Abstract
We have recently identified Np9 as a novel nuclear protein produced by the human endogenous retrovirus K and were able to document the exclusive presence of np9 transcript in tumors and transformed cells. With the aim of studying whether Np9 has a role in tumorigenesis, a systematic search for interacting proteins was performed. Here, we identify the RING-type E3 ubiquitin ligase LNX (ligand of Numb protein X) as an Np9-interacting partner. We furthermore show that the interaction involves N- and C-terminal domains of both proteins and can affect the subcellular localization of LNX. LNX has been reported to target the cell fate determinant and Notch antagonist Numb for proteasome-dependent degradation, thereby causing an increase in transactivational activity of Notch. We document that LNX-interacting Np9, like Numb, is unstable and degraded via the proteasome pathway and that ectopic Numb can stabilize recombinant Np9. Combined, these findings point to the possibility that Np9 affects tumorigenesis through the LNX/Numb/Notch pathway.
Up to 8% of the human genome consists of retrovirus-derived sequences (19). They are the result of numerous infections and subsequent integration of proviral elements into the germ line of human ancestors during the past 40 million years (2). Most of the recent human endogenous retrovirus (HERV) elements are defective due to the accumulation of mutations. Nonetheless, retroviral genes are frequently expressed in normal and transformed human tissues. The function of these genes in normal human cell physiology is still unclear (32). So far, only the expression of the env gene of HERV-W is known to have a crucial function in normal organismal physiology, namely human trophoblast cell fusion and differentiation (5, 12, 23). In contrast, retroviral gene expression has been linked to several human diseases, including tumorigenesis. In particular, expression of HERV-K seems to be strongly associated with transformation. Early hints pointing in this direction came from the observation that expression of the HERV-K proteins Gag and Env, as well as antibodies directed against these proteins, could be specifically detected in patients suffering from germ cell tumors (17, 24, 30, 31). Other studies implicate a contribution of HERV-K10 gag RNA expression to the development of leukemia (9) or show that env as well as the expression of subgenomic env transcripts correlates with human breast cancer (35, 36).
It is unclear, at the present, how these proteins may support tumorigenesis. The Rev-like regulatory protein Rec (20, 21) (formerly cORF) of HERV-K(HML-2.HOM) may exert a transforming potential through its interaction with the promyelocytic leukemia (PML) zinc finger protein PLZF (6). Recently, we have identified a novel protein translated from the HERV-K env reading frame that shares its first 14 amino acids with HERV-K Rec. Remarkably, the np9 transcript is exclusively expressed in tumor tissues and transformed cell lines (1): 52% of mammary carcinoma biopsies, 37% of germ cell tumor biopsies, and 33% of leukemia blood lymphocytes tested positive for np9 transcripts, while only 10, 25, and 0% of these were positive for rec. This led us to hypothesize that Np9 constitutes an oncoprotein. Here, we document that Np9 interacts with the E3 ubiquitin ligase LNX and that cellular Np9 levels are highly regulated, suggesting the involvement of Np9 in important functions.
Murine LNX was originally identified in searches for proteins interacting with the phosphotyrosine-binding domain of the cell fate determinant Numb (10). Two isoforms of LNX—p80LNX, carrying an N-terminal RING finger domain, and p70LNX, spliced to lack the RING finger—have been characterized. A human homolog with 88% sequence identity to p70LNX was reported by Xie et al. (38), and a human p80LNX isoform was published in the National Center for Biotechnology Information database (accession number BC022983). p80LNX functions as a RING-type E3 ubiquitin ligase that targets the p72 and p66, but not the p71 and p65, isoforms of Numb for proteasome degradation and thereby causes an increase of gene transactivation by the Numb-target, nuclear Notch (22, 25, 26). Consequently, inhibition of the proteasome pathway results in an increase in the levels of p72 and p66 Numb. LNX is furthermore able to form a complex with the coxsackievirus and adenovirus receptor (CAR) (33). The cellular function of CAR is still unclear but is suggested to be tumor suppressive (28). LNX and CAR colocalize in tight junctions (33). In contrast, Numb localizes predominantly in early endosomes (29). Here, we show that LNX-interacting Np9, like Numb, is subject to ubiquitin-dependent degradation, that ectopic Numb can stabilize recombinant Np9, and that Np9 can affect the subcellular localization of LNX.
MATERIALS AND METHODS
Plasmids.
pEG202-Np9 was constructed by amplification of np9 with the primer pair A (5′-CAT TTG Aga att cCC AAT GAA CCC ATC AGA G-3′) and B (5′-GAC AAA ACC ACC ctc gag ATC ATG GCC CGT-3′) to provide EcoRI and XhoI restriction sites, respectively (restriction sites for all primers discussed in this paragraph are represented by lowercase letters). The amplified fragment was EcoRI and XhoI digested and inserted into the EcoRI and XhoI sites of pEG202 (Clontech). The pEGFP-LNX-C and pEGFP-LNX plasmids were constructed by amplification of the LNX-C fragment with primers C (5′-GCG CGC gga tcc AGC TTT CAT GTG ATT CTC AAC AAA-3′) and D (5′-GCG CGC gga tcc ATT GAT TCT ATA AAA AAG-3′), the full-length LNX fragment was amplified with primers E (5′-GCG CGC gga tcc ATC ATG AAC CAG CCA GAG-3′) and D, all providing BamHI restriction sites. The amplified fragments were BamHI digested and inserted into the BamHI site of the pEGFP-C1 vector (Clontech). For the generation of the Np9-Dsred vector construct, np9 was amplified with primers F (5′-CGC GCg gat ccA TGA ACC CAT CGG AGA TGC AA-3′) and G (5′-CGC Tgg atc cGG GCA CAT AAC AAA ATG GAG-3′) to provide BamHI restriction sites and to remove the np9 stop codon. The PCR fragment was BamHI digested and inserted into the BamHI site of pDsred-N1 (Clontech). pSG5-Np9 and pSG5-Np9NLS1 were constructed by inserting the np9 sequence from EGFP-Np9, previously described in Armbruester et al. (1), and from EGFP-Np9NLS1 (see “PCR-directed mutagenesis,” below) into the BamHI sites of pSG5 (Stratagene). pGEX-Np9 and pGEX-Np9NLSmut1 were constructed by amplification of np9 from pGEM-np9 (1) or EGFP-Np9NLS1 with the primers H (5′-CGC GCg gat ccA TGA ACC CAT CGG AGA TGC AA-3′) and I (5′-CGC GCg gat ccA ACA GAA TCT CAA GGC AGA AG-3′) to provide BamHI restriction sites. The fragments were BamHI digested and inserted in frame into the BamHI site of the pGEX-4T-1 vector (Pharmacia-Biotech). pGEX-np9-N was constructed by amplification of the np9 sequence from pGEX-np9 with primers H and J (5′-GCG CGC gga tcc TTA ATC GTC ATC ATG GCC-3′), from pGEX-np9-C with the primers K (5′-GCG GCg gat ccA TGG CGG TTT TGT CGA AAA G-3′) and I, from pGEX-np9-Cx with primers K and G (to remove the stop codon), and from pGEX-np9-ΔC with primers H and L (5′-CGC gga tcc TTA ACT TCT TTC TAC ACA GAC-3′). All fragments were BamHI digested and inserted into the pGEX-4T-1 vector. LNX-pSG5-HA plasmids were constructed as follows: the LNX-F fragment was amplified from EGFP-LNX with primers M (5′-CGC GCG gaa ttc ATG AAC CAG CCA GAG-3′) and N (5′-AGG GTA gga tcc TAA AAA AGT GCC AGG-3′), the LNX-N fragment was constructed with the primers M and O (5′-AGG GTA gga tcc GTC ATC TCG GGG TCT G-3′), LNX-C was constructed with the primers P (5′-CGC gaa ttc ATG AGC TTT CAT GTG ATT-3′) and N, and LNX-ΔPDZ was constructed with the primers M and Q (5′-GCG CGC gga tcc ACC ATC TGG AAT CAG GTG-3′). All fragments were EcoRI and BamHI digested and inserted in a modified EcoRI- and BamHI-digested pSG5 vector construct, encoding a hemagglutinin-Tag downstream of the BamHI site. Numb-pSG5-HA was obtained by amplification of a Numb full-length fragment of the Numb p65 isoform from Numb-pCDNA3 (a gift of Moshe Oren) with primers R (5′-CGC GCG gaa ttc ATG AAC AAA TTA CGG-3′) and S (5′-AGG GTA aga tct AAG TCC AAT TTC AAA CG-3′); the amplified fragment was EcoRI and BglII digested and inserted into the EcoRI- and BamHI-digested pSG5-HA vector.
Yeast two-hybrid screen.
A human testis cDNA library (MATCHMAKER, LexA Libraries; Clontech) was screened with the yeast strain EGY48 and plasmid pEG202-Np9 as a bait according to the manufacturer's protocol. Plasmids from yeast clones showing specific activation of the reporter genes leu and lacZ were recovered and transfected into Escherichia coli. Isolated clones carrying pJG4-5 plasmids and encoding putative Np9-interacting sequences were transformed with pEG202-Np9 to confirm reporter gene activation. Same plasmids were also transmitted into EGY48 to verify lack of DNA binding. Yeast clones were classified positive only when both reporter genes were activated in the presence of galactose, but not glucose, which inhibits expression from the pJG4-5 plasmids (internal control of the MATCHMAKER system). Yeast strain EGY48 as well as reporter and control plasmids were generous gifts of Roger Brent.
GST pulldown assay.
Glutathione S-transferase (GST) proteins or GST fusion proteins were generated by transforming E. coli with plasmids pGEX, pGEX-np9, pGEX-np9NLSmut1, pGEX-np9-N, pGEX-np9-ΔC, pGEX-np9-C, or pGEX-np9-Cx. Exponentially growing cultures were induced with isopropyl-1-thio-β-d-galactopyranoside (final concentration, 100 nM) for 4 h at 37°C, and cell pellets were resuspended in GST-low-salt buffer (20 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 1% NP-40, 2 mM dithiothreitol, 0.2 mM phenylemethyle-sulfonylfluoride, 20 μg of aprotinin/ml) with lysozyme in a final concentration of 50 μg/ml. The extracts were sonicated for 1 min and cleared by centrifugation, and supernatants were then added to glutathione-Sepharose beads (1/10 volume) and incubated for 3 h at 4°C with gentle shaking. Beads were collected by centrifugation and washed four times in GST-low-salt buffer. 35S-labeled methionine (or translabeled cysteine and methionine) LNX-F, LNX-N, LNX-C, LNX-ΔPDZ, or Numb was synthesized in vitro with the encoding pSG5-HA constructs in the rabbit reticulocyte lysate-based TNT T7-coupled in vitro transcription-translation system (Promega) according to the manufacturer's protocol. Fifty microliters of protein-coated agarose beads was incubated overnight at 4°C with 500 μl of GST-low-salt buffer and 10 μl of radiolabeled protein. Pellets were then washed five times with GST-high-salt buffer (50 mM Tris-HCl [pH 7.5], 200 mM NaCl, 2 mM EDTA, 0.1% NP-40, 2 mM dithiothreitol, 0.2 mM phenylemethyle-sulfonylfluoride, 20 μg of aprotinin/ml) and boiled for 10 min in sodium dodecyl sulfate (SDS)-gel loading buffer. The supernatants were loaded onto a SDS-polyacrylamide gel. The gel was fixed (50% [vol/vol] methanol, 10% [vol/vol] acetic acid) for 30 min, washed three times (each, 10 min) in H2O, and incubated for 1 h in 1 M Na-salicylate before being dried. Ten percent of in vitro-translated protein input (1 μl) in 10 μl of SDS-gel loading buffer was used as an input control.
Cell culture, transfection, and MG132 and CHX treatments.
All cells except Tera-1 cells, were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum and 1% sodium pyruvate at 37°C and in a 5% CO2 atmosphere. Tera-1 cells were grown in supplemented McCoy's 5A medium. Transfections were performed with FuGene-6 (Roche) or Lipofectamine 2000 (Invitrogen), according to the manufacturer's recommendations. MG132 treatment was carried out as follows. Twenty-four hours after transfection or passaging of cells, MG132 (final concentration, 5 μM in dimethyl sulfoxide [DMSO]; Sigma), CHX (final concentration, 25 μg/ml of DMSO; Sigma), or the same volume of DMSO was added.
SDS-polyacrylamide gel electrophoresis (PAGE) gradient, Western blot analysis, immunoprecipitation, and antiserum.
All SDS-PAGE gradients contained 9.5 to 20% acrylamide and were produced following standard procedures. Western blot analysis was carried out as described previously (6). Np9-specific signals were detected with the rabbit polyclonal anti-Np9 antiserum K82 (1) at a dilution of 1:100. The rabbit polyclonal anti-Numb antiserum, a gift of Moshe Oren, was used at a dilution of 1:500. The monoclonal mouse anti β-actin antibody (Sigma) was used at a dilution of 1:1,000. Immunoprecipitation was performed with the anti-Np9 antiserum K82 and protein G-Sepharose (Amersham) in extracts dissolved and washed in lysis buffer (10 mM Tris [pH 7.5], 0.14 mM NaCl, 3 mM MgCl2, 0.5% NP-40).
Subcellular localization and immunofluorescence assays.
For subcellular localization or colocalization assays, cells were grown to about 20% density on glass coverslips and were then transfected with FuGene-6 (Roche) with 2 to 10 μg of each DNA construct, according to supplied protocols. Cells were fixated in paraformaldehyde (4% in phosphate-buffered saline) at 48 h after transfection, and DNA was stained with 200 ng of 4′,6′-diamidino-2-phenylindole (DAPI)/ml of methanol to visualize nuclei. Intracellular localization and colocalization were studied by fluorescence microscopy or confocal laser-scanning microscopy as outlined in the figure legends. Immunofluorescence studies were carried out as described previously (30).
PCR-directed mutagenesis.
The mutants expressing enhanced green fluorescent protein and the nuclear localization signal (EGFP-NLS mutants) were generated by PCR-directed mutagenesis. Primers with partly altered sequences were designed to change lysines and arginines to alanines within the predicted NLSs. Construction of the Np9-NLS1 and Np9-NLS2 mutants were performed by three amplification steps. In step one, two fragments with an overlapping region containing the altered sequences were amplified with the EGFP-Np9 construct as template DNA. In a second PCR step without primers (3 min at 94°C; 4 cycles of 1 min at 94°C, 70 s at 58°C, and 3 min at 72°C; and 10 min at 72°C), the two overlapping fragments were annealed and single strands were completed on the full-length double-strand fragment. A third PCR step was used to amplify the mutated sequence. To generate the NLS1 mutant, the following primer combinations were used (changed nucleotides listed below are underlined). For the first PCR step, primer pair T (5′-GTT CGT GAC CGC CGC CGG GAT-3′) and U (5′-CGC CGA TGG CGC CTG CGC CGC CGG AGC TGT TGG GTA CAC-3′) was used for Np9-NLS1 construction. Primers V (5′-CCG GCG GCG CAG GCG CCA TCG GCG ACG GGC CAT GAT GAC GAT-3′) and W (5′-CAT TTT ATG TTT CAG GTT CAG-3′) were used for Np9-NLS2 construction. The third PCR step was performed with primers S and W. The NLS2 mutant was made with primers S and X (5′-CCC ACA CGC CCC CGC CGC CTT TTC GAC AAA ACG GCC ATC-3′) and Y (5′-GAA AAG GCG GCG GGG GCG TGT GGG GAA AAG CAA GAG AGA-3′) and W, respectively. The third amplification step was also performed with primers S and W.The NLS3 mutant was generated in one PCR step with primers S and Z (5′-GGA TCC GCA CAT AAC AAA ATG GAG CGC CGC ATG CGC ACT TCT TTC TAC ACA GAC ACA-3′). (The full-length fragments were digested with BamHI and reinserted into the BamHI sites of pEGFP-C1. To generate the EGFP-GST Np9NLS, and EGFP-GST constructs, an EcoRI-SalI-digested GST-encoding sequence was inserted into EcoRI- and SalI-digested plasmids. Amplification of the GST sequence was performed with primers AA (5′-G CGC GCg aat tcT ATG TCC CCT ATA CTA GGT-3′) and BB (5′-GCG CGC gtc gac ACG CGG AAC CAG ATC CGA-3′), which provide the EcoRI and SalI restrictions sites (indicated by lowercase letters). The pGEX-4T-1 vector from Pharmacia-Biotech served as a template for the GST sequence.
Subcellular fractionation.
Approximately 5 × 107 exponentially growing, MG132-treated Tera-1 cells were scraped into 5 ml of phosphate-buffered saline at 4°C, pelleted at 1,500 × g for 5 min, and resuspended in 1 ml of RSB-8 buffer (10 mM Tris [pH 7.5], 10 mM NaCl, 8 mM Mg-acetyl [Ac]) for 30 min on ice to allow cell swelling. Fifty microliters of suspension was dissolved in Laemmli buffer and analyzed as total cell protein. Cells were then spun down for 30 s in an Eppendorf centrifuge, the pellet was gently resuspended in 0.5 ml of RSB-NP-40 buffer (10 mM Tris [pH 7.5], 10 mM NaCl, 1.5 mM Mg-Ac, 0.5% NP-40, protease inhibitors), and the cells were Dounce homogenized with 20 strokes. Cell debris was centrifuged at 800 × g for 5 min. The supernatant was harvested and centrifuged at 14,000 × g for 10 min to remove residual debris, and the clear supernatant was saved as a soluble cytoplasmic protein fraction. The pellet from the first centrifugation was resuspended in 1 ml of RSB-NP-40 buffer, and centrifugation at 800 × g was repeated. Cell nuclei were then resuspended in 0.5 ml of sucrose solution 1 (0.34 M sucrose, 0.5 mM Mg-Ac, protease inhibitors); 1/10 volume was saved for the preparation of the nuclear protein fraction. The remaining resuspended nuclei were sonicated three times on ice at 120 W for 10 s, with 15 s of intermittent cooling time. The release of the nucleoli was followed microscopically. The suspension was then loaded onto a 1-ml sucrose cushion of solution 2 (0.88 M sucrose) in a 2-ml Eppendorf tube and centrifuged for 10 min at 3,000 × g. The supernatant was saved as soluble nuclear extract, and the pellet was saved as a nucleolus fraction. All proteins were analyzed by SDS-PAGE.
RESULTS
Identification of LNX as an interacting partner of Np9.
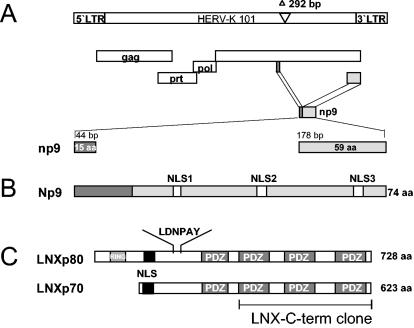
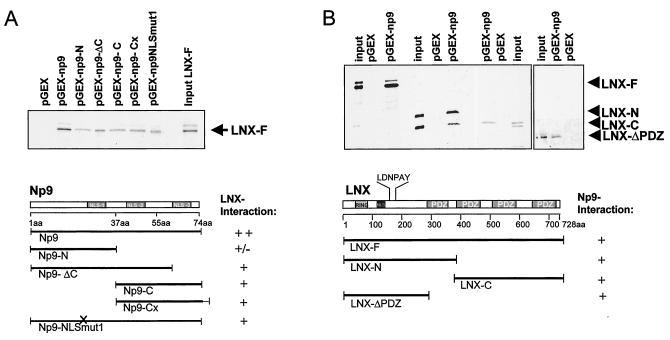
Recently, the discovery of a novel transcript, np9, encoded by human endogenous retrovirus K type 1 (Fig. 1A and B) and its expression exclusively in transformed cells was described (1). So far, there have been a total of six HERV-K type 1 sequences characterized (HERV-K101, HERV-K102, HERV-K103, HERV-K107, HERV-K110, and HERV-KII) (2, 34) which all contain intact open reading frames for np9 and the relevant splice donor site. The np9 transcripts found in tumors and transformed cells all originated from the provirus types HERV-K101 and HERV-KII (1). To begin to understand the function of Np9 protein, we screened a human testis cDNA library for Np9-interacting partners. A full-length np9 construct fused with the DNA-binding domain of the LexA-transactivating complex (pEG202-Np9) served as bait. The two-hybrid screen identified four Np9-interacting proteins, three of them containing one or more PDZ domains (where PDZ is a designation for postsynaptic density protein-disc large protein-zo-1 protein) (18). Four cDNA clones showed 98 to 99% homology to the C terminus of the RING-type E3 ubiquitin ligase LNX. The interacting LNX-C-term clones harbored three of the four PDZ domains present in full-length LNX, which suggested that association with Np9 is mediated by at least one of these domains (Fig. 1C). The interaction passed several tests for specificity. Yeast transformed with pJG4 to 5-LNX-C-term and the DNA-binding domain (pEG202) alone, or pJG4-5-LNX-C-term alone, were not able to transactivate the LexA promoter construct.
FIG. 1.
(A) Schematic representation of the HERV-K101 provirus. The open reading frames encoding the viral proteins Gag, Prot, and Pol are indicated. The env reading frame encodes Np9. The two exons of the np9 gene are highlighted. The shown provirus type fails to code for Rec, due to the type 1-specific deletion of 292 bp depicted. (B) Np9 protein. The positions of the three putative NLSs are depicted. (C) Schematic representation of the two human LNX splice variants, LNXp80 and LNXp70. The RING finger domain and the four PDZ domains are designated. The depicted LNX-C-term clone represents the Np9-interacting domain. A putative NLS and the N-terminal-binding site for Numb protein (LDNPAY), are indicated.
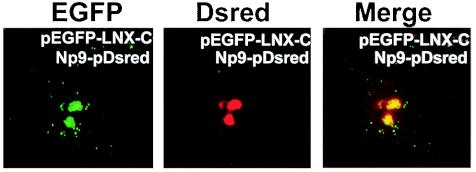
Np9Dsred and EGFP-LNX-C fusion products colocalize within the nucleus.
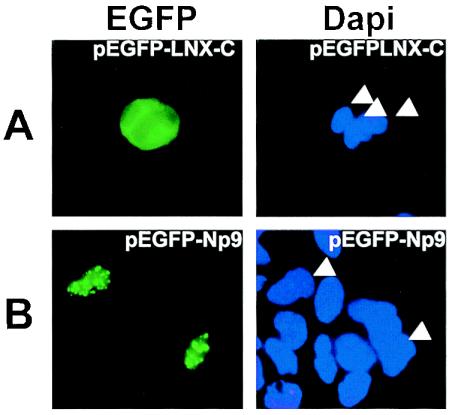
Physical association between Np9 and LNX in vivo would demand colocalization. Confocal laser-scanning microscopy of cells cotransfected with an Np9-pDsred fusion construct and an EGFP-LNX-C construct revealed that the red fluorescent Np9 and the green fluorescent LNX-C-term proteins colocalize in spotted structures within the nucleus 48 h after transfection (Fig. 2). Notably, transfection of pEGFP-LNX-C alone revealed a predominantly cytoplasmic distribution (Fig. 3A), in agreement with the presence of a putative nuclear localization signal within the N terminus of LNX (Fig. 1C). In contrast, EGFP-Np9 alone produced the same spotted pattern observed upon cotransfection of Np9-pDsred with EGFP-LNX-C (Fig. 3B). This indicated that Np9-pDsred is able to not only bind EGFP-LNX-C but direct it into distinct structures in the cell nucleus. Dsred alone was unable to translocate the EGFP-LNX-C fusion protein (data not shown).
FIG. 2.
Fluorescence of Cos-1 cells transiently cotransfected to produce the fusion proteins EGFP-LNX-C (green fluorescence) and Np9Dsred (red fluorescence). Images were obtained by confocal laser-scanning microscopy. The righthand panel shows the colocalization sites in the merged images.
FIG. 3.
Fluorescence of Cos-1 cells transfected with pEGFP-LNX-C (A) or pEGFP-Np9 (B). Cell nuclei were stained with DAPI as a control. DAPI-stained relevant nuclei are marked by arrowheads.
Colocalization of Np9 with full-length EGFP-LNX and nuclear localization of Np9.
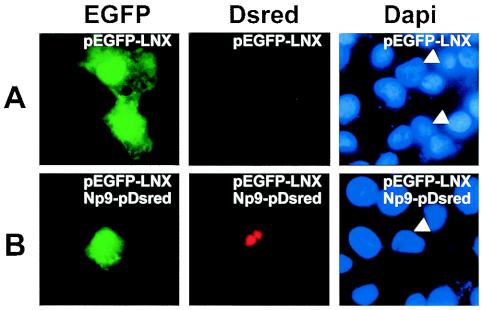
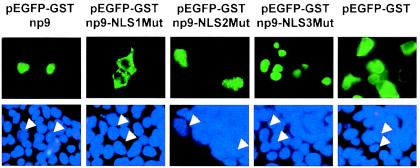
We next studied the colocalization of Np9 with a full-length p80LNX-EGFP fusion construct (pEGFP-LNX). The full-length LNX construct was obtained from the RZPD (Deutsches Ressourcenzentrum für Genomforschung GmbH) and subcloned into the pEGFP fusion vector. Transfection into Cos-1 cells of pEGFP-LNX alone resulted in both cytoplasmic and diffuse nuclear staining (Fig. 4A), revealing that full-length LNX can enter the nucleus independently of Np9 but, importantly, cannot translocate into the nuclear structures targeted by Np9. Notably and as with the C-terminal fragment of LNX, full-length EGFP-LNX and Np9-Dsred colocalized into the punctate nuclear structures in cotransfections (Fig. 4B), substantiating that Np9 and LNX can interact in vivo. It was likely that nuclear localization of Np9 requires one of the three putative NLS sites within the protein (Fig. 1B). However, since the nuclear pore permits some diffusion up to a molecular mass of 60 kDa (27) and since the EGFP-Np9 fusion product possesses a calculated molecular mass of 36 kDa, we inserted the GST protein (26 kDa) between EGFP and the Np9 mutants to obtain EGFP-GST-Np9 fusion products with an approximate molecular mass of 62 kDa. The EGFP-GST control protein was excluded from the nuclei and diffusely distributed in the cytoplasm, documenting that GST alone cannot direct nuclear localization. Transfection of the pEGFP-GST-Np9wt contructs led to the characteristic Np9-induced spotted patterns within the nucleus (Fig. 5). Mutation of the K and R residues to A revealed that NLS1 was crucial for nuclear localization of Np9. In contrast, mutation of NLS2 failed to produce detectable effects on nuclear localization. Remarkably, mutation of NLS3 allowed nuclear localization but generated a diffuse pattern, instead of the punctate pattern produced by intact Np9. Thus, NLS1 of Np9 is critical for efficient nuclear localization, and NLS3 is involved in directing the protein to subnuclear compartments.
FIG. 4.
(A) Localization of full-length LNX in Cos-1 cells transfected with pEGFP-LNX. (B) Colocalization of LNX and Np9. Cos-1 cells were cotransfected with pEGFP-LNX and Np9pDsred constructs. The relevant cell nucleus is marked by arrowheads in the DAPI-stained control.
FIG. 5.
Intracellular localization of EGFP-GST-Np9, EGFP-GST-Np9 NLS mutants (EGFP-Np9NLS1Mut, EGFP-Np9NLS2Mut, and EGFP-Np9NLS3Mut), and EGFP-GST alone in transiently transfected 293gp cells. Cell nuclei were stained with DAPI as a control, and relevant nuclei are marked by arrowheads.
Mapping of the Np9/LNX interaction domains.
GST pulldown assays were performed to map the interaction between Np9 and LNX. While LNX failed to associate with GST alone, it bound strongly to full-length Np9 protein and, to a lesser degree, to the various truncated forms of Np9 (Fig. 6A), indicating that the interaction is not limited to one domain of the Np9 protein. Furthermore, different truncated variants of LNX bound to full-length Np9 (Fig. 6B), indicating the existence of different interaction domains within the N and C termini of LNX and furthermore documenting that LNX/Np9 interaction in vitro does not require a PDZ domain. By contrast, Np9 failed to associate directly with the Numb and Notch-1 proteins in GST pulldowns (see Fig. 9B and data not shown), documenting that the interaction between Np9 and the truncated variants of LNX is not the result of a general stickiness of the protein.
FIG. 6.
Mapping of the Np9-LNX interaction domains by GST pulldown assays. (A) Full-length Np9 and variants with the indicated N- and C-terminal truncation, additional C-terminal amino acid residues (Cx), or a mutated NLS1 (NLSmut1) were employed as GST fusion products and incubated with in vitro-translated radiolabeled full-length LNX (LNX-F). A GST-only construct (pGEX) served as a control. (B) GST-full-length Np9 was incubated with the indicated variants of LNX. A putative NLS and the N-terminal-binding site for Numb (LDNPAY) are shown.
FIG. 9.
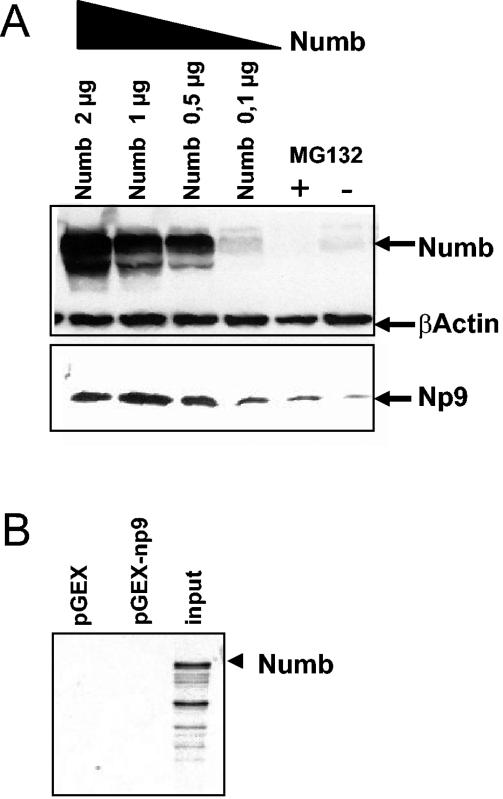
(A) Changes in steady-state levels of Np9 in dependence of Numb. Cos-1 cells were transiently transfected with pSG5-Np9 plus Numb-pSG5-HA at increasing quantities (0.1 to 2 μg) or with pSG5-Np9 and empty vector only. MG132 (5 μM) was added to the latter, and protein extracts were prepared at 48 h after transfection. Numb was detected with anti-Numb polyclonal antibody at a 1:500 dilution; β-actin was detected with anti-β-actin monoclonal antibody at a 1:1,000 dilution, and Np9 was detected with anti-Np9 polyclonal antibody at a 1:100 dilution. (B) Results of a GST pulldown assay documenting that Numb fails to directly interact with Np9.
Ectopic Np9 protein is stabilized by the proteasome inhibitor MG132.
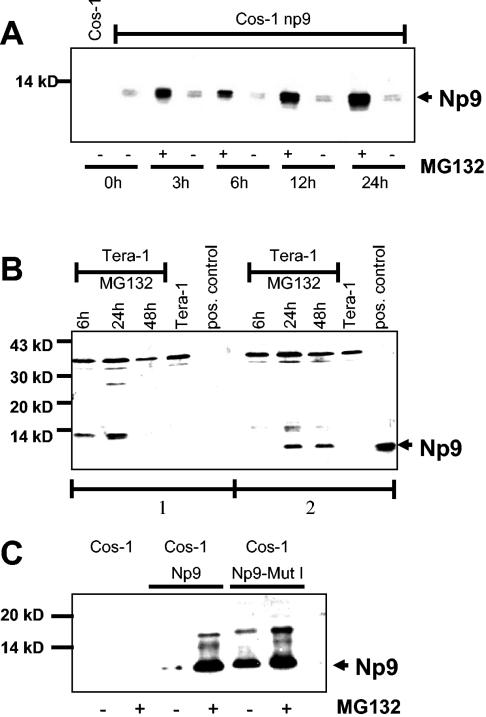
P80LNX was recently described as an E3 ubiquitin ligase that targets Numb for proteasome-dependent degradation. Inhibition of the proteasome pathway by MG132 treatment resulted in stabilization of endogenous Numb (25). Ubiquitination requires the RING finger domain of p80LNX, which recruits the ubiquitin machinery. The truncated p70LNX, which lacks the RING finger domain (Fig. 1C), is not able to target proteins for degradation and may exert other functions. The mapping data suggested that Np9 can interact with both forms of LNX. With this in mind and in light of the previous difficulties detecting Np9 protein, we next asked whether Np9 is subject to proteasome-dependent degradation. When Cos-1 cells were transiently transfected with pSG5-Np9 for 24 h and were subsequently treated with 5 μM MG132 for 3, 6, 12, or 24 h, Np9 protein was clearly detectable in Western blot analyses at 3 h posttreatment. Np9 protein levels increased by at least a factor of 10 in treated cells (Fig. 7A). The polyclonal Np9 antiserum detected two Np9-specific signals in the transfected Cos-1 cells, suggesting that Np9 is posttranslationally modified. Inspection revealed that Np9 harbors a putative casein kinase II phosphorylation site (1).
FIG. 7.
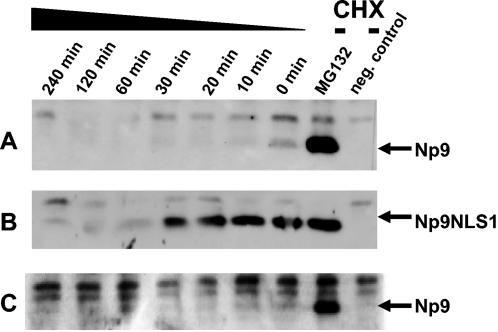
(A) Western blot analysis of protein extracts prepared from Cos-1 cells transiently transfected with pSG5-Np9. Twenty-four hours posttransfection, cells were treated with 5 μM MG132 (MG132 +), and extracts were harvested at the designated time points posttreatment with MG132. Untreated transiently transfected cells (MG132 −) and Cos-1 cells served as controls. Extracts were separated on a SDS-9.5 to 20% PAGE gradient and immunoblotted. Np9 was detected with the anti-Np9 polyclonal antiserum K82. (B) Western blot analysis of Tera-1 cell extracts partly treated with 5 μM MG132 for the times shown. A transiently pSG5-Np9-transfected Cos-1 cell extract served as a positive control. The total cellular protein extracts were separated on a SDS-9.5 to 20% PAGE gradient. Panel 1 shows a blot stained with anti-Np9 serum K82 preincubated with bacterially expressed Np9 protein. Panel 2 shows a blot stained with anti-Np9 serum K82 preincubated with the corresponding bacterial vector control. Np9-specific signals are marked with an arrowhead. (C) Western blot analysis of a cytolplasmic Np9 mutant (pSG5-Np9NLS1) in transiently transfected Cos-1 cells. Parallel cultures of Cos-1 cells were transiently transfected with pSG5-Np9 or a pSG5-Np9NLS1 mutant. One culture was treated 24 h posttransfection with 5 μM MG132 for an additional 24 h (MG132 +); the other culture remained untreated (MG132 −). Untransfected Cos-1 cells, with and without MG132 treatment, served as controls. Total cellular protein extracts were separated on a SDS-9.5 to 20% PAGE gradient. The arrowhead marks Np9-specific signals detected with the anti-Np9 polyclonal antiserum K82.
Endogenous Np9 is expressed in Tera-1 cells.
To investigate whether proteasome inhibition will also stabilize endogenous Np9, we examined the human teratocarcinoma cell line Tera-1, a line strongly expressing np9 transcript, following MG132 treatment. After exposure of exponentially growing cultures to 5 μM MG132 for 6, 24, and 48 h, a signal of 9 kDa became visible in Western blot analyses after 24 and 48 h. In contrast, untreated Tera-1 cell extracts as well as cell extracts from cultures treated for only 6 h failed to show Np9 expression. To confirm the authenticity of the signal, we preincubated the Np9-specific serum once with bacterially expressed Np9 or with empty vector extract. The Np9 extract successfully competed with the Tera-1 cell extracts, whereas the vector extract was unable to do so (Fig. 7B), confirming that the signal was indeed produced by endogenous Np9. In contrast to ectopic Np9 in transfected Cos-1 cells, endogenous Np9 (as well as recombinant Np9) (Fig. 8) was present only in the higher-molecular-weight form in Tera-1 cells. Thus, endogenous Np9 produced in Tera-1 cells is an unstable protein subject to degradation by the proteasome pathway.
FIG. 8.
(A) Half-life of ectopically expressed Np9 in Tera-1 cells. Tera-1 cultures were transfected for 24 h with pSG5-Np9. The cultures were then treated either with MG132 (5 μM) for another 24 h or with CHX (25 μg/ml) for the indicated times at 48 h after transfection. Np9 protein was subjected to immunoprecipitation with K82 antiserum and analyzed by immunoblotting. Untreated Tera-1 extracts served as negative controls. (B) Half-life of transfected cytoplasmic Np9NLS1 in Tera-1 cells. Tera-1 cultures were treated and analyzed as described in panel A but were transfected with plasmid pSG5-Np9NLS1. (C) Stability of transfected Np9 in the presence of transfected p65 Numb. Tera-1 cultures were transfected and analyzed as outlined in panel A, except that pSG5-Np9 was cotransfected with Numb-pSG5-HA. Transfection efficiency was controlled by Western blotting (results not shown).
Cytoplasmic Np9 is stable in transiently transfected cells.
Np9 with a defective NLS1 was exclusively localized in the cytoplasm. We therefore asked whether cytoplasmic Np9 is subject to degradation to the same extent as nuclear. When Cos-1 cells were transfected to produce Np9 or Np9NLS1 for 24 h and were either mock treated or treated with 5 μM MG132 for another 24 h, a strong Np9-specific signal was observed in the mock-treated cultures only with the Np9NLS1 mutant, whereas intact Np9 produced a strong signal exclusively in the presence of the proteasome inhibitor MG132 (Fig. 7C). The signals produced by Np9 in the presence of MG132 and by Np9NLS1 were approximately equally strong. When Tera-1 cells instead of Cos-1 were transfected to express Np9 or Np9NLS1, Np9 protein was again very unstable, with a half-life of less than 15 min, whereas the mutant was relatively stable, with a half-life of more than 30 min (Fig. 8A and B). Together, this shows that cytoplasmic Np9 is protected from proteasome-mediated degradation despite Np9 and Np9NLSmut1 still being able to associate with LNX in vitro (Fig. 6A).
Np9 is stabilized by Numb.
Np9 protein levels were difficult to detect in cells unless the proteasome inhibitor MG132 was present, and the Np9- and Numb-interacting LNX ubiquitin ligase seems to be widely expressed (10). Furthermore, the p72 and p66 isoforms of Numb are known targets of LNX-mediated proteasome degradation (25, 26). The p71 and p65 forms of Numb, although not subjected to proteasome degradation, are nonetheless able to physically associate with LNX (26). Combined, this suggested that Np9 and Numb are able to compete for LNX, and that as a consequence, Numb may affect Np9 stability. To study the stability of Np9 in the presence of ectopically expressed Numb, Cos-1 cells were transiently cotransfected with 0.1 μg of pSG5-Np9 and increasing quantities of Numb-pSG5-HA, and the steady-state protein levels were determined by immunoblotting at 48 h after transfection. Transient transfections of Cos-1 cell cultures with FuGene-6 (Roche) routinely produced 70 to 80% transfected cells. While the levels of β-actin remained unchanged, the Np9 steady-state levels increased in the presence of ectopic Numb (Fig. 9A); GST pulldowns revealed that Numb does not directly interact with Np9 (Fig. 9B). In contrast, neither ectopic Notch-1 nor HERV-K Gag expression had any detectable effect on the Np9 levels (data not shown), showing that the stabilization of Np9 in the presence of ectopic Numb is specific. However, it should be noted that we were unable to show, vice versa, the stabilization of endogenous Numb by ectopic Np9 (not shown). This might be because only two of the four isoforms of Numb are subject to LNX-mediated degradation (26), whereas the remaining forms of Numb are stable in spite of LNX interaction and Numb is targeted by at least one other ubiquitin ligase, Mdm-2 (39). A more-direct study of the Np9 destabilization by LNX (for instance, through knockdown of LNX expression via RNA interference), was not feasible due to the lack of anti-human LNX antibodies and LNX-deficient cells. Furthermore, we were not able to show stabilization of endogenous Np9 upon overproduction of ectopic Numb (data not shown). One reason could be that the endogenous Np9 is degraded via several different pathways in Tera-1 cells. This was compatible with the observation that even transiently overproduced, ectopic Np9 was surprisingly unstable in the presence of cooverproduced Numb in Tera-1 cells, whereas overproduced Np9 was clearly stabilized by Numb in Cos-1 cells (Fig. 8C and 9A). Combined, these data thus point to the possibility that the LNX/Numb/Notch pathway can regulate Np9 expression and is dependent on cellular context.
Np9 localizes to the nucleoli and other subnuclear structures.
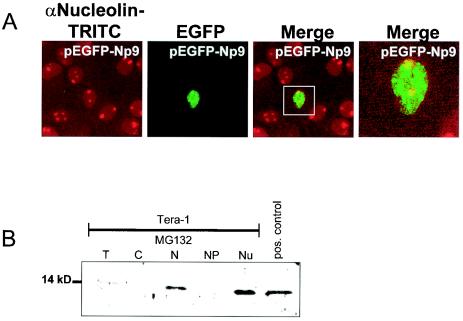
Transfection of Cos-1 cells with EGFP-LNX showed that the full-length protein is localized almost exclusively in the cell nucleus, independently of Np9, and produces diffuse staining (Fig. 4A). Remarkably, when Np9-Dsred was coexpressed with EGFP-LNX in these cells, the EGFP-LNX signal was predominantly present in subnuclear structures that were most likely identical with the nucleoli on the basis of the characteristic exclusion of these structures by the DAPI stain (Fig. 4B), suggesting that in the presence of Np9, LNX translocates to the nucleoli. To further substantiate that Np9 is present in the nucleoli independently of cell type, HeLa cells were transfected to produce EGFP-Np9 fusion protein, and the nucleoli were subsequently stained with a rabbit polyclonal antibody directed against human nucleolin. As shown in Fig. 10 A and in agreement with the nucleolar staining of Np9-Dsred in Cos-1 cells (Fig. 4B), EGFP-Np9 strongly stained the nucleoli; however, the fusion protein was also present to some extent in as-yet-unidentified dot-like nuclear structures in HeLa cells. Staining with antibodies against the PML or survival of motor neuron (SMN) proteins revealed that these nuclear structures are not identical with PML oncogenic domains and Geminis of coiled bodies (data not shown). To confirm the observations obtained by immunofluorescence and to examine whether endogenous, cellular Np9 is also present in nucleoli, a subcellular fractionation of Tera-1 cells treated with MG132 was carried out, and the fractions were analyzed by Western immunoblotting. As indicated by the fluorescence data, Np9 was predominantly present in the nucleoli fraction and was essentially absent from the nucleoplasm (Fig. 10B). Combined, the data suggest that Np9 and LNX can colocalize in the nucleoli.
FIG. 10.
(A) Combined immunofluorescence and autofluorescence analysis of nucleolin and EGFP-Np9. HeLa cells were transiently transfected with EGFP-Np9 to express Np9 hybrid protein. At 48 h posttransfection, cell nucleoli were immunostained with an anti-nucleolin polyclonal antibody and visualized with tetramethyl rhodamine isocyanate (TRITC) anti-rabbit secondary antibody. Images were produced by confocal laser-scanning microscopy. Colocalization of Np9 and nucleolin within the nucleoli is shown in the merged image. (B) Western immunoblot identifying Np9 in subcellular fractions. Subcellular fractions of exponentially growing Tera-1 cells were subjected to SDS-PAGE and were analyzed with the rabbit anti-Np9 polyclonal antibody K82 at a 1:100 dilution. T, total cell protein; C, free cytoplasmic protein; N, nuclear protein; NP, nucleoplasm without nucleoli fraction; Nu, nucleoli fraction; pos. control, total protein from Np9-transfected Cos-1 cells.
DISCUSSION
Although previous work has shown that proteins encoded by human endogenous retroviral sequences can contribute to normal tissue function and development (5, 12, 23), current results imply that the majority of these proteins are expressed in the context of disease, in particular of deregulated proliferation. Provided that these proteins do not just represent tumor markers but contribute to tumorigenesis, one would expect them to interact and interfere with biochemical pathways previously identified as contributing to neoplastic transformation. Along this line, it was recently documented that HERV-K protein Rec (formerly cORF), preferentially expressed in tumors of the germ line, can physically associate with the PML zinc finger protein PLZF (6), which was originally identified in the context of leukemia (8, 13) but was then linked to abnormal spermatogenesis in the course of transgenic knockout studies (N. Hawe, V. Soares L. Niswander, G. Cattoretti, and P. Pandolfi, abstract from the 38th American Society of Hematology Annual Meeting and Exposition 1996, Blood 88[Suppl.]:291, 1996). Moreover, it has been shown that the novel splice variant of the rec gene of HERV-K type 1, the np9 transcript, is expressed in carcinomas of the breast, leukemias, germ line tumors, and tumor cell lines but not in normal, untransformed cells (1). Here, we identified the E3 ubiquitin ligase LNX as an interaction partner of Np9 and presented data indicating that the interaction between the two proteins can affect the subcellular localization of LNX and that the LNX interaction partner Numb can affect Np9 steady-state levels. LNX and Numb are part of the differentiation-proproliferative Numb/Notch pathway (25, 26).
The mapping of the interaction domains on Np9 and LNX by GST pulldown assays revealed the presence of at least two N- and C-terminally localized interaction domains. A similarly complex distribution of contact domains has been described previously for the interaction between LNX and Numb, with Numb binding to the N-terminal LDNPAY motif and the first PDZ domain of LNX (25, 26). Since Np9 bound in yeast two-hybrid screens to a C-terminal fragment of LNX containing essentially only the PDZ domains and furthermore bound to a number of unrelated proteins that shared only the PDZ domains with LNX (data not shown), Np9 and LNX most likely interact via LNX PDZ domains. However, the interaction of Np9 with PDZ domain-containing proteins was nevertheless specific as, for instance, no interaction was observed between Np9 and the PDZ domain protein NHERF (also known as SIP) (data not shown). PDZ domains have been reported to help establish and stabilize larger protein complexes involved in many cellular processes, including signal transduction and cell surface channel protein functions (4, 14). The domains can interact with themselves as well as with protein domains frequently harboring C-terminal hydrophobic amino acid residues (14). Among the binding partners of PDZ domain proteins are the N-type Ca2+ channel and L6 antigen that contain the C-terminal motif X-X-C typical for class III PDZ domain interaction partners (14). Notably, the X-X-C motif also terminates the Np9 protein. However, elimination of X-X-C as the C-terminal motif (i.e., construct Np9-Cx) (Fig. 6A) failed to affect binding in pulldown assays, thus calling into question the assertion that Np9 is a classical class III PDZ domain interaction partner of LNX.
While LNX and Numb seem to colocalize predominantly in early endosomes (25), Np9 was able to direct truncated cytoplasmic LNX that lacked the putative NLS, as well as full-length LNX, to the nucleoli. Although it is unclear at present whether LNX, Numb, and Np9 can form trimeric complexes or whether Np9 can sequester LNX away from Numb, this suggests that Np9 can regulate LNX and possibly Numb through translocation. Like Numb, Np9 is an unstable protein degraded via the proteasome pathway, and LNX functions as an ubiquitin ligase. We do not know whether Np9 is exclusively degraded through its interaction with LNX or whether other degradation pathways are employed as well. Numb, for instance, is also proteasome degraded through the interaction with the ubiquitin ligase Mdm-2 (39). Our preliminary studies with GST pulldown assays have so far failed to show interaction of Np9 and Mdm-2 (data not shown). Nonetheless, the outlined interrelations link Np9 to the LNX/Numb/Notch pathway.
np9 transcript is expressed in tumors of the breast, leukemias, and germ cell cancers (1). It is precisely these tumors with which the Numb/Notch pathway has recently been associated (3, 7, 15). For instance, the notch-4 gene has been reported to be a common integration site for the mouse mammary tumor virus genome. Integration entails a deregulated, ectopic expression of the proproliferative intracellular domain of Notch-4, linking Notch to mammary tumorigenesis (7). The recent discovery that Notch is located downstream of p21Ras and constitutes an essential part of the pro-proliferative Ras-signaling pathway underscores its possible role in tumorigenesis (37). Aberrant expression of the intracellular domain of Notch, as the result of chromosomal translocation, has also been documented in human T-cell acute lymphoblastoid leukemia (3). Finally, Notch signaling has been reported to be essential for gametogenesis in the germ line of Caenorhabditis elegans and has been implicated in rat and human spermatogenesis as well (15). It is now well established that abnormal spermatogenesis can predispose humans to the development of the carcinoma-in situ, which is recognized as a precursor lesion for all testicular germ cell tumors (16). Not least, Numb protein has been reported to promote the degradation of the Notch receptor as well as the Notch intracellular domain, which both correlate with a loss of Notch-dependent transcriptional activation (22). Although Numb can subject Notch to degradation and Notch has proproliferative functions, at least certain isoforms of Numb seem nonetheless to be associated with proliferation as well (11). The p71 and p72 isoforms were predominantly expressed in proliferating cells of the embryo, testes, and the majority of transformed cells but not in normal resting cells. Likewise, Np9 was exclusively expressed in transformed cells (1). Further studies will clear up whether Np9 can modulate the proproliferative effects of Numb and Notch and whether this modulation or other functions of Np9 are important for tumorigenesis of the breast, lymphoid line cells, or germ line cells.
Acknowledgments
We are grateful to H. Stahl, Institute for Biochemistry and Molecular Biology, Hamburg, Germany, for providing the anti-nucleolin antibody; to M. Oren, Weizmann Institute, Rehovot, Israel, for providing the rabbit polyclonal anti-Numb serum; and to S. Schwarz and S. Huth for excellent technical assistance. We thank J. Mayer, Institute for Human Genetics, Hamburg, Germany, for helpful and stimulating discussions.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG-Mu 452/5).
REFERENCES
- 1.Armbruester, V., M. Sauter, E. Krautkraemer, E. Meese, A. Kleiman, B. Best, K. Roemer, and N. Mueller-Lantzsch. 2002. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 8:1800-1807. [PubMed] [Google Scholar]
- 2.Barbulescu, M., G. Turner, M. I. Seaman, A. S. Deinhard, K. K. Kidd, and J. Lenz. 1999. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr. Biol. 9:861-868. [DOI] [PubMed] [Google Scholar]
- 3.Beverly, L. J., and A. J. Capobianco. 2003. Perturbation of Ikaros isoform selection by MLV integration is a cooperative event in Notch(IC)-induced T cell leukemogenesis. Cancer Cell 3:551-564. [DOI] [PubMed] [Google Scholar]
- 4.Bezprozvanny, I., and A. Maximov. 2001. PDZ domains: more than just a glue. Proc. Natl. Acad. Sci. USA 98:787-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blond, J. L., D. Lavillette, V. Cheynet, O. Bouton, G. Oriol, S. Chapel-Fernandes, B. Mandrand, F. Mallet, and F. L. Cosset. 2000. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 74:3321-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boese, A., M. Sauter, U. Galli, B. Best, H. Herbst, J. Mayer, E. Kremmer, K. Roemer, and N. Mueller-Lantzschz. 2000. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene 19:4328-4336. [DOI] [PubMed] [Google Scholar]
- 7.Callahan, R., and A. Raafat. 2001. Notch signaling in mammary gland tumorigenesis. J. Mammary Gland Biol. Neoplasia 6:23-36. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Z., N. J. Brand, A. Chen, S. J. Chen, J. H. Tong, Z. Y. Wang, S. Waxman, and A. Zelten. 1993. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO 12:1161-1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Depil, S., C. Roche, P. Dussart, and L. Prin. 2002. Expression of a human endogenous retrovirus, HERV-K, in the blood cells of leukemia patients. Leukemia 16:254-259. [DOI] [PubMed] [Google Scholar]
- 10.Dho, S. E., S. Jacob, C. D. Wolting, M. B. French, L. R. Rohrschneider, and C. J. McGlade. 1998. The mammalian numb phosphotyrosine-binding domain. Characterization of binding specificity and identification of a novel PDZ domain-containing numb binding protein, LNX. J. Biol. Chem. 273:9179-9187. [DOI] [PubMed] [Google Scholar]
- 11.Dho, S. E., M. B. French, S. A. Woods, and J. C. McGlade. 1999. Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J. Biol. Chem. 274:33097-33104. [DOI] [PubMed] [Google Scholar]
- 12.Frendo, J. L., D. Olivier, V. Cheynet, J. L. Blond, O. Bouton, M. Vidaud, M. Rabreau, D. Evain-Brion, and F. Mallet. 2003. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell. Biol. 23:3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guidez, F., W. Huang J. H. Tong, C. Dubois, N. Balitrand, S. Waxman, J. L. Michaux, P. Martiat, L. Degos, and Z. Chen. 1994. Poor response to all-trans retinoic acid therapy in a t(11;17) PLZF/RAR alpha patient. Leukemia 8:312-317. [PubMed] [Google Scholar]
- 14.Harris, B. Z., and W. A. Lim. 2001. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114:3219-3231. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi, T., Y. Kageyama, K. Ishizaka, G. Xia, K. Kihara, and H. Oshima. 2001. Requirement of Notch 1 and its ligand jagged 2 expressions for spermatogenesis in rat and human testes. J. Androl. 22:999-1011. [DOI] [PubMed] [Google Scholar]
- 16.Heimdal, K., H. Olson, S. Tretli, S. D. Fossa, A. L. Borresen, and D. T. Bishop. 1997. A segregation analysis of testicular cancer based on Norwegian and Swedish families. Br. J. Cancer 75:1084-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herbst, H., M. Sauter, and N. Mueller-Lantzsch. 1996. Expression of human endogenous retrovirus K elements in germ cell and trophoblastic tumors. Am. J. Pathol. 149:1727-1735. [PMC free article] [PubMed] [Google Scholar]
- 18.Hung, A. Y., and M. Sheng. 2002. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 277:5699-5702. [DOI] [PubMed] [Google Scholar]
- 19.Lander, E. S., L. M. Linton, B. Birren, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 20.Löwer, R., R. R. Tönjes, C. Korbmacher, R. Kurth, and J. Löwer. 1995. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 69:141-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magin, C., R. Löwer, and J. Löwer. 1999. cORF and RcRE, the Rev/Rex and RRE/RxRE homologues of the human endogenous retrovirus family HTDV/HERV-K. J. Virol. 73:9496-9507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGill, M. A., and J. C. McGlade. 2003. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278:23196-23203. [DOI] [PubMed] [Google Scholar]
- 23.Mi, S., X. Lee, X. Li, G. M. Veldman, H. Finnerty, L. Racie, E. La Vallie, X. Y. Tang, P. Edouard, S. Howes, J. C. Keith, Jr., and J. M. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785-789. [DOI] [PubMed] [Google Scholar]
- 24.Mueller-Lantzsch, N., M. Sauter, A. Weiskircher, K. Kramer, B. Best, M. Buck, and F. Graesser. 1993. Human endogenous retroviral element K10 (HERV-K10) encodes a full-length gag homologous 73-kDa protein and a functional protease. AIDS Res. Hum. Retrovir. 9:343-350. [DOI] [PubMed] [Google Scholar]
- 25.Nie, J., M. A. McGill, M. Dermer, S. E. Dho, C. D. Wolting, and C. J. McGlade. 2002. LNX functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J. 21:93-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nie, J., S. Shawn, C. Li, and C. J. McGlade. 2004. A novel PTB-PDZ domain interaction mediates isoform-specific ubiquitylation of mammalian Numb. J. Biol. Chem. 279:20807-20815. [DOI] [PubMed] [Google Scholar]
- 27.Ohno, M., M. Fornerod, and I. W. Mattaj. 1998. Nucleocytoplasmic transport: the last 200 nanometers. Cell 92:327-336. [DOI] [PubMed] [Google Scholar]
- 28.Okegawa, T., R. C. Pong, Y.Li, J. M. Bergelson, A. I. Sagalowsky, and J. T. Hsieh. 2001. The mechanism of the growth-inhibitory effect of coxsackie and adenovirus receptor (CAR) on human bladder cancer: a functional analysis of car protein structure. Cancer Res. 61:6592-6600. [PubMed] [Google Scholar]
- 29.Santolini, E., C. Puri, A. E. Salcini, M. C. Gagliani, P. G. Pelicci, C. Tacchetti, and P. P. Di Fiore. 2000. Numb is an endocytic protein. J. Cell Biol. 151:1345-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sauter, M., S. Schommer, E. Kremmer, K. Remberger, G. Dölken, I. Lemm, M. Buck, B. Best, D. Neumann-Haefelin, and N. Mueller-Lantzsch. 1995. Human endogenous retrovirus K10: expression of Gag protein and detection of antibodies in patients with seminomas. J. Virol. 69:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sauter, M., K. Roemer, B. Best, M. Afting, S. Schommer, G. Seitz, M. Hartmann, and N. Mueller-Lantzsch. 1996. Specificity of antibodies directed against Env protein of human endogenous retroviruses in patients with germ cell tumors. Cancer Res. 56:4362-4365. [PubMed] [Google Scholar]
- 32.Shiroma, T., J. Sugimoto, T. Oda, Y. Jinno, and F. Kanaya. 2001. Search for active endogenous retroviruses: identification and characterization of a HERV-E gene that is expressed in the pancreas and thyroid. J. Hum. Genet. 46:619-625. [DOI] [PubMed] [Google Scholar]
- 33.Sollerbrant, K., E. Raschperger, M. Mirza, U. Engström, L. Philipson, P. O. Ljungdahl, and R. F. Pettersson. 2003. The Coxsackievirus and adenovirus receptor (CAR) forms a complex with the PDZ domain-containing protein ligand-of-numb protein-X (LNX). J. Biol. Chem. 278:7439-7444. [DOI] [PubMed] [Google Scholar]
- 34.Sugimoto, J., N. Matsuura, Y. Kinjo, N. Takasu, T. Oda, and Y. Jinno. 2001. Transcriptionally active HERV-K genes: identification, isolation, and chromosomal mapping. Genomics 72:137-144. [DOI] [PubMed] [Google Scholar]
- 35.Wang-Johanning, F., A. R. Frost, G. L. Johanning, M. B. Khazaeli, A. F. LoBuglio, D. R. Shaw, and T. V. Strong. 2001. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin. Cancer Res. 7:1553-1560. [PubMed] [Google Scholar]
- 36.Wang-Johanning, F., A. R. Frost, B. Jian, L. Epp, D. W. Lu, and G. L. Johanning. 2003. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene 22:1528-1535. [DOI] [PubMed] [Google Scholar]
- 37.Weijzen, S., P. Rizzo, M. Braid, R. Vaishnav, S. M. Jonkheer, A. Zlobin, B. A. Osborne, S. Gottipati, J. C. Aster, W. C. Hahn, M. Rudolf, K. Siziopikou, M. Kast, and L. Miele. 2002. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat. Med. 8:979-986. [DOI] [PubMed] [Google Scholar]
- 38.Xie, Y., W. Zhao, W. Wang, S. Zhao, R. Tang, K. Ying, Z. Zhou, and Y. Mao. 2001. Identification of a human LNX protein containing multiple PDZ domains. Biochem. Genet. 39:117-126. [DOI] [PubMed] [Google Scholar]
- 39.Yogosawa, S., Y. Miyauchi, R. Honda, H. Tanaka, and H. Yasuda. 2003. Mammalian Numb is a target protein of Mdm2, ubiquitin ligase. Biochem. Biophys. Res. Commun. 302:869-872. [DOI] [PubMed] [Google Scholar]