, L., R. S. Russell, J. Hu, Y. Guan, L. Kleiman, C. Liang, and M. A. Wainberg. 2001. Hydrophobic amino acids in the human immunodeficiency virus type 1 p2 and nucleocapsid proteins can contribute to the rescue of deleted viral RNA packaging signals. J. Virol. 75:7230-7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rong, L., R. S. Russell, J. Hu, M. Laughrea, M. A. Wainberg, and C. Liang. 2003. Deletion of stem-loop 3 is compensated by second-site mutations within the Gag protein of human immunodeficiency virus type 1. Virology 314:221-228. [DOI] [PubMed] [Google Scholar]
- 35.Russell, R. S., J. Hu, V. Beriault, A. J. Mouland, M. Laughrea, L. Kleiman, M. A. Wainberg, and C. Liang. 2003. Sequences downstream of the 5′ splice donor site are required for both packaging and dimerization of human immunodeficiency virus type 1 RNA. J. Virol. 77:84-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakuragi, J. I., S. Ueda, A. Iwamoto, and T. Shioda. 2003. Possible role of dimerization in human immunodeficiency virus type 1 genome RNA packaging. J. Virol. 77:4060-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakuragi, J. I., A. Iwamoto, and T. Shioda. 2002. Dissociation of genome dimerization from packaging functions and virion maturation of human immunodeficiency virus type 1. J. Virol. 76:959-967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen, N., L. Jette, C. Liang, M. A. Wainberg, and M. Laughrea. 2000. Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J. Virol. 74:5729-5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shubsda, M. F., A. C. Paoletti, B. S. Hudson, and P. N. Borer. 2002. Affinities of packaging domain loops in HIV-1 RNA for the nucleocapsid protein. Biochemistry 41:5276-5282. [DOI] [PubMed] [Google Scholar]
- 40.Skripkin, E., J. C. Paillart, R. Marquet, B. Ehresmann, and C. Ehresmann. 1994. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA 91:4945-4949. [DOI] [PMC free article] [PubMed] [Google Scholar]