Abstract
The release of retroviruses from the plasma membrane requires host factors that are believed to be recruited to the site of budding by the late (L) domain of the virus-encoded Gag protein. The L domain of Rous sarcoma virus (RSV) has been shown to interact with a ubiquitin (Ub) ligase, and budding of this virus is dependent on Ub. RSV is similar to other retroviruses in that it contains ∼100 molecules of Ub, but it is unique in that none of these molecules has been found to be conjugated to Gag. If transient ubiquitination of RSV Gag is required for budding, then replacement of the target lysine(s) with arginine should prevent the addition of Ub and reduce budding. Based on known sites of ubiquitination in other viruses, the important lysines would likely reside near the L domain. In RSV, there are five lysines located just upstream of the L domain in a region of the matrix (MA) protein that is dispensable for membrane binding, and replacement of these with arginine (mutant 1-5KR) reduced budding 80 to 90%. The block to budding was found to be on the plasma membrane; however, the few virions that were released had normal size, morphology, and infectivity. Budding was restored when any one of the residues was changed back to lysine or when lysines were inserted in novel positions, either within this region of MA or within the downstream p10 sequence. Moreover, the 1-5KR mutant could be rescued into particles by coexpression of budding-competent Gag molecules. These data argue that the phenotype of mutant 1-5KR is not due to a conformational defect. Consistent with the idea that efficient budding requires a specific role for lysines, human T-cell leukemia virus type 1, which does not bud well compared to RSV and lacks lysines close to its L domain, was found to be released at a higher level upon introduction of lysines near its L domain. This report strongly supports the hypothesis that ubiquitination of the RSV Gag protein (and perhaps those of other retroviruses) is needed for efficient budding.
Ubiquitin (Ub) is a small protein that cells use to modify other proteins to accomplish a remarkably diverse set of tasks that are important at nearly all levels of cellular activity, including DNA replication, transcription, translation, protein folding, sorting, signaling, and degradation. Modification occurs on the ɛ-amino group of lysines of target proteins and results in either monoubiquitination or polyubiquitination (i.e., chains of Ub). While polyubiquitination usually targets proteins to the proteasome for degradation, monoubiquitination provides a signal that alters or regulates the function of the modified protein (16, 42, 48, 51, 54).
Over the past few years, numerous observations have been made that together seem to indicate an important role for monoubiquitination in the late stages of retrovirus budding on the plasma membrane. First, all retroviruses that have been examined have been found to contain ∼100 molecules of Ub, an amount that is much higher than can be explained by random trapping during budding (30-32, 41). Second, some of these Ub molecules are individually linked to Gag, the viral protein that drives particle assembly and budding (30-32). Third, the identified sites of monoubiquitination are invariably near the viral late (L) domains (30, 31), which are responsible for recruiting host factors needed for the late steps in budding (12). Fourth, virus release has been shown to correlate with the recruitment of Ub ligase activity by L domains (2, 21, 55, 64). Fifth, Tsg101, a host factor recruited by the L domains of some retroviruses, is a Ub-binding protein (38, 39). Sixth, the release of some retroviruses has been shown to be reduced by proteasome inhibitors, which lower the levels of free Ub in the cytoplasm (2, 32, 34, 49). Seventh, in the case of Rous sarcoma virus (RSV), the effects of proteasome inhibitors can be counteracted by overexpressing Ub in trans or by fusing it to the C terminus of Gag (34). Eighth, it has been noted (34) that retrovirus budding bears a topological resemblance to the cellular budding events that lead to the formation of multivesicular bodies (MVBs), and the movement of cellular proteins into these nascent buds on late endosomes requires transient monoubiquitination (11). Ninth, there is increasing evidence that retrovirus budding makes use of host factors involved in MVB formation (13, 26, 40, 56, 57).
Although the evidence for a role of Ub in retrovirus budding is extensive, there are several observations that would seem to argue against this idea. First, all of the Ub present in RSV is unconjugated (41). This could be due to rapid deubiquitination of Gag, either naturally during virus budding or artifactually during lysis of cells or virions. In this regard, it is interesting that Ub-mediated sorting of proteins into MVBs is accompanied by rapid deubiquitination (11), and on the plasma membrane, the photoreceptor of the Drosophila eye is not endocytosed until after Ub is removed, unlike most examples where Ub promotes endocytosis (7). Alternatively, it is possible that RSV Gag is never modified, but instead Ub conjugation might be required on a cellular protein that is essential for budding. Although this is what is believed to be the case for endocytosis of the growth hormone receptor (14), a trans role for Ub is less likely to be the case for RSV given that a Gag-Ub fusion protein can bud even in the presence of proteasome inhibitors, which inhibit wild-type Gag (34). Second, the release of some retroviruses (e.g., equine infectious anemia virus and mouse mammary tumor virus) is not affected by proteasome inhibitors even though their Gag proteins are modified by Ub (31-33). Such viruses might make use of alternative budding mechanisms or carry redundant “Ub-like” information that enables budding when Ub is limiting (33). Third, elimination of known sites of ubiquitination on the Gag proteins of human immunodeficiency virus type 1 (HIV-1) and murine leukemia virus (MLV) has no effect on budding or infectivity (30). The significance of this observation is difficult to ascertain because the Ub literature contains many examples of secondary sites of ubiquitination being used when the primary sites are eliminated (25, 36, 45). Moreover, in light of the many examples of transient monoubiquitination, the sites of Ub modification that have been mapped on Gag proteins could correspond to secondary sites that happen to be more resistant to deubiquitination. Alternatively, Ub modification of some Gag proteins might truly be unimportant for budding and may occur on occasion merely because these viral proteins pass by Ub ligases on their way out of the cell.
This study addresses the importance of lysines for the late steps of RSV budding. Although direct evidence for a Ub-modified Gag protein remains elusive for this virus, the experiments described below unequivocally show that a cluster of five lysines near the L domain are important for budding.
MATERIALS AND METHODS
Expression vectors.
The wild-type RSV gag gene was originally derived from the Prague C proviral vector, pATV-8 (19, 52). To analyze budding, gag alleles were transferred into either proviral vector pRC.V8 (10) or pGag.GFP (4), the latter of which contains a gag-egfp fusion. For quantitative infectivity studies, gag alleles were cloned into pRS.V8.eGFP (5), which also carries the coding sequence for enhanced green fluorescent protein (eGFP) in a nonessential region of the genome. For mutagenesis of the human T-cell leukemia virus type 1 (HTLV-1) gag gene, it was subcloned into the pMH vector (Boehringer Mannheim, Indianapolis, Ind.) to construct pMH-HTLVGag (58).
Mutagenesis.
