Abstract
In vitro studies have described the synthesis of an alternative reading frame form of the hepatitis C virus (HCV) core protein that was named F protein or ARFP (alternative reading frame protein) and includes a domain coded by the +1 open reading frame of the RNA core coding region. The expression of this protein in HCV-infected patients remains controversial. We have analyzed peripheral blood from 47 chronically or previously HCV-infected patients for the presence of T lymphocytes and antibodies specific to the ARFP. Anti-ARFP antibodies were detected in 41.6% of the patients infected with various HCV genotypes. Using a specific ARFP 99-amino-acid polypeptide as well as four ARFP predicted class I-restricted 9-mer peptides, we show that 20% of the patients display specific lymphocytes capable of producing gamma interferon, interleukin-10, or both cytokines. Patients harboring three different viral genotypes (1a, 1b, and 3) carried T lymphocytes reactive to genotype 1b-derived peptides. In longitudinal analysis of patients receiving therapy, both core and ARFP-specific T-cell- and B-cell-mediated responses were documented. The magnitude and kinetics of the HCV antigen-specific responses differed and were not linked with viremia or therapy outcome. These observations provide strong and new arguments in favor of the synthesis, during natural HCV infection, of an ARFP derived from the core sequence. Moreover, the present data provide the first demonstration of the presence of T-cell-mediated immune responses directed to this novel HCV antigen.
An estimated 170 million people are infected with hepatitis C virus (HCV) worldwide. In developed countries, HCV infection accounts for 40% of end-stage cirrhosis and 60% of hepatocellular carcinomas and has become the leading cause of liver transplantations (36). Mounting evidence indicates that evolution towards a chronic carrier state, a prerequisite to the development of cirrhosis and hepatocellular carcinomas, is associated with impaired T-cell-mediated immune responses (29, 39, 43). Similar to other viruses, in particular those causing viral persistence, HCV has been shown to express proteins harboring the potential to inhibit viral elimination by the infected host. HCV antigens such as NS5A, E2, and more recently, NS3 may interfere with the alpha/beta interferon (IFN-α/β)-mediated antiviral resistance (11-13, 38). Finally, multiple studies have implicated the HCV core in the dysfunction of cells from the immune system, including dendritic cells (10, 34). The core has been shown to perturb the function of cellular proteins (such as tumor necrosis factor [TNF] receptor 1, lymphotoxin β receptor, and C1q receptor) which play an important role in T-cell proliferation and Fas/TNF-α-induced apoptosis (15, 17, 24). In a transgenic mouse model, the HCV core leads to immune suppression and liver damage (37).
Interestingly, recent reports indicate that the HCV genome contains an overlapping +1 reading frame encoding alternative core antigens (4, 41, 42, 44). Evidence for the existence of such proteins results, so far, from in vitro studies, with a limited description of the presence of specific antibodies (Abs) in HCV patients (4, 42, 44). Furthermore, no consensus exists on the molecular mechanisms at the origin of the synthesis of such an alternative form of core. For genotype 1a-derived alternate protein, referred to as F protein or ARFP (alternative reading frame protein), the frameshifting would take place at or near codon 11 (42, 44) and the protein ends at codon 161. In contrast, in genotype 1b, alternate proteins seems to result from a +1 frameshift at codon 42, which could be, for some of them, followed by a rephasing in the normal open reading frame at the stop codon 144 (4). Although the shift junction and the length of the proteins seem to be different, both genotype 1a and 1b ARFP exhibit a common central frameshifted domain of 101 residues starting at codon 43 and ending at codon 144. Definite demonstration of the existence of an alternative core protein (which we have named ARFP here) during natural HCV infection and its potential role in the development of chronicity and virus-associated pathogenesis remain mainly unanswered questions. We describe in this study the presence of B-cell- and T-cell-mediated immune responses specific to the ARFP in individuals suffering from an ongoing or a past infection by HCV. These data provide further and novel evidence supporting the natural synthesis of an alternative HCV core antigen.
MATERIALS AND METHODS
Subjects.
Forty-seven HCV-seropositive patients were included in the study. Their clinical characteristics are summarized in Table 1. These patients were divided in four groups according to their treatment and response to therapy at the time of blood collection (20 to 40 ml): patients who were not treated at the time of blood collection (NT, n = 11), patients receiving antiviral therapy either responding (R, n = 12) or not responding (NR, n = 12), sustained virological responders defined as patients who have cleared viremia for at least 6 months after the end of therapy (SVR, n = 12). Blood samples from healthy HCV-seronegative individuals were obtained from the French blood center (Etablissement Français du Sang, Lyon, France). Informed consent was obtained from all subjects before inclusion in the study. Patients who were enrolled in longitudinal studies (patients 5 to 10) received a dual therapy (IFN-α and ribavirin) for either 6 (patient 5) or 12 (patients 6 to 10) months.
TABLE 1.
Clinical characteristics of patients at time of study
| Characteristic | Result for chronic HCV patient type (n):
|
|||
|---|---|---|---|---|
| NT (11) | NR (12) | R (12) | SVR (12) | |
| Mean age (yr) (range) | 54 (39-74) | 53 (40-62) | 45 (34-72) | 45 (32-61) |
| No. of patients with HCV genotype: | ||||
| 1 | 9 | 7 | 6 | 7 |
| 2 | 0 | 0 | 0 | 0 |
| 3 | 0 | 1 | 6 | 3 |
| 4 | 1 | 1 | 0 | 1 |
| NDa | 1 | 3 | 0 | 1 |
| No. of viremic patients | 11 | 12 | 0 | 0 |
| Mean ALTb (IU/ml) (range) | 89 (48-158) | 57 (48-163) | 33.5 (12-80) | 26 (9-85) |
| No. of patients receiving therapy regimen | ||||
| Ribavirin | 0 | 3 | 0 | 0 |
| IFN + ribavirin | 0 | 4 | 7 | 0 |
| PEG-IFNc + ribavirin | 0 | 5 | 3 | 0 |
| None (not treated) | 11 | 0 | 2 | 12 |
ND, not determined.
ALT, alanine amino transferase.
PEG-IFN, pegylated interferon.
HLA class I and II typing were performed at the Etablissement Français du Sang.
HCV RNA quantification.
For 36 of 47 patients, HCV RNA was quantified from either plasma or serum samples by real-time PCR after reverse transcription of the 5′ noncoding region of the genome as described elsewhere (21). Briefly, RNA was extracted from 50 μl of serum and plasma and reverse transcribed by using the Thermoscript reverse transcriptase kit from GibcoBRL (Cergy Pontoise, France) and the RC21 primer (nucleotides [nt] −20 to −39). cDNA (220 bp) was then quantified by real-time PCR by using RC1 sense (nt −251 to −268) and RC21 antisense primers, the LC DNA Master SYBR Green I kit from Roche Diagnostics (Meylan, France), and the LightCycler (Roche Diagnostics) according to a standard curve constructed from serial dilutions of synthetic HCV RNA. The sensitivity of the assay is 1,200 copies/ml.
Peptides and antigens. (i) Cellular immune responses.
For evaluation of cellular immune responses, genotype 1b peptides and antigens were derived from the HCV-JA isolate (EMBL entry code D89872) (20). The G97A synthetic peptide of ARFP (99 amino acids [aa] in length and located between aa 42 and 141 of the core sequence) was synthesized by Clonestar Ltd., Bristol, United Kingdom. The full sequence of the peptide is GWVCARLGRLPSGRNLVEGDNLSPRLAGPRVGPGLSPGTLGPSMATRVWGGQDGSCHPVALGLVGAPQTPGVGRVIWVRSSIPLHAASPTSWGTFRLSA. Four 9-mer peptides (W7L, A7L, G7L, R7V) (see Fig. 3B) predicted to bind HLA.A2 or HLA.B7 class I molecules (http://www-bimas.dcrt.nih.gov/molbio/hla_bind/) were purchased from Neosystem (Strasbourg, France). The peptides mapped at positions 40 to 48 (A7L), 43 to 51 (W7L), 50 to 58 (R7V), and 73 to 81 (G7L) of the ARFP sequence. Their theoretical binding scores to HLA molecules were 240 for A7L (HLA.B7), 84 and 20 for W7L (HLA.A2 and HLA.B7, respectively), 69 for R7V (HLA.A2), and 80 for G7L (HLA.B7).
FIG. 3.
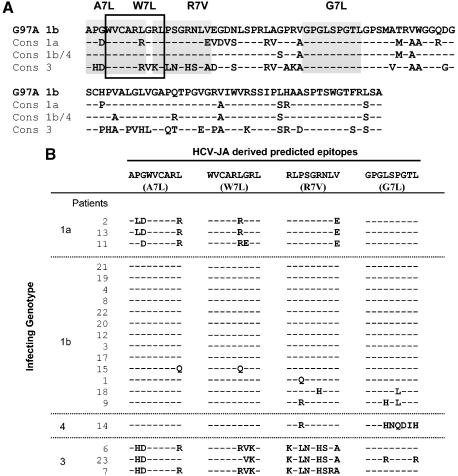
Conservation of ARFP sequences between HCV-JA and genotype 1a, 1b/4, and 3 consensus sequences. (A) The amino acid sequence of the G97A peptide derived from the HCV-JA isolate (genotype 1b) was aligned with the consensus sequences derived from the dominant circulating viral variants carried by the different patients enrolled in the study. The positions of the 4 class I restricted epitopes used in the study (W7L, G7L, A7L, and R7V) are indicated by shaded boxes. Differing amino acids are indicated in boldface type. (B) The 20 sequences of the 4 class I restricted epitopes derived from each patient are aligned with the genotype 1b sequence from which these 9-mer epitopes were derived. Common amino acids between these patients' derived sequences and the corresponding synthetic genotype 1b peptide are represented by dashes while differing amino acids are specified.
A genotype 1a recombinant core protein produced in Escherichia coli was also used in proliferation assays (aa 2 to 169). The 9-mer peptides, the G97A peptide, and the core were used, respectively, at concentrations of 10 μM, 1 μg/ml, and 2 μg/ml in the various assays.
The peptides and antigens used as positive controls in enzyme-linked immunospot (ELISPOT), cytometric bead array (CBA), and proliferative assays were the tetanus toxoid (TT; Aventis-Pasteur, Marcy l'Etoile, France) at 2 μg/ml, a panel of 23 major histocompatibility complex class I-restricted viral peptides at 10 μM, polyclonal activators including phytohemagglutinin M (Sigma, St. Louis, Mo.) at a 1/100 dilution, lipopolysaccharide (Sigma) at 1 μg/ml, and phorbol myristate acetate-ionomycin (Sigma) at 1 and 500 ng/ml, respectively.
(ii) Humoral immune responses.
For the evaluation of humoral immune responses, core proteins and ARFP were produced as described elsewhere (21a). Briefly, amplification of ARFP and core sequences was achieved by PCR from a genotype 1a HCV sequence (22). For amplification of the ARFP sequence, an A deletion at nucleotide position 31 was introduced to obtain an ARFP open reading frame of 161 aa containing the first 10 aa of the core protein. Core sequences were amplified to obtain a core PCR product corresponding to aa 1 to 119 of the HCV core. F and core PCR products were cloned into the expression vector pET21b (Novagen) upstream of a hexahistidine tail, allowing overexpression in E. coli and purification on an Ni-nitrilotriacetic acid agarose column.
ELISPOT assays.
Peripheral blood mononuclear cells (PBMCs) were isolated as previously described (1). Interleukin-10 (IL-10)- and IFN-γ-producing cells were quantified by using ELISPOT kits from Diaclone (Besançon, France) according to the manufacturer's instructions. Briefly, PBMC were stimulated with peptides, polyclonal activators, or RPMI complete medium (RPMI 1640 [Invitrogen, Cergy Pontoise, France] supplemented with 10% fetal calf serum [FCS; Sigma], 2 mM l-glutamine [Invitrogen], 50 IU of penicillin [Invitrogen]/ml, 50 μg of streptomycin [Invitrogen]/ml) as a negative control for 36 h. When stimulated with antigens (TT, core, or G97A), cells were first preincubated with these antigens in microtubes (Micronic Systems) overnight and then transferred for another 24-h incubation to the ELISPOT plates (35). Spots were counted by computer-assisted analysis with KS ELISPOT software (Zeiss, Jena, Germany). The number of spots observed in the negative control wells was subtracted from the number of spots observed in the experimental wells. A response was considered positive if the number of spots for 106 PBMCs was greater than 50 spots and at least twice that in control wells.
Proliferation assays.
PBMCs were incubated at 5 × 105 cells/ml in 200 μl of complete RPMI medium alone or supplemented with peptides or proteins in triplicate in 96-well U-bottom plates for 6 days at 37°C with 5% CO2. Proliferative responses were evaluated after a 16-h [3H]thymidine incorporation as previously described (1). Stimulation indices (defined as the ratio of [3H]thymidine incorporation in stimulated cultures to than in unstimulated cultures) greater than four were regarded as positive for antigen-specific T-cell stimulation.
In the longitudinal experiments, proliferation in response to HCV antigens and TT was measured by CFSE {5-(and-6)-carboxyfluorescein diacetate, succinimidyl ester [5 (6)-CFDA, SE] mixed isomers} staining. Four million PBMCs at a concentration of 20 million cells/ml were stained with 1 μM CFSE (Molecular Probes, Eugene, Oreg.) in RPMI containing 2% FCS for 13 min at 37°C in the dark with shaking every 3 min. The reaction was stopped with a large volume of RPMI-2% FCS, and the cells were washed two times in RPMI-2% FCS. Two million CFSE-labeled cells were then stimulated with proteins in 24-well plates for 7 days, stained with anti-CD3-phycoerythrin (BD PharMingen, San Diego, Calif.), and analyzed by flow cytometry.
CBA immunoassay.
PBMCs were cultured at 2 × 106 cells/ml in the presence of antigens or medium alone in 96-well U-bottom plates for 48 h and then restimulated with phorbol myristate acetate-ionomycin (200 and 20 μg/ml, respectively), and supernatants were recovered after centrifugation and frozen at −20°C until analysis. IL-2, IL-4, IL-5, IL-10, TNF-α, and IFN-γ were simultaneously quantified by using the Th1/Th2 CBA kit and analyzed by flow cytometry with CBA software as recommended by the manufacturer (BD Biosciences).
Detection of anti-core protein and anti-ARFP Abs.
The presence of Abs specific for the core protein and ARFP was evaluated in the serum or the plasma of 36 of 47 patients with an in-house enzyme-linked immunosorbent assay (ELISA) as described elsewhere (21a).
Briefly, microtiter plates were coated with 100 μl of core protein (0.5 μg/ml) or ARFP (1 μg/ml) in 50 mM sodium carbonate buffer (pH 9.5) by overnight incubation at room temperature. After washing and saturation, a 1:100 dilution of each sample serum was added and incubated at 37°C for 1 h. After washing, wells were incubated at 37°C for 1 h with peroxidase-conjugated affinipure goat anti-human immunoglobulin G whole Ab (Jackson ImmunoResearch Laboratories) diluted 1:10,000. Finally, the color reaction, started by adding O-phenylenediamine dihydrochloride-H2O2 buffer, was stopped after 30 min by adding 1.8 N H2SO4. Absorbance was measured at 490 nm in a microplate reader (Molecular Devices). For each experiment, the cutoff was determined as the mean plus three standard deviations of the results from three HCV-negative serum samples. A serum samples was considered positive when the absorbance was equal or superior to 0.1 plus the cutoff.
Amplification and sequencing of ARFP circulating sequences.
Viral RNA was extracted from 140 μl of serum or plasma by using the QIAamp viral RNA mini spin kit as recommended by the manufacturer (Qiagen, Hilden, Germany) and resuspended in 30 μl of elution buffer. HCV RNA was amplified by reverse transcription (RT)-PCR with the one-step RT-PCR kit (Qiagen) and two primers: NC4R (nt 286 to 312) and 13R (nt 852 to 874). Ten microliters of RNA was used for RT-PCR. Conditions were as follows: prehybridization at 70°C for 10 min; reverse transcription at 50°C for 30 min; activation of PCR at 95°C for 15 min; 45 cycles of 95°C for 1 min, 51°C for 1 min, and 72°C for 1 min; and a final extension at 72°C for 10 min. Eight microliters of the RT-PCR product was analyzed on an agarose gel, and the remainder was purified by using the mini elute gel extraction kit (Qiagen). The concentration and purity of the PCR products were determined by measuring the absorbance at 260 nm and the A260/A280 absorbance ratio. Sequencing was performed by Genome Express (Meylan, France). Nucleotide sequences were translated in the +1 frame on the website http://www.infobiogen.fr/services/analyseq/cgi-bin/traduc_in.pl and aligned with CLUSTALW (http://pbil.ibcp.fr/html/pbil_index.html).
Statistical analysis.
Because of sample sizes, nonparametric statistics were used. Comparisons between all groups were achieved with the Kruskal-Wallis test, which is a nonparametric analysis of variance.
RESULTS
HCV-seropositive patients display cellular and humoral immune responses specific to ARFP.
To document the existence of cellular immune responses against the ARFP, we used five different specific peptides: G97A polypeptide (99-mer), synthesized from a genotype 1b sequence and including aa 42 to 140 of the +1 reading frame sequence, which represents a domain shared by all frame-shifted core proteins described to date, and four 9-mer peptides (W7L, A7L, G7L, and R7V) contained within G97A or overlapping core and G97A and predicted to bind HLA.A2 (W7L and R7V) or HLA.B7 (A7L, G7L, and W7L) class I molecules. We measured cell proliferation and quantified cytokine-producing cells after in vitro stimulation of PBMC with the different peptides. PBMC were isolated from different cohorts of HCV-seropositive individuals (Table 1), including nontreated chronic HCV patients (NT, n = 11), chronic HCV patients not responding to therapy (NR, n = 12), chronic HCV patients responding to therapy (R, n = 12), sustained virological responders (SVR, n = 12), and HCV-seronegative healthy individuals (S, n = 12). The presence of ARFP-specific Abs was also evaluated for 36 of 47 patients for which serum or plasma were available. Reactivity against the ARFP was assessed by using an in-house ELISA as described in Materials and Methods. Results of the overall analyses performed are illustrated in Fig. 1 and 2.
FIG. 1.
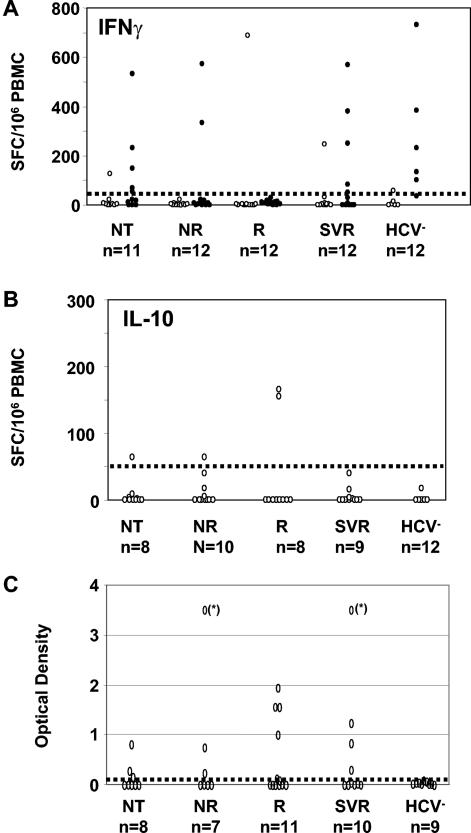
Detection of IFN-γ- and IL-10-producing cells specific to the ARFP. ELISPOT assays were performed with PBMC from different cohorts of patients chronically infected with HCV and control subjects including nontreated carriers (NT), nonresponding patients (NR), responding patients (R), sustained virological responding patients (SVR), and HCV-seronegative subjects (HCV−). The number of IFN-γ (A)- and IL-10 (B)-producing cells per 106 PBMC following in vitro stimulation by the G97A peptide (empty circles) and of IFN-γ-producing cells in response to TT (black dots) (A) were evaluated by ELISPOT on the different cohorts of patients. (C) The presence of anti-ARFP Abs in the serum or plasma of 36 of these 47 patients was assessed and is reported as OD values on the graph (saturating OD values are marked by asterisks on the graph). The dotted line on each graph represents the cutoff value of each assay, determined as described in Materials and Methods.
FIG. 2.
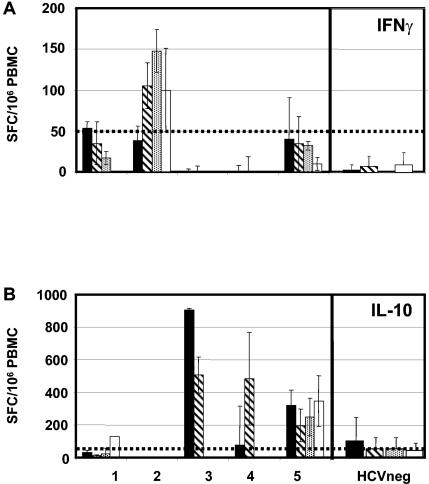
IFN-γ- and IL-10-producing cells are detected in response to class I predicted 9-mer epitopes derived from ARFP. The presence of IFN-γ-producing (A) and IL-10-producing (B) cells in the peripheral blood of 5 chronic HCV patients (patient 1, NT, genotype 1, HLA.A1, HLA.A24, HLA.B14, HLA.B44; patient 2, NT, genotype 1b, HLA.A1, HLA.A3, HLA.B7, HLA.B8; patient 3, NR, genotype 1, HLA.A2, HLA.B18, HLA.B35; patient 4, R, genotype 1b, HLA.A2, HLA.B18, HLA.B35; patient 5, SVR, genotype 1a, HLA.A24, HLA.A69, HLA.B51) was determined following in vitro stimulation with the following class I predicted HLA-A2- or HLA-B7-restricted peptide epitopes: peptide R7V (black bars), W7L (hatched bars), A7L (dotted bars), and G7L (white bars). The reactivity of PBMC from HCV-negative individuals against these same epitopes was evaluated and represented as the mean ± standard deviation of the results from nine and seven experiments with IFN-γ-producing cells and IL-10-producing cells, respectively. The dotted line on each graph represents the cutoff value of each assay determined as described in Materials and Methods.
T cells from infected or recovered patients proliferate poorly but produce IFN-γ or IL-10 in response to G97A polypeptide.
Although all patients mounted a normal proliferative response to phytohemagglutinin, PBMC from only 1 patient of 47 showed a significant proliferative response to G97A polypeptide stimulation (data not shown).
The numbers of IFN-γ- and IL-10-producing cells in response to G97A polypeptide for the various patients are reported in Fig. 1A and B. Cytokine-producing cells were detected in 6 of 47 HCV patients. The overall number of specific IFN-γ- or IL-10-producing cells ranged from 55 to 700 and 60 to 160 per 106 PBMC, respectively. Different patterns were observed. For 3 of 6 patients (2R and 1 NR), only IL-10-producing cells were detected. For 2 of 6 patients (1 R and 1 SVR), G97A-specific T cells produced IFN-γ without IL-10. Finally, for 1 of 6 patients (NT), both IFN-γ- and IL-10-secreting cells could be detected. We could exclude, for 12 of 43 patients, a functional defect of the ARFP-specific T cells in IFN-γ production, since IFN-γ-producing cells were induced in response to stimulation by TT. Of note, PBMC from the one patient that proliferated in response to G97A produced neither IFN-γ nor IL-10 when stimulated by this polypeptide.
There was no obvious link between the detection of specific T cells and the clinical status of the patients, as G97A-specific T cells were detected in all four cohorts of HCV-seropositive patients (P > 0.05). Furthermore, although G97A polypeptide was derived from a genotype 1b sequence, cytokine-producing cells were detected in HCV-seropositive patients infected with various viral genotypes (1a, 1b, and 3).
CD8+ memory T cells specific for HLA.A2- and/or HLA.B7-predicted epitopes derived from ARFP are present in HCV-seropositive patients.
Twenty-two of the 47 patients were also tested for reactivity of their PBMC to HLA.A2- and HLA.B7-predicted epitopes. Five of 22 patients (patients 1 to 5) had circulating T cells capable of producing either IFN-γ or IL-10 when stimulated with the peptide epitopes (Fig. 2). Only two of these patients (patients 2 and 5) also had detectable responses to the G97A polypeptide (data not shown), suggesting that processing of the 99-mer peptide for class I presentation is not optimal.
Cells from 2 of the 5 patients were mainly biased toward the production of IFN-γ (patients 1 and 2, both NR) while cells from the 3 other patients produced more significant levels of IL-10 (patients 3 [NT], 4 [R], and 5 [SVR]). The HLA restriction of the patients did not systematically guide the reactivity of the patients' cells. For example, while patient 2 reacted only to stimulation with the HLA.B7-predicted epitopes, in accordance with his HLA typing (HLA.A1, HLA.A3, HLA.B7, and HLA.B8), patient 5 displayed significant IL-10-secreting cells in response to both HLA.A2- and HLA.B7-restricted epitopes, although this patient (HLA.A24, HLA.A69, and HLA.B51) presented neither an HLA.A2 nor an HLA.B7 restriction.
It is interesting that, in contrast to the response specific to G97A which was observed in patients infected with various genotypes (1a, 1b, and 3), the responses specific to the predicted HLA epitopes were essentially restricted to patients infected with genotype 1a or 1b viruses (patients 1 and 3 harbored genotype 1 viruses, patients 2 and 4 harbored genotype 1b viruses, and patient 5 harbored a genotype 1a virus).
Overall (Fig. 1 and 2), 10 of 47 (21%) HCV-seropositive patients display ARFP-specific T-cell responses. Proliferating cells were detected for 1 patient only, IFN-γ- and/or IL-10-producing cells in response to the 99-mer peptide were present for 6 patients (among whom 2 also responded to 9-mer peptides), and IFN-γ- and/or IL-10-producing cells in response to 9-mer peptides were detected for only three additional patients.
Abs specific to ARFP are detected in HCV-seropositive patients.
For 36 of 47 HCV-infected patients, we searched for the presence of anti-ARFP-specific Abs in the serum or plasma by using an in-house ELISA (Fig. 1C). As expected, none of the HCV-seronegative healthy individuals tested (n = 9) showed detectable anti-ARFP Abs in their sera. In contrast, for 15 of 36 patients (41.6%), we could document the presence of anti-ARFP-specific Abs with optical density (OD) values above the cutoff, as illustrated in Fig. 1C. Such a humoral immune response was detected independently of the infecting viral genotype (P > 0.05) and of the clinical status (P > 0.05), since 3 of 8 NT, 3 of 7 NR, 5 of 11 R, and 4 of 10 SVR patients displayed anti-ARFP Abs. All 47 patients displayed anti-core protein Abs (data not shown).
Among the 15 patients displaying anti-ARFP Abs, 2 patients also had a detectable G97A-specific T-cell response and 3 patients displayed T cells reactive to minimal epitopes (combined data not shown).
Overall, our data provide the first evidence of cellular immune responses against an alternative reading frame form of the HCV core protein in chronic or previously HCV-infected patients. Furthermore, we confirm previous reports describing the presence of anti-ARFP Abs in HCV patients.
ARFP sequences from genotypes 1 and 3 display divergence.
To determine whether the absence of anti-ARFP-specific T-cell responses in the majority of the patients tested could result from mutations of the viral genome within this particular sequence, ARFP sequences derived from the dominant circulating species from 20 of 47 patients, including 10 NT, 7 NR and 4 R patients, were identified and compared. They included sequences from all patients whose sera reacted with the G97A polypeptide and/or the HLA.A2- and HLA.B7-predicted epitopes and for whom viral RNA was present in the plasma. For the 4 patients having cleared viremia during therapy (i.e., responders), viral RNA was extracted from plasma samples obtained before clearance. Consensus sequences from the various genotypes represented in the 20 patients (3 genotype 1a strains, 13 genotype 1b strains, 3 genotype 3 strains, and 1 genotype 4 strain) were aligned with that of the synthetic G97A peptide (genotype 1b, HCV-JA isolate), which includes the sequences of the 9-mer synthetic peptides (Fig. 3A).
A comparison of all variant sequences indicated that the sequence of the ARFP polypeptide was highly conserved within genotype 1a and 3 strains (>92% identity) and somewhat less within genotype 1b isolates (>60%) (data not shown). A comparison of consensus sequences from genotype 1a and 1b isolates shows 86 and 79% identity, respectively, with the G97A polypeptide sequence (Fig. 3A). Figure 3B shows the alignment of the 20 sequences of the class I-restricted epitopes derived from each patient in comparison with the genotype 1b sequence from which these 9-mer epitopes were derived. When comparing the sequences of the small class I-restricted epitopes derived from genotype 1a and 1b isolates, 3 of 4 epitopes showed 80 to 100% identity with the synthetic genotype 1b sequence (G7L, W7L, and R7V). The remaining epitope (A7L) was almost totally conserved among genotype 1b sequences but was more variable when considering the genotype 1a sequences, which displayed, for 2 of 3 of them, 3 mutations compared with the synthetic sequence. By contrast, sequences derived from genotype 3 isolates were more divergent from the HCV-JA sequence, with only 60% identity at the level of the whole G97A polypeptide. A poor homology (between 22 and 77%) was also observed for 3 of 4 of the consensus 9-mer peptides compared to the synthetic genotype 1b sequences used in our assays (A7L, W7L, and R7V). The G7L peptide sequence was much more conserved, as 2 of 3 sequences are identical to the HCV-JA-derived epitope.
Overall, these data indicate that (i) the absence of cytokine-producing or -proliferating T cells in response to G97A for 79% of HCV-seropositive patients cannot be due to sequence diversity between the patients' viral sequences and that of synthetic peptides used in our assays and (ii) the lack of detection of T cells specific to the minimal peptide epitopes in patients infected by genotype 3 virus may be related to the poor sequence conservation compared to that of genotype 1b synthetic peptides.
Vigor and kinetics of ARFP-specific T-cell- and B-cell-mediated responses are different from those specific to the core protein in patients receiving therapy.
For 6 of 47 patients, blood samples were obtained before and at different time points during combined IFN-α and ribavirin therapy. These patients were infected by genotype 1 (patients 5, 8, and 9 [1a] and 10 [1b]) or genotype 3 (patients 6 and 7) isolates. T-cell responses specific for core protein, ARFP, and TT were evaluated over time by measuring both the proliferative capacity and the cytokine secretion pattern of PBMC in response to these antigens (Fig. 4A and B). The presence of Abs specific for core protein and ARFP in the serum of these patients before and during treatment was also assessed by using an in-house ELISA (Fig. 4C). The fluctuation of viremia was concomitantly analyzed for all patients by HCV-specific real-time PCR (Fig. 4C).
FIG. 4.
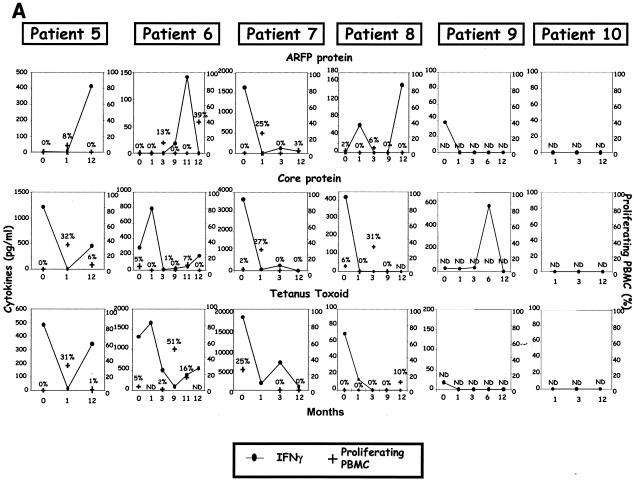
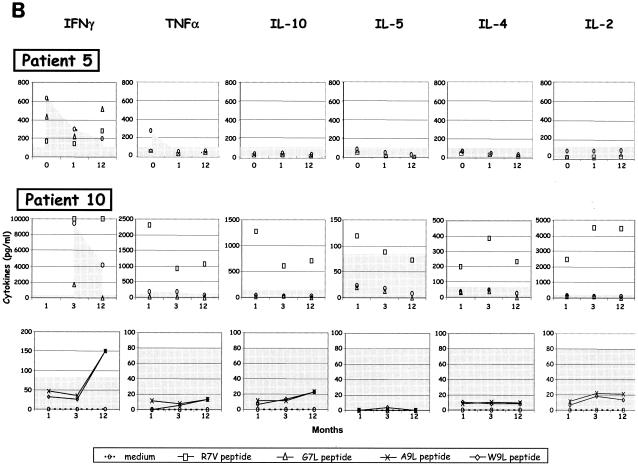
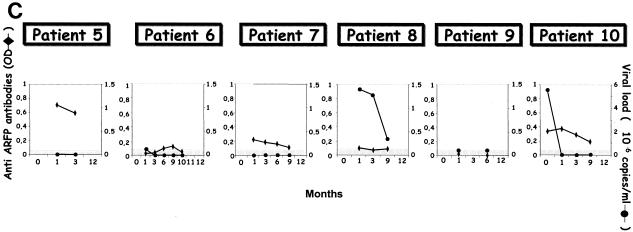
Evolution of core- and ARFP-specific cytokine profiles, T-cell proliferation, Abs, and viremia in chronic HCV patients receiving therapy. Immune and virologic readouts were analyzed for six chronically infected HCV patients (patients 5 to 10) from the time of therapy onset (T0) up to month 12 (M12) following therapy initiation. Patient 5 received only a 6-month course of therapy while all others received a 12-month course. (A) Six cytokines were measured (CBA test) in the supernatants of peripheral blood cells after in vitro stimulation with the G97A peptide, core antigen, or TT. Only cytokines produced at significant levels after antigen stimulation (sensitivity threshold, >80 pg/ml) are represented on the graph (left hand axis). T-cell proliferation, measured by flow cytometry after a 6-day incubation in the presence of CFSE, is reported as the percentage of proliferating T cells (right hand axis) and is represented by bold black crosses. (B) For patients 5 (HLA.A25, HLA.A69, and HLA.B51) and 10 (HLA.A2, HLA.A29, HLA.B7, and HLA.B51), cytokines produced in the supernatant of PBMC stimulated with the corresponding predicted 9-mer peptides were measured with the Th1/Th2 CBA kit. (C) Evolution of anti-ARFP Abs measured by ELISA (OD values are shown on the left hand axis) is reported along with the viremia levels (right hand axis) during antiviral therapy.
For all patients, we could document ARFP-specific T-cell responses at least at one time point during follow-up (Fig. 4A and B). Similarly, core-specific T-cell responses were detected for all but one patient (patient 10). These responses were essentially characterized by IFN-γ secretion or proliferation, with these two functions being generally uncoupled (Fig. 4A). A striking feature of these detected T-cell responses is that both HCV-specific and nonspecific (TT) T-cell responses fluctuate considerably over time, with an overall decrease in cytokine production at month 1 after the onset of therapy (Fig. 4A). Another one is that the vigor of the ARFP-specific response was systematically lower than the core-specific T-cell response (mean IFN-γ production in response to core and ARFP was 1,300 and 470 pg/ml, respectively). ARFP-specific T-cell responses did not appear to follow the same kinetics as the core-specific T-cell responses, with the latter usually detectable at the initiation of therapy while the former seemed to be delayed in time for most of the patients.
The HLA-restricted epitopes derived from the G97A domain were tested with cells from patients 5 and 10. For patient 5, the reactivity to the 9-mer peptides was quite limited. Only one time point was found positive (month 12) (Fig. 4B), at which IFN-γ was detected in response to G7L and, to a lesser extent, R7V peptides. For this patient, at this same time point (month 12), IFN-γ was also secreted in response to G97A (Fig. 4A). PBMC from patient 10 (HLA.A2, HLA.A29, HLA.B7, and HLA.B51), although not reactive to G97A or core stimulation, produced not only IFN-γ in response to 3 of 4 minimal epitopes (R7V, A7L, and W7L) (Fig. 4B) but also, in the case of epitope R7V, significant levels of the other five cytokines analyzed, i.e., TNF-α, IL-2, IL-4, IL-5, and IL-10 (cytokine production ranging from 80 to 4,500 pg/ml) (Fig. 4B).
For these 6 patients, we also evaluated the presence of Abs specific to the core protein and ARFP in the serum during follow-up. All patient sera contained high levels of anti-core protein-specific Abs (saturating OD values) that were maintained throughout treatment (data not shown). The detection of anti-ARFP Abs is shown on Fig. 4C along with the quantification of viremia performed at the same time points. Four of 6 patients cleared the virus from their serum 1 to 3 months after the initiation of therapy (patients 5, 7, and 10 and patient 6, respectively) while the other 2 patients remained viremic during therapy. Significant detection of anti-ARFP Abs (OD > 0.100, i.e., cutoff value) could be documented for 5 of 6 patients with OD values ranging from 0.103 (patient 8; M1) to 0.711 (patient 5; M1) (Fig. 4C).
DISCUSSION
We evaluated in this study the existence of immune responses specific to the newly described alternative core protein during ongoing or therapeutically resolved HCV infection. Although we did not directly search for expression of the ARFP antigen in the infected host, we were able to identify the signature of a past or present existence of such an antigen by documenting the presence of immunological markers specific to ARFP determinants. We could document, for the first time, the presence of both T-cell-mediated and B-cell-mediated immune responses specific for ARFP antigen in subjects chronically infected or having resolved the infection. First, T lymphocytes specific to a 99-mer polypeptide derived from the alternative reading frame of the core protein were detected in about 15% of the tested patients. These cells were mainly characterized by their capacity to produce cytokines such as IFN-γ and/or IL-10 but poorly proliferated in response to the polypeptide. Only 1 of 47 of the patients from the single time point cohort (Fig. 1) and 3 of 4 patients from the longitudinal cohort (Fig. 4) displayed significant proliferative T-cell responses to G97A. For these latter 3 patients, only cells from a few scattered time points were found capable of in vitro proliferation. This fluctuating reactivity was not only observed for the proliferation response to G97A but was a general feature of both proliferative and cytokine secretion responses to both HCV-specific (core protein and ARFP) nonspecific (such as the recall TT) antigens. The fluctuation of the T-cell-based response that we observed contrasts with previous longitudinal studies describing an enhancement of HCV-specific CD4+-T-cell responses, including core-specific responses, during antiviral therapy (increase in frequency, strength, and breadth) (2, 19). The mechanisms responsible for these fluctuations are unclear. They may be linked to a direct effect of therapy but could as well be unrelated to antiviral treatment. Indeed, fluctuating CD8+-T-cell responses have been previously observed in a longitudinal follow-up of untreated chronic HCV patients (33). As previously shown with HCV infection (2), our longitudinal follow-up also reveals that IFN-γ secretion and proliferation are rarely concomitant. Discordance between the frequency of IFN-γ production and lymphoproliferation is also a characteristic feature of other viral infections such as human immunodeficiency virus type 1 infection (32). In human immunodeficiency virus type 1-infected patients, CD4+-T-cell proliferation and IL-2 production, independent of the production of IFN-γ, seem to be associated with good clinical outcome (3, 28). Our present study does not allow us to link the proliferative ability of HCV-specific T lymphocytes in response to ARFP with the clinical outcome. Although controversial to some extent (6, 39), it is widely admitted that a low or absent CD4+-T-cell-mediated response is one of the major features of chronic HCV infection, whereas a vigorous and maintained HCV-specific CD4+-T-cell response, in contrast, is typically found to be associated with viral clearance (7, 8, 14, 16, 29).
The fact that G97A-specific T cells have a dramatically altered capacity to expand upon antigenic stimulation suggests that they could be functionally impaired. Impairment of HCV-specific T lymphocytes (whether CD4+ or CD8+) during chronicity is classic (39, 43). While such impairments play, without much ambiguity, a role in the development of HCV chronicity, the existence of regulatory T cells, recently identified in response to the core antigen, may also participate in such development (23). High levels of IL-10 production by the regulatory type 1 subset of CD4 T cells, typically associated with the production of low amounts of IFN-γ, IL-5, and transforming growth factor β, can suppress protective T-cell responses (9, 27). At that stage, we can report that ARFP-specific lymphocytes have the capacity to produce IL-10, in association or not with IFN-γ. Whether or not these two cytokines are produced by the same cell and whether or not such lymphocytes display the phenotype and function of regulatory cells remain to be further analyzed.
Another original finding from our study is that of the presence, during chronic infection as well as during or after resolution, of T cells specific to ARFP minimal class I-restricted epitopes (Fig. 2). These T cells, presumed to be CD8+ T lymphocytes, were characterized by the production of IFN-γ and/or IL-10 as well as, for one patient analyzed longitudinally (Fig. 4), a whole array of other cytokines (TNF-α, IL-10, IL-5, IL-4, and IL-2). Although puzzling, nontraditionally derived cytotoxic T lymphocyte epitopes have been documented in various infectious and tumorigenic models (for a review, see reference 25). Influenza virus and retroviruses have been shown to encode cytotoxic T lymphocyte epitopes from alternative open reading frames (5, 18, 26). Such cryptic, frameshift-derived epitopes appear to be infrequent and found at low concentrations, but their existence is now formally recognized, even though their physiological relevance remains mostly unclear. Indeed, different mechanisms for nontraditional epitope generation do not necessarily involve the expression of the frameshifted protein. Only a few examples of viruses have been shown to express proteins through a ribosomal frameshift during translation. This event usually occurs at a low frequency, such as the translation of the