Construction of pT10C.GFP was described previously (34). pΔMA3.GFP was made by cutting pSV.ΔMA3 (63) with SstI and BspEI and moving the 5′ gag fragment into the equivalent location in pGag.GFP. pGag.3h.GFP was created by amplifying gag sequence from pRC.V8 with a forward primer (5′-GATCTCGAGCTCTACTGCAGG-3′) spanning the SstI site upstream of the initiation codon and a reverse primer (5′-CCTAACCAAGGGGGGCCCGAGATGTTCCAT-3′) in the PR coding sequence, thereby generating the “3h” deletion mutant described previously (59) while simultaneously creating an ApaI site at the 3′ site for subsequent fusion of gag with egfp in pCMV.N2-eGFP (Clontech). The PCR products and recipient plasmid were digested with SstI and ApaI and then ligated. To create pGag.3h, the egfp coding sequence was removed from this recombinant by digestion with ApaI and NotI, followed by incubation with Klenow fragment and ligation. This manipulation resulted in the introduction of two foreign amino acids (arginine and leucine) in the place of eGFP. Lysine-to-arginine substitutions in the NC domain were created by three sequential rounds of mutagenesis by the QuikChange method (Stratagene, La Jolla, Calif.), using pGag.3h.GFP as the template. In the first round, the K36, K37, and K39 codons (AAA, AAA, and AAG, respectively) were changed to CGG, AGA, and AGG, respectively, and as a result a new SstII site overlapping the K36 codon was introduced. The second round of mutagenesis changed the K58 (AAA) and K62 (AAG) codons to CGC and AGG, respectively, and at the same time, codon A57 was silently changed from GCT to GCG to introduce a BssHII site. Finally, the G72 and K73 codons were changed from GGA AAA to GGC CGC, creating another SstII site and the final lysine-to-arginine substitution. Following mutagenesis, the entire gag gene was sequenced after screening presumptive clones for the new restriction sites. Lysine-to-arginine, arginine-to-lysine, and glutamate-to-lysine substitutions in MA, p10, and CA were carried out by either M13 mutagenesis with MGAG-S as a template (5, 62) or by PCR mutagenesis with pGag.GFP or p1-5KR.GFP as template. All lysine codons were changed to the arginine codon CTG. Arginine and glutamate codons were changed to the AAA lysine codon. Mutations were transferred into pGag.GFP, pRC.V8, or pRS.V8.eGFP with SstI and FseI, thereby replacing the 5′ half of gag. To create the L domain substitution construct, pPPPY-A.GFP, the proline and tyrosine codons were changed to four alanine codons (GCT) by PCR mutagenesis. The mutant allele was then transferred to pGag.GFP with SstI and FseI. To create Gag(−), 1-5KR(−), and T10C(−), the egfp sequences were removed from pGag.GFP, p1-5KR.GFP, and pT10C.GFP, respectively, by digesting the vectors with ApaI and NotI, followed by incubation with Klenow fragment (to create blunt ends) and ligation. This manipulation inserts two foreign amino acids (arginine and leucine) in the place of eGFP. All of the newly constructed RSV gag alleles described here were sequenced to confirm that only the desired mutations were present.
HTLV-1 substitutions at matrix (MA) amino acids 102 and 109 (P102K and D109K) and the double mutant R97K/P102K were made by mutagenesis of pMH-HTLVGag by the QuikChange method, as reported previously (58). The following oligonucleotides were used in the mutagenesis reactions: for P102K, 5′-CCCGTCCCGCGCCACCGAAGCCGTCATCCCCCACC-3′ and 5′-GGTGGGGGATGACGGCTTCGGTGGCGCGGGACGGG-3′; for P109K, 5′-CCGTCATCCCCCACCCACAAACCCCCGGATTCTGATCC-3′ and 5′-GGATCAGAATCCGGGGGTTTGTGGGTGGGGGATGACGG-3′; and for R97K/P102K, 5′-GCCCAGATCCCGTCCAAACCCGCGCCACCGAAGCCGTCATCCCCCACC-3′ and 5′-GGTGGGGGATGACGGCTTCGGTGGCGCGGGTTTGGACGGGATCTGGGC-3′. For expression of HTLV-1 Gag, the mutant alleles were transferred from pMH-HTLVGag into pCMV-HT1 on a DraIII-NheI fragment. All clones were sequenced to confirm the presence of the desired substitutions and to be certain that no undesired substitutions had occurred.
Budding assays.
To quantitate RSV budding, QT6 cells (28) were transfected by the CaPO4 method, as previously described (10), and 24 h later, they were metabolically radiolabeled with l-[35S]methionine or an l-[35S]methionine-cysteine mix (50 μCi, >1,000 Ci/mmol). Cells transfected with plasmids expressing Gag proteins which lack PR (e.g., Gag.GFP) were radiolabeled for 2.5 h, and viral proteins were immunoprecipitated from detergent-lysed cells and particles with a polyclonal rabbit serum against whole RSV, as previously described (59). Immunoprecipitates were separated in sodium dodecyl sulfate (SDS)-12% polyacrylamide gels, which were subsequently dried and exposed to Kodak X-Omat AR5 X-ray film. Gag proteins were also quantitated by PhosphorImager (Molecular Dynamics) analysis. The budding proficiency was calculated as the amount of Gag in the medium divided by the total amount in the cell lysate and medium.
Cells transfected with proviral constructs (which encode active proteases) were done in duplicate. One plate was pulse-labeled for 5 min, at which time only unprocessed Gag is detected, while the other plate was radiolabeled for 3 h. Gag proteins were immunoprecipitated from detergent-lysates and analyzed by SDS-polyacrylamide electrophoresis. Unprocessed Gag from the 5-min pulse-labeled cells and mature CA protein from the 3-h-labeled medium fractions were quantitated by PhosphorImager analysis. Budding proficiency was calculated as the amount of CA in the medium divided by the amount of Gag in the pulse-labeled cell lysate.
For the complementation experiments, QT6 cells were cotransfected with equal amounts of the two Gag constructs to be tested while a parallel plate was cotransfected with pGag.GFP and pGag(−) to provide a normal-budding control for comparison. Proteins were radiolabeled, immunoprecipitated, analyzed, and quantitated as described above.
To analyze HTLV-1 budding, 293T cells were transfected by the CaPO4 precipitation method. Methods for preparing cell lysates and fractions of media have been detailed previously (58). Briefly, transfected cells were lysed in radioimmunoprecipitation assay (RIPA) buffer and immunoprecipitated with anti-HTLV1-p19 monoclonal antibody (Zeptometrix, Buffalo, N.Y.). Medium fractions collected from transfected cells were subjected to ultracentrifugation for 1 h and 40,000 × g to obtain particles. Western blotting analysis was performed with anti-HTLV1-p19 monoclonal antibody as primary antibody and horseradish peroxidase-conjugated anti-mouse immunoglobulin as secondary antibody with the ECL enhanced chemiluminescence Western analysis kit (Amersham, Arlington Heights, Ill.). The efficiency of particle production was normalized for cell-associated gp46. Quantitation of band intensities was done with the Quantity One software package with the Chemi Doc 2000 documentation system (Bio-Rad, Richmond, Calif.).
Sucrose gradient sedimentation.
The size distribution of 1-5KR mutant particles was evaluated as described previously with only minor variations (22). Briefly, three plates of 106 QT6 cells in 60-mm-diameter dishes were transfected by the CaPO4 method with pRC.V8 (wild-type RSV genome), pRC.1-5KR (mutant), or pGag.3h (normal-size control). At 18 h posttransfection, the cells were labeled with l-[35S]methionine/cysteine for 5 h. Three hundred microliters of either wild-type or 1-5KR particles was mixed with 100 μl of the normal-size control, and the two samples were centrifuged at 26,000 rpm (90,000 × g) for 30 min through 10 to 30% sucrose gradients in an SW41 rotor at 4°C. Fractions were collected from each tube by dripping from the bottom, and the Gag proteins were then immunoprecipitated with anti-RSV rabbit serum, electrophoresed in an SDS-12% polyacrylamide gel, and quantitated by PhosphorImager analysis.
Infectivity assays.
QT6 cells were transfected with pRS.V8.eGFP or pRS.1-5KR.eGFP by the CaPO4 transfection, and virions were allowed to accumulate in the medium for 24 h. The medium fraction was centrifuged at 1,000 × g for 5 min to remove any cells, and half of the cell-free medium was pelleted through a 25% sucrose cushion at 126,000 × g for 40 min at 4°C in a TLA100.4 rotor. Pelleted virions were resuspended in phosphate-buffered saline and analyzed by a reverse transcriptase assay as previously described (10) to determine the amount of virions in the other half of each medium sample. Equal concentrations of virions were placed on DF-1 cells (17, 47) for 24 h, after which new medium was added. The numbers of infected (i.e., green) cells present at subsequent time points were counted by fluorescence-activated cell sorter (FACS) analysis.
Confocal microscopy.
Live QT6 cells were washed with Tris-buffered saline at 24 h posttransfection and overlaid with a glass coverslip. The subcellular locations of GFP-tagged Gag proteins were observed by confocal microscopy with a Zeiss laser-scanning microscope following excitation with a helium-argon laser (488-nm peak excitation).
EM.
QT6 cells were seeded in 60-mm-diameter Permanox dishes (Electron Microscopy Sciences, Ft. Washington, Pa.) at 3 × 106 per plate, transfected with proviral plasmids by the CaPO4 method, and processed for electron microscopy (EM) as described previously (10).
RESULTS
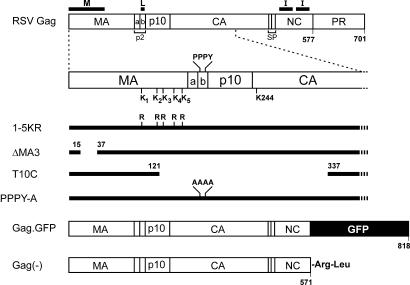
The RSV Gag protein (Fig. 1) contains 31 lysines, and the functions of many of these are known. The MA sequence is rich in lysines, containing nearly 50% of the total. Of the nine lysines in the first half of MA, all are critical for the interaction of the membrane-binding (M) domain with phospholipids in the plasma membrane; however, binding appears to be dependent on charge rather than lysines per se (5, 6). The second half of MA contains five lysines positioned between the M and L domains. Small deletions in this region have no effect on budding or infectivity, suggesting this region contains little structure. However, some large deletions result in budding defects, but this could have been due to conformational defects of surrounding regions (29). There are no lysines within p2 (which contains the L domain) or p10. Capsid (CA) contains eight lysines, but this part of Gag can be deleted in its entirety with minimal effects on budding (60). The six lysines in nucleocapsid (NC) are thought to be involved in RNA binding events that promote tight interactions among Gag proteins, but functionality of the interaction (I) domain appears to be dependent more on charge than lysines per se (3), and replacement of all six of these residues with arginine had no effect of budding (data not shown). Protease (PR) contains three lysines, but deletion of PR has little effect on budding (60).
FIG. 1.
RSV Gag mutants and chimeras. Sites of cleavage, cleavage products, and domains required for budding are indicated. The C-terminal half of MA contains five lysine residues (amino acids 95, 115, 124, 138, and 148) that are potential sites of ubiquitination. For simplicity, the lysines are referred to as 1, 2, 3, 4, and 5 respectively. The lysines were all changed to arginine (1-5KR), changed in combinations (e.g., 1,2,3,5KR) or changed individually (e.g., 2KR). ΔMA3, an M domain mutant, has a deletion in the N-terminal half of MA which does not allow membrane binding. T10C deletes the L domain plus a portion of MA and CA, resulting in a “late” phenotype. Gag.GFP is a C-terminal fusion between Gag and eGFP. The resulting fusion occurs in NC and eliminates the last seven amino acids of NC. Gag(−) deletes eGFP, and therefore the resulting Gag protein is truncated 7 amino acids from the end of NC, but the deletion resulted in the addition of an arginine and leucine at the C terminus.
Given what is known about the lysines in RSV Gag and the location of known sites of ubiquitination in other retroviruses, the lysines most likely to play a role in the late steps of budding seemed to be the those located in the second half of MA. To ascertain their importance, these five residues were changed to arginine to eliminate the potential for ubiquitination while preserving structure.
Budding of mutant 1-5KR.
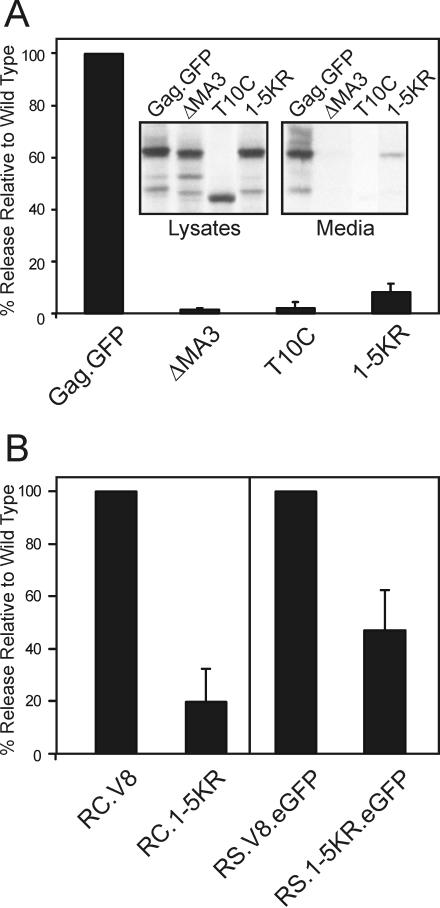
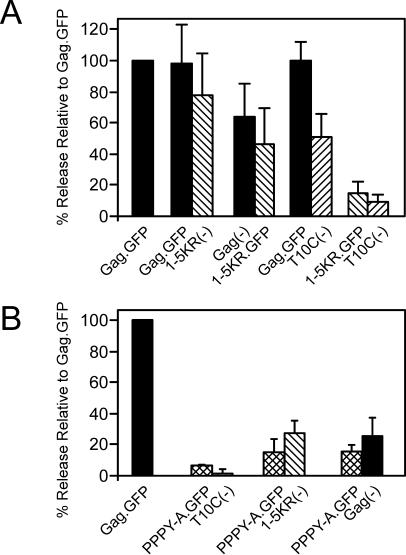
The first mutant made was 1-5KR in which all five of the lysines in the second half of MA were replaced with arginine in a budding-competent derivative of Gag that has eGFP linked to its C terminus (Gag.GFP; Fig. 1). QT6 cells were transfected with plasmids encoding the parental Gag.GFP (wild type), ΔMA3.GFP (M domain deletion mutant), T10C.GFP (L domain deletion mutant), or 1-5KR.GFP. As expected from previous studies (61, 63), ΔMA3.GFP and T10C.GFP were released at levels <5% of wild type (Fig. 2A). Release of mutant 1-5KR.GFP was also reduced but slightly higher than the M and L domain mutants at 8% of the wild-type level. Replacement of the closest downstream lysine in CA (K244R), which resides in a consensus sumoylation site (44, 46), had no effect on budding either alone or in the context of the 1-5KR mutant (data not shown). These results suggest that one or more of the lysines immediately upstream of the L domain are critical for budding.
FIG. 2.
Requirement of lysines 95, 115, 124, 138, and 148 for particle release. (A) Transfected QT6 cells were metabolically labeled for 2.5 h 24 h posttransfection. Gag proteins were immunoprecipitated from the cell lysates and the labeling media with an anti-RSV serum. Gag proteins were visualized by autoradiography and quantitated by PhosphorImager analysis (see Materials and Methods). Mutants were compared to the wild type, which was normalized to 100%. The graph represents three independent experiments. (Inset) An autoradiograph with cell lysates and medium fractions shows that 1-5KR.GFP migrates at the expected size of 88 kDa. (B) The 1-5KR substitutions were introduced into two proviral constructs (RC.V8 and RS.V8.eGFP) to create RC.1-5KR and RS.1-5KR.eGFP, respectively. Gag proteins were expressed, radiolabeled, immunoprecipitated, visualized, and quantitated (see Materials and Methods). The experiments using RC.V8 and RC.1-5KR were repeated three times, whereas those using RS.V8.eGFP and RS.1-5KR.eGFP were repeated nine times.
The effect of the 1-5KR substitution on budding was also examined in the context of two different proviral genomes. pRC.V8 is a vector in which the nonessential v-src gene of RSV has been replaced with a gene for hygromycin resistance, whereas in pRS.V8.eGFP, v-src has been replaced with the gene for eGFP. The gag genes in both vectors encode active PR. Upon transient transfection of QT6 cells, Gag protein processing for the 1-5KR derivatives was unaffected (data not shown), but budding was once again seen to be reduced compared to the wild type: 80% reduction for RC.1-5KR and 53% reduction for RS.1-5KR.eGFP (Fig. 2B). The apparent difference between proviral mutants was observed only in transient transfection experiments; cells infected with RS.1-5KR.eGFP exhibited an 80% reduction in budding compared to the wild type (see the description of the infectivity experiments below). Therefore, the budding defect originally seen for 1-5KR.GFP appears not to be an artifact of expression of Gag in the absence of the other viral genes.
Subcellular localization of 1-5KR.
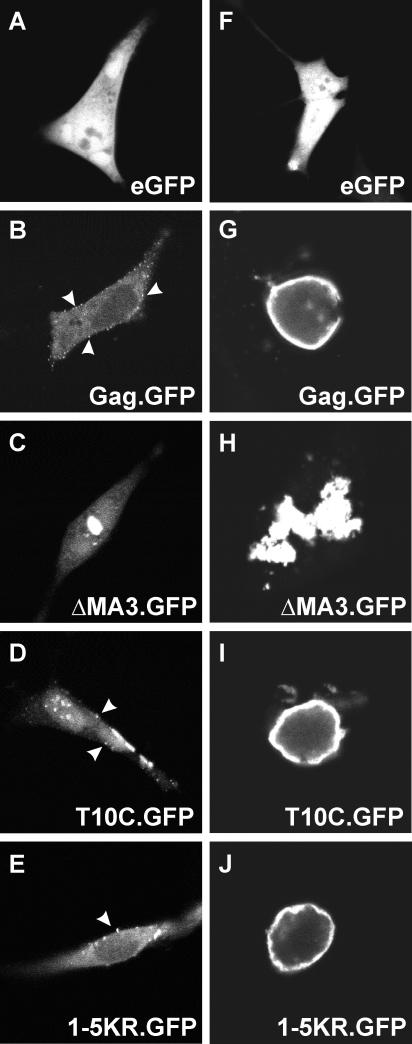
In cells treated with proteasome inhibitors, Gag proteins accumulate at the plasma membrane (34). If one or more of the five lysines in MA are involved in a ubiquitination event that is important for budding, then the 1-5KR mutant should also accumulate at the plasma membrane. On the other hand, due to their close proximity to the M domain, substitutions at these five residues could conceivably interfere with transport to the membrane. To ascertain where the mutant proteins accumulate, QT6 cells were transfected with small amounts (1 μg) of the various pGag.GFP constructs and analyzed by confocal microscopy (Fig. 3A to E). Wild-type Gag.GFP was seen in punctate fluorescence at the plasma membrane (arrowheads), while ΔMA3.GFP, unable to bind to the plasma membrane, was found in large aggregates in the cytoplasm, and an L domain mutant, T10C.GFP, was found at the plasma membrane. 1-5KR.GFP, like the wild type and the L domain mutant, was also localized to the plasma membrane. This phenotype was seen more clearly in cells transfected with larger amounts (10 μg) of DNA (Fig. 3F to J); however, under these conditions, the cells typically round up. It is clear from all of these results that the 1-5KR mutant has no obvious membrane-targeting defect and the substitutions likely affect a later step in budding.
FIG. 3.
Intracellular localization of mutant 1-5KR. peGFP, pGag.GFP, p1-5KR.GFP, pΔMA3.GFP, and pT10C.GFP were transfected into QT6 cells with 1 μg (A to E) or 10 μg (F to J) of DNA and examined by confocal microscopy 24 h after transfection.
Analysis of 1-5KR virions.
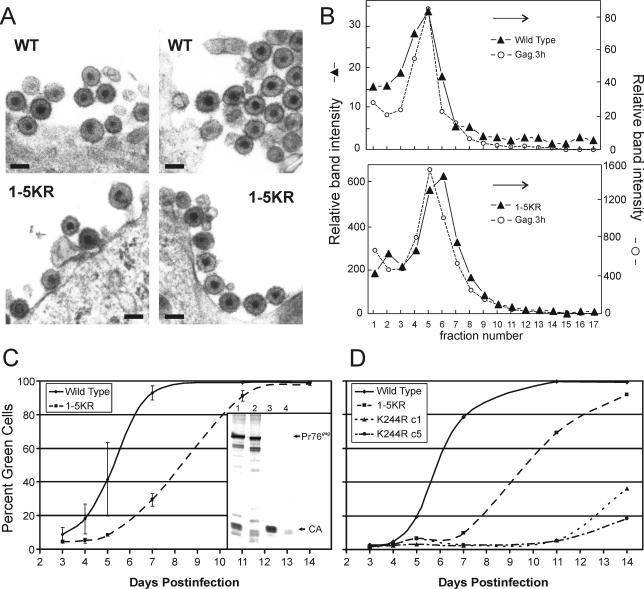
Retroviruses lacking their L domains form particles that accumulate at the plasma membrane, unable to be released efficiently (12). Similarly, when RSV-infected cells are treated with proteasome inhibitors to deplete free Ub levels, large clusters of virions are often seen stuck to the cell surface (34). Surprisingly, analysis of RC.1-5KR-infected cells by transmission EM revealed no hints of clusters tethered to the surface, but instead only particles of normal size and morphology were seen (Fig. 4A), even though parallel budding assays revealed the expected defect in particle release (data not shown). Scanning EM of cells expressing 1-5RK Gag also showed that clusters of particles do not form on the cell surface, but there was an increased number of normal-size particles blocked to budding compared to the wild type, similar to an L domain mutant (M. Johnson and V. Vogt, personal communication). To test the possibility that particle clusters were rapidly released, medium from RC.1-5KR-transfected cells was subjected to sucrose gradient centrifugation. A small population of 1-5KR particles indeed were found to sediment faster than normal; however, most were of normal size (Fig. 4B). If the 1-5KR mutations affect ubiquitination, then it may be the case that low-level usage of alternative lysines provides sufficient activity to destabilize any clusters that form on the cell surface (see Discussion).
FIG. 4.
Characterization of 1-5KR virions. (A) QT6 cells infected with RC.V8 or RC.1-5KR were thin sectioned and examined by EM. 1-5KR virions are visible with a morphology similar to that of the wild type. (Magnification, ×50,000; bar, 100 nm) (B) RC.V8 and RC.1-5KR particle size was determined by sedimentation of virions through a 10 to 30% sucrose gradient for 30 min. Fractions were collected and proteins were separated by SDS-PAGE. For RC.V8 and RC.1-5KR, the amount of CA present in each fraction was determined by PhosphorImager analysis and is shown on the left y axis as arbitrary units. The amount of internal control (Gag.3h) present in each fraction was also determined by PhosphorImager analysis and is shown as arbitrary units on the right y axis. The graph is representative of three independent experiments. (C) QT6 cells were transfected with pRS.V8.eGFP or pRS.1-5KR.eGFP, and virions were collected in the medium for 24 h. Equal amounts of virions were added to DF-1 cells for 24 h. The cells were analyzed by FACS at various times postinfection to determine the percentage of green cells, indicating an infection. (Inset) An autoradiograph showing Gag proteins from cells infected for 14 days, obtained as in Fig. 2. Lanes 1 and 2 are cell lysate fractions from the wild type and 1-5KR, respectively, and lanes 3 and 4 are medium fractions from the wild type and 1-5KR, respectively. The graph shows the average of three independent experiments. (D) Lysine 244 is part of the consensus sumoylation sequence, ΨxKE, where Ψ is a hydrophobic residue and x is any residue. To destroy the potential sumoylation site, lysine 244 was changed to arginine (K244R) and cloned into RS.V8.eGFP. Two K244R clones were analyzed for infectivity as described above and were compared to the wild type and 1-5KR. A representative graph of the results of three experiments is shown.
To determine whether 1-5KR particles have any infectivity, QT6 cells were transfected with pRS.V8.eGFP, pRS.1-5KR.eGFP, or pRS.K244R.eGFP, and particles were allowed to accumulate in the media for 24 h. Equal amounts of released reverse transcriptase activity were used to infect fresh cultures of DF-1 cells, and at various days postinfection, these were trypsinized and sorted by FACS to count the number of green (i.e., infected) cells. The half-time of spreading with wild-type virus was 5.3 days, while the 1-5KR half-time was 8.3 days (Fig. 4C). In contrast, two clones of mutant K244R were found to be largely noninfectious (Fig. 4D), even though they bud with the same efficiency as the wild type (data not shown). At the end of the experiment, when the cells were fully infected with mutant 1-5KR (day 14), budding assays revealed that particle release was still only ∼20% the wild-type efficiency (Fig. 4C, inset), and thus, the ability of this mutant to spread through the culture was not due to reversion or suppressor mutations. All together, these experiments show that the 1-5KR mutant is infectious and suggest that its slower rate of spread is simply due to a reduced rate of budding.
Complementation rescue of 1-5KR.
The L domain mutant T10C can heteromultimerize with and be rescued into particles by coexpressing budding-competent Gag molecules from RSV or MLV (1, 61). If the 1-5KR mutant is blocked at a late step in budding, then it too should be rescued by complementation; however, if it instead has a severe conformational defect or has been directed into a different pathway, then rescue might not be likely. To be able to distinguish the Gag derivatives used in the complementation assays, three constructs were made that lack the eGFP tag (Fig. 1) and hence are detectably smaller than their counterparts: Gag(−), 1-5KR(−), and T10C(−). Control experiments showed that each of these alone behaved as expected: i.e., the first buds like the wild type, while the latter two are defective (data not shown).
Cells were cotransfected with the pairs of DNAs to be tested, while a control plate was cotransfected with pGag.GFP and pGag(−). In this control, release of Gag.GFP was monitored to provide a measure of normal-budding, and pGag(−) was included in the transfection to keep the total amount of DNA equal to that used in the test plates. As expected, release of the L domain mutant, T10C(−), was greatly increased to50% of the control when coexpressed with Gag.GFP, and an inhibitory effect was not exerted upon the rescuing molecule (Fig. 5A). Mutant 1-5KR(−) was even more efficiently rescued (80%). Switching the eGFP tag from the rescuing Gag construct to the 1-5KR mutant reduced the efficiency, but complementation still occurred. Coexpression of T10C(−) and 1-5KR.GFP did not result in rescue of either defective mutant. Together with the ability of 1-5KR to reach the plasma membrane, these results suggest that conformational defects do not account for the block to budding.
FIG. 5.
Complementation of the 1-5KR defect with wild-type Gag. Wild-type (WT) and mutant Gag proteins were coexpressed. (A) To differentiate between the two proteins, eGFP was deleted from Gag.GFP and 1-5KR.GFP [Gag(−) and 1-5KR(−), respectively] and the proteins were coexpressed with 1-5KR.GFP and Gag.GFP, respectively. Gag.GFP was also coexpressed with T10C(−), and 1-5KR.GFP was also coexpressed with T10C(−). (B) PPPY-A.GFP was coexpressed with Gag(−), 1-5KR(−), or T10C(−). Cotransfected cells were metabolically radiolabeled, and Gag proteins were immunoprecipitated, separated by SDS-PAGE, and quantitated as in the legend to Fig. 2. The percent release of Gag proteins was determined by normalizing the release of Gag.GFP when coexpressed with Gag(−) to 100% (single solid bar) and comparing the release of Gag protein in wild-type-mutant (paired solid and hatched bars) and mutant-mutant (paired hatched bars) coexpressions. Solid bars represent release of wild-type Gag proteins, and hatched bars represent release of mutant Gag proteins. The results of three independent experiments are shown.
An L domain point mutant is dominant negative for budding.
To ascertain whether the inability of T10C and 1-5KR to rescue each other was due to the size of the deletion in T10C, the complementation experiment was repeated with the L domain point mutant, PPPY-A.GFP. As expected, budding of PPPY-A.GFP was low when expressed alone (15%) (data not shown) or when coexpressed with the L domain deletion mutant, T10C(−) (∼7%) (Fig. 5B). Likewise, mutant 1-5KR(−) could not be rescued by PPPY-A.GFP. Surprisingly, budding of Gag(−) was strongly inhibited (75%) and PPPY-A.GFP budding was not increased. Elucidation of the mechanism behind this dominant-negative phenotype will require further experimentation (see Discussion).
Addition of lysines to 1-5KR.
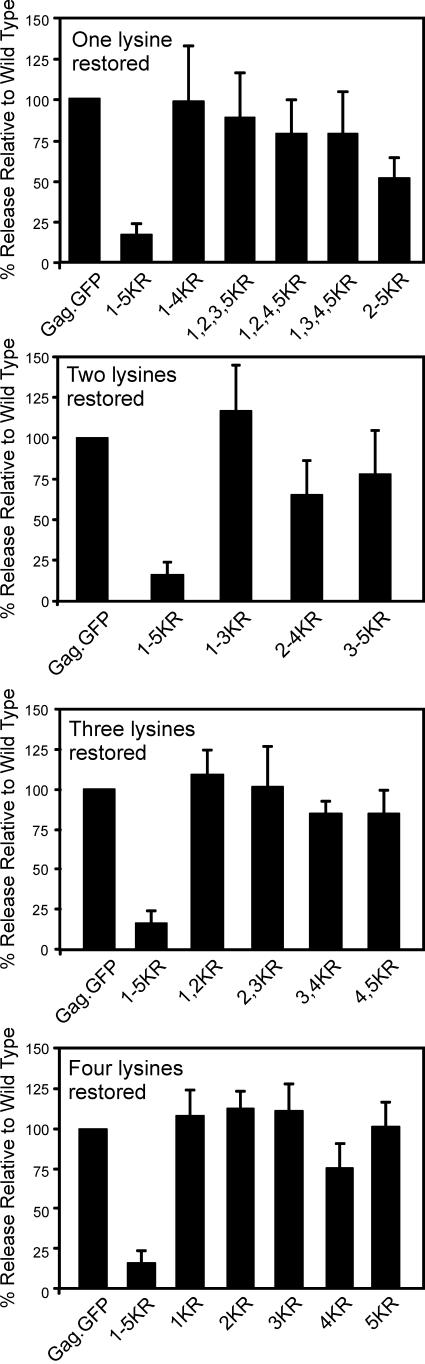
Further evidence that loss of budding is not the result of conformational defects caused by one or more of the five substitutions was obtained by adding lysines back into mutant 1-5KR. This was done in two ways. First, lysines were reintroduced at each of the five original positions, either singly or in groups of two, three, or four (Fig. 6). The reintroduction of just one lysine at any one of the five positions brought budding back to levels that are similar to wild type, although the effect seemed to diminish somewhat with changes furthest from the L domain. Lysines reintroduced in various combinations of two or more also restored budding and showed that no single lysine is indispensable. Moreover, several lysine-restoration mutants (1-4KR, 2-4KR, 2-3KR, and 2KR) were cloned into the provirus, and again budding was found to be restored (data not shown). Because the original five substitutions are scattered through a large region of the protein (54 amino acids), it is difficult to imagine how a gross conformational defect could be corrected simply by restoring any of the positions back to lysine.
FIG. 6.
Restoration of budding with same-site revertants. Mutants were created that restore one, two, three, or four of the five lysines. The percent release of mutants compared to the wild type was determined as in the legend to Fig. 2. A graphical representation of the results of three independent experiments is shown.
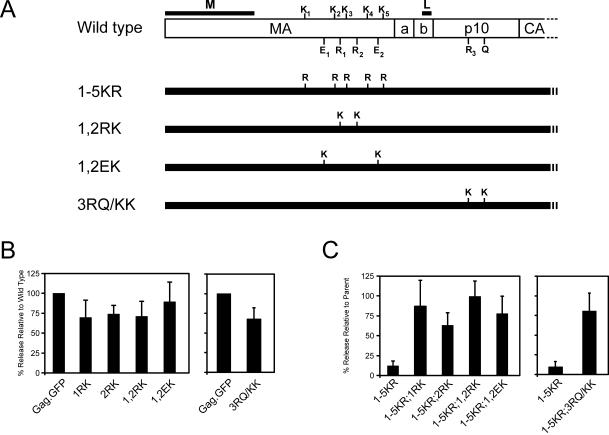
The second way that lysines were added back was by targeting novel positions within the 1-5KR mutant, both upstream and downstream of the L domain (Fig. 7A). Some of these were conservative changes in which arginines at positions 121 and 135 were replaced with lysine, both individually (1RK and 2RK) and together (1,2RK). Two sets of nonconservative changes also were made by inserting lysines in place of glutamates 111 and 143 (1,2EK) upstream of the L domain or in place of arginine 205 and glutamine 213 downstream of the L domain (3RQ/KK). By themselves, these new lysines had only minor effects on budding (Fig. 7B), but all suppressed the defect of the 1-5KR mutant (Fig. 7C). Although novel lysines were not introduced at positions more distant to the L domain, it is clear that the presence of eGFP and its 20 lysines to the C terminus of Gag does not suppress the 1-5KR phenotype (Fig. 2). Collectively, these results demonstrate that one or more lysines must be present in the vicinity of the L domain for efficient particle release. Whatever the role that lysines play (e.g., ubiquitination), it appears to require the L domain to be present on the same molecule because 1-5KR.GFP, which contains an L domain, was unable to rescue T10C(−), which retains two lysines that would normally be sufficient for budding (Fig. 5).
FIG. 7.
Restoration of budding with second-site revertants. (A) Lysines were substituted for arginines at positions 121 (1RK), 135 (2RK), or both (1,2RK); for glutamates at amino acid positions 111 and 143 (1,2EK); and downstream of the L domain in p10 at R205 and Q213 (3RQ/KK). Substitutions were also created in the context of 1-5KR. (B) The percent release of mutants with additional lysines in otherwise wild-type Gag compared to Gag.GFP was determined as in the legend to Fig. 2. (C) The budding of 1-5KR with additional lysines was determined as percent release of each construct relative to its parent containing the five lysines: e.g., the percent release of 5KR;1,2EK was relative to 1,2EK normalized as 100%. Transfection, metabolic radiolabeling, immunoprecipitation, and quantitation of Gag proteins were done as in the legend to Fig. 2.
Lysine insertion mutants of HTLV-1 Gag.
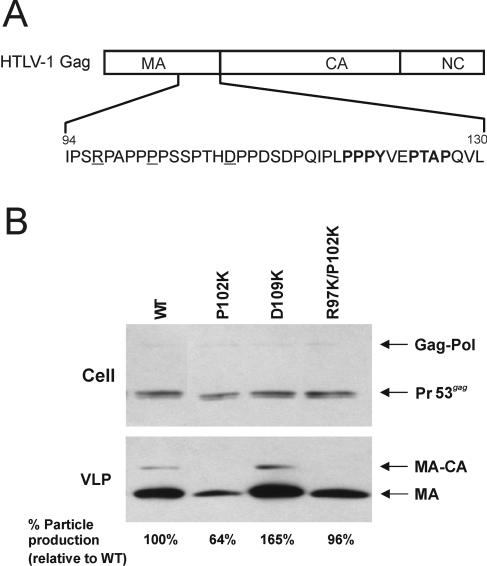
If lysines near L domains are important for efficient release of retroviruses, then the scarcity of such lysines might explain why HTLV-1 does not produce particles as well as RSV. The nearest lysine upstream from the L domain (PPPY) of this virus is 44 residues away (K74). Based on the three-dimensional structure of the highly similar MA sequence of HTLV-2 (8), this residue is predicted to be part of the M domain (23). The nearest downstream lysine is only 27 residues away (K148), but it resides in the CA domain rather than an unstructured region of Gag (9, 20).
The poor release properties of HTLV-1 provided an opportunity for exploring the need for lysines in a gain-of-function approach. For this, lysines were introduced at positions R97, P102, and D109K upstream of the L domain (Fig. 8A). Consistent with the hypothesis, insertion of a single lysine at a position close to the L domain (D109K) resulted in a substantial (64%) increase of particle release over that of the wild-type control. For unknown reasons, budding of mutant P102K was reduced to nearly half that of the wild type; however, this phenotype was also suppressed by the insertion of another lysine to create mutant R97K/P102K (Fig. 8B). Although attempts to create single-substitution mutant R97K were unsuccessful, these results nevertheless offer further support for the conclusion that lysines near the L domain are required for efficient retrovirus budding.
FIG. 8.
Introduction of lysines in HTLV-1. (A) HTLV-1 Gag is shown with the MA, CA, and NC domains indicated. The amino acids at the C terminus of MA are shown with the residues changed to lysine (i.e., residues R97, P102, and D109) underlined and the PPPY and PTAP motifs in boldface letters. (B) Immunoprecipitation and Western blot analysis of cell- and virus-like particle (VLP)-associated proteins. 293T cells were transfected with the wild type (WT) or derivatives containing the indicated HTLV MA mutations. VLPs were pelleted by ultracentrifugation and then subjected to immunoprecipitation-Western blot analysis. Cell-associated material was immunoprecipitated with an antibody to HTLV-1 p19 (MA) prior to Western blot analysis (see Materials and Methods). The relative levels of VLP-associated p19 (normalized for cell-associated Pr53gag) are indicated under each lane of the VLP panel. The positions of Pr53gag, Gag-Pol, p19, and incompletely processed intermediate MA-CA are shown. The experiment was repeated three times, and representative data are shown.
DISCUSSION
Lysines serve many roles in the functions of proteins. Structurally, they may contribute to proper folding by providing hydrophilic, positive charges that interface with the aqueous environment or interact with negatively charged moieties. In general, these structural roles can also be satisfied by arginine. In RSV, for example, interaction of the M domain with membranes requires a critical number of basic residues but not lysines per se (5), and all six of the lysines in NC could be replaced with arginine without affecting budding (data not shown). In contrast, the studies described here revealed a strict requirement for lysines in the vicinity of the L domain for efficient budding of RSV. The precise location and number of lysines were not important factors, suggesting that the block to budding seen in the absence of lysines (mutant 1-5KR) is not due to conformational or structural defects in the structure of Gag. The increase in budding seen when additional lysines were inserted near the L domain of HTLV-1 provides further support for a nonstructural role for these residues.
Lysines can also provide sites for at least five different types of modification to modulate the functions of proteins. For example, acetylation of lysines is known to regulate the activity of histones, HMG (high-mobility group) proteins, nuclear import factors, and transcription factors, including the Tat protein of HIV-1 (18, 37). Methylation of specific lysines in histones further regulates chromatin structure and gene expression (15). Hydroxylation of lysines in collagen is needed for cross-linking of triple helices, and some of the resulting hydroxylysines are subsequently glycosylated (24). The identification of phospholysine phosphatases suggests that phosphorylation of lysines can also occur (27). Finally, modification of lysines by Ub or Ub-like molecules serves a multitude of functions for a variety of different proteins (16, 42, 48, 51, 54). The data presented here do not exclude any of these various posttranslation modifications with regard to the lysines near the L domain, but the hypothesis that monoubiquitination is involved seems most likely given numerous other lines of supporting evidence (see the introduction). Although a previous report showed that known sites of ubiquitination in the p6 protein of HIV-1 can be eliminated without affecting budding or infectivity (30), it did not take into account the presence of two additional lysines located just 13 and 19 residues upstream from the L domain. In light of the redundancy and positional independence of lysines described here for RSV, the HIV-1 data should be interpreted with caution.
While it is clear that lysines near the L domain are important for RSV budding, there are reasons for doubting that they are involved in ubiquitination. Ub modification has never been detected for Gag or MA from this virus (35, 41). Moreover, in the course of this study, attempts were made to prevent postlysis removal of Ub from Gag molecules present in cell extracts, using strong denaturants such as guanidinium chloride or guanidinium thiocyanate in the presence of N-ethymaleimide to inactivate deubiquitination enzymes, but modified Gag products still could not be detected by immunoblotting with Ub-specific antisera (data not shown). Nevertheless, this absence of evidence does not eliminate the possibility that transient Ub modifications take place on Gag, as is the case for proteins that bud into multivesicular bodies (11). Attempts were also made to rescue mutant 1-5KR by placing Ub at its C terminus, but the chimera was equally defective for budding (data not shown). This result is especially perplexing since a similar Ub fusion was capable of repressing the negative effects of proteasome inhibitors on RSV budding (34). Attempts to rescue mutant 1-5KR by fusing the HIV-1 p6 sequence to its C terminus were unsuccessful too. While an explanation for this remains to be found, one possibility is that host factors bound to the L domain, waiting to modify lysines that are no longer present, result in a dominant-negative effect over p6. Further investigation of the role of the lysines near the L domain is needed.
Whatever the role of the lysines near the RSV L domain, the block to budding observed when they are missing appears to be at a late step. The evidence for this is severalfold. First, cell-associated proteolytic processing of the 1-5KR mutant was similar to that of the wild type with no buildup of Pr76gag or intermediate cleavage products. The decreased levels of mature cleavage products seen with the mutant are possibly the result of degradation following aborted budding attempts (Fig. 4C) (data not shown). This is unlike M domain mutants of RSV and MLV in which proteolysis is reduced, presumably because dimerization of the viral protease is limited in the absence of membrane binding (43, 50, 62, 63) but similar to RSV L domain mutants (61). Second, confocal microscopy showed that the mutant is present at the plasma membrane. Third, EM showed that mutant virus particles are formed on the cell surface. Unlike what has been seen in cells treated with proteasome inhibitors (34), an increased number of particles or clusters of particles compared to the wild type was not observed by transmission EM. This could be due either to the limited selection of electron micrographs that were observed or to collapse of nascent particles as a result of viral protease activity. Studies done by scanning EM found 1-5KR blocked at the cell surface, similar to an L domain mutant, but not in clusters (M. Johnson and V. Vogt, personal communication). The fact that no particle clusters like those seen in Ub-depleted cells were observed may indicate an additional requirement for ubiquitination of a cellular protein during budding. In any case, the ability to rescue mutant 1-5KR into particles using budding-competent Gag proteins suggests that not every molecule in the population requires lysines near the L domain. The inability of 1-5KR and mutant T10C to rescue each other suggests that lysines and L domains may be required on the same Gag protein. The lack of complementation is not likely due to a failure to heteromultimerize with T10C, since even MLV Gag can rescue this L domain deletion mutant when the M domains are the same (1). Thus, it appears that the RSV L domain requires lysines to function.
During the course of this study, it was found that a budding-competent Gag molecule can rescue mutant 1-5KR and the L domain deletion mutant T10C, but it cannot rescue the point mutant PPPY-A. Indeed, the rescuing Gag molecule is strongly inhibited when coexpressed with the point mutant. The dominant-negative activity of mutant PPPY-A is puzzling, but may be due to the binding of previously unrecognized host factors on regions of Gag that are missing in the large deletion mutant. For example, there is a YPSL motif just downstream of the RSV L domain, in the p10 sequence, and this is remarkably similar to the YPDL motif in equine infectious anemia virus, which provides L domain activity and binds the host protein AIP1 (56, 57). Perhaps binding of AIP1 to the RSV Gag protein results in a dominant-negative phenotype when the L domain is absent. Further experimentation will be required to determine the nature of the block.
The studies described here also revealed a potential involvement of SUMO-1 in RSV replication. This Ub-like protein has been shown to play roles in protein translocation, subnuclear structure formation, and modulation of transcriptional activity and has an antagonistic role against Ub (53). The only lysine in the RSV Gag protein that resides in a clear sumoylation consensus sequence (ΨKxE), where Ψ is a hydrophobic residue and x is any residue (44, 46) is the one at position 244 in CA (IK244TE). Although this residue was found to be unimportant for budding, it proved to be critical for infectivity. Further experimentation will be required to ascertain whether the loss of infectivity seen with mutant K244R is due to loss of sumoylation or is the result of structural defects.
Acknowledgments
We thank Marc Johnson and Volker Vogt for providing their scanning EM data to us prior to publication. We thank Eric Callahan for construct pΔMA3.GFP, Roland Meyers for thin-sectioning and EM, and Nate Sheaffer of the PSU Cell Science/Flow Cytometry Core Facility for assistance with the FACS analyses.
This work was supported by National Institutes of Health grants CA47482 to J.W.W. and AI053155 to L.M.M.
REFERENCES
- 1.Bennett, R. P., and J. W. Wills. 1999. Conditions for copackaging Rous sarcoma virus and murine leukemia virus Gag proteins during retroviral budding. J. Virol. 73:2045-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bouamr, F., J. A. Melillo, M. Q. Wang, K. Nagashima, S. M. de Los Santos, A. Rein, and S. P. Goff. 2003. PPPYEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101. J. Virol. 77:11882-11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowzard, J. B., R. J. Visalli, C. B. Wilson, J. S. Loomis, E. M. Callahan, R. J. Courtney, and J. W. Wills. 2000. Membrane targeting properties of a herpesvirus tegument protein-retrovirus Gag chimera. J. Virol. 74:8692-8699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan, E. M., and J. W. Wills. 2000. Repositioning basic residues in the M domain of the Rous sarcoma virus Gag protein. J. Virol. 74:11222-11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Callahan, E. M., and J. W. Wills. 2003. Link between genome packaging and rate of budding for Rous sarcoma virus. J. Virol. 77:9388-9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carthew, R. W., and C. Xu. 2000. Endocytosis: why not wait to deubiquitinate? Curr. Biol. 10:R532-R534. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, A. M., M. A. Massiah, B. G. Turner, W. I. Sundquist, and M. F. Summers. 1996. Three-dimensional structure of the HTLV-II matrix protein and comparative analysis of matrix proteins from the different classes of pathogenic human retroviruses. J. Mol. Biol. 264:1117-1131. [DOI] [PubMed] [Google Scholar]
- 9.Cornilescu, C. C., F. Bouamr, X. Yao, C. Carter, and N. Tjandra. 2001. Structural analysis of the N-terminal domain of the human T-cell leukemia virus capsid protein. J. Mol. Biol. 306:783-797. [DOI] [PubMed] [Google Scholar]
- 10.Craven, R. C., A. E. Leure-duPree, R. A. Weldon, Jr., and J. W. Wills. 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J. Virol. 69:4213-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dupre, S., C. Volland, and R. Haguenauer-Tsapis. 2001. Membrane transport: ubiquitylation in endosomal sorting. Curr. Biol. 11:R932-R934. [DOI] [PubMed] [Google Scholar]
- 12.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 14.Govers, R., T. ten Broeke, P. van Kerkhof, A. L. Schwartz, and G. J. Strous. 1999. Identification of a novel ubiquitin conjugation motif, required for ligand-induced internalization of the growth hormone receptor. EMBO J. 18:28-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hampsey, M., and D. Reinberg. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429-432. [DOI] [PubMed] [Google Scholar]
- 16.Hicke, L. 2001. Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 17.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 18.Kaehlcke, K., A. Dorr, C. Hetzer-Egger, V. Kiermer, P. Henklein, M. Schnoelzer, E. Loret, P. A. Cole, E. Verdin, and M. Ott. 2003. Acetylation of Tat defines a cyclin T1-independent step in HIV transactivation. Mol. Cell 12:167-176. [DOI] [PubMed] [Google Scholar]
- 19.Katz, R. A., C. A. Omer, J. H. Weis, S. A. Mitsialis, A. J. Faras, and R. V. Guntaka. 1982. Restriction endonuclease and nucleotide sequence analyses of molecularly cloned unintegrated avian tumor virus DNA: structure of large terminal repeats in circle junctions. J. Virol. 42:346-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khorasanizadeh, S., R. Campos-Olivas, C. A. Clark, and M. F. Summers. 1999. Sequence-specific 1H, 13C and 15N chemical shift assignment and secondary structure of the HTLV-I capsid protein. J. Biomol. NMR 14:199-200. [DOI] [PubMed] [Google Scholar]
- 21.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for Gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krishna, N. K., S. Campbell, V. M. Vogt, and J. W. Wills. 1998. Genetic determinants of Rous sarcoma virus particle size. J. Virol. 72:564-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Blanc, I, A. R. Rosenberg, and M.-C. Dokhélar. 1999. Multiple functions for the basic amino acids of the human T-cell leukemia virus type 1 matrix protein in viral transmission. J. Virol. 73:1860-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lodish, H., A. Berk, L. Zipursky, P. Matsudaira, D. Baltimore, and J. Darnell. 2000. Molecular cell biology. W. H. Freeman and Company, New York, N.Y.
- 25.Lohrum, M. A. E., D. B. Woods, R. L. Ludwig, É. Bálint, and K. H. Vousden. 2001. C-terminal ubiquitination of p53 contributes to nuclear export. Mol. Cell. Biol. 21:8521-8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Serrano, J., A. Yaravoy, D. Perez-Caballero, and P. D. Bieniasz. 2003. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc. Natl. Acad. Sci. USA 100:12414-12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matthews, H. R. 1995. Protein kinases and phosphatases that act on histidine, lysine, or arginine residues in eukaryotic proteins: a possible regulator of the mitogen-activated protein kinase cascade. Pharmacol. Ther. 67:323-350. [DOI] [PubMed] [Google Scholar]
- 28.Moscovici, C., M. G. Moscovici, H. Jimenez, M. M. Lai, M. J. Hayman, and P. K. Vogt. 1977. Continuous tissue culture cell lines derived from chemically induced tumors of Japanese quail. Cell 11:95-103. [DOI] [PubMed] [Google Scholar]
- 29.Nelle, T. D., and J. W. Wills. 1996. A large region within the Rous sarcoma virus matrix protein is dispensable for budding and infectivity. J. Virol. 70:2269-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 31.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, K. Nagashima, and U. Schubert. 2002. Equine infectious anemia virus and the ubiquitin-proteasome system. J. Virol. 76:3038-3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ott, D. E., L. V. Coren, R. C. Sowder II, J. Adams, and U. Schubert. 2003. Retroviruses have differing requirements for proteasome function in the budding process. J. Virol. 77:3384-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pepinsky, R. B., I. A. Papayannopoulos, S. Campbell, and V. M. Vogt. 1996. Analysis of Rous sarcoma virus Gag proteins by mass spectrometry indicates trimming by host exopeptidase. J. Virol. 70:3313-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 37.Polevoda, B., and F. Sherman. 30 April 2002, posting date. The diversity of acetylated proteins. Genome Biol. 3:reviews0006.1-reviews0006.6. [Online.] http://genomebiology.com. [DOI] [PMC free article] [PubMed]
- 38.Pornillos, O., S. L. Alam, D. R. Davis, and W. I. Sundquist. 2002. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat. Struct. Biol. 9:812-817. [DOI] [PubMed] [Google Scholar]
- 39.Pornillos, O., S. L. Alam, R. L. Rich, D. G. Myszka, D. R. Davis, and W. I. Sundquist. 2002. Structure and functional interactions of the Tsg101 UEV domain. EMBO J. 21:2397-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 41.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 42.Reed, S. I. 2003. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat. Rev. Mol. Cell Biol. 4:855-864. [DOI] [PubMed] [Google Scholar]
- 43.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez, M. S., C. Dargemont, and R. T. Hay. 2001. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276:12654-12659. [DOI] [PubMed] [Google Scholar]
- 45.Roth, A. F., and N. G. Davis. 2000. Ubiquitination of the PEST-like endocytosis signal of the yeast α-factor receptor. J. Biol. Chem. 275:8143-8153. [DOI] [PubMed] [Google Scholar]
- 46.Sampson, D. A., M. Wang, and M. J. Matunis. 2001. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J. Biol. Chem. 276:21664-21669. [DOI] [PubMed] [Google Scholar]
- 47.Schaefer-Klein, J., I. Givol, E. V. Barsov, J. M. Whitcomb, M. VanBrocklin, D. N. Foster, M. J. Federspiel, and S. H. Hughes. 1998. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology 248:305-311. [DOI] [PubMed] [Google Scholar]
- 48.Schnell, J. D., and L. Hicke. 2003. Non-traditional functions of ubiquitin and ubiquitin-binding proteins. J. Biol. Chem. 278:35857-35860. [DOI] [PubMed] [Google Scholar]
- 49.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with Gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schultz, A. M., and A. Rein. 1989. Unmyristylated Moloney murine leukemia virus Pr65gag is excluded from virus assembly and maturation events. J. Virol. 63:2370-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwartz, D. C., and M. Hochstrasser. 2003. A superfamily of protein tags: ubiquitin, SUMO and related modifiers. Trends Biochem. Sci. 28:321-328. [DOI] [PubMed] [Google Scholar]
- 52.Schwartz, D. E., R. Tizard, and W. Gilbert. 1983. Nucleotide sequence of Rous sarcoma virus. Cell 32:853-869. [DOI] [PubMed] [Google Scholar]
- 53.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 54.Spence, J., R. R. Gali, G. Dittmar, F. Sherman, M. Karin, and D. Finley. 2000. Cell cycle-regulated modification of the ribosome by a variant multiubiquitin chain. Cell 102:67-76. [DOI] [PubMed] [Google Scholar]
- 55.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 57.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 58.Wang, H., K. M. Norris, and L. M. Mansky. 2002. Analysis of bovine leukemia virus gag membrane targeting and late domain function. J. Virol. 76:8485-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weldon, R. A., Jr., C. R. Erdie, M. G. Oliver, and J. W. Wills. 1990. Incorporation of chimeric Gag protein into retroviral particles. J. Virol. 64:4169-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weldon, R. A., Jr., and J. W. Wills. 1993. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J. Virol. 67:5550-5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wills, J. W., R. C. Craven, and J. A. Achacoso. 1989. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J. Virol. 63:4331-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wills, J. W., R. C. Craven, R. A. Weldon, Jr., T. D. Nelle, and C. R. Erdie. 1991. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J. Virol. 65:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yasuda, J., E. Hunter, M. Nakao, and H. Shida. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep. 3:636-640. [DOI] [PMC free article] [PubMed] [Google Scholar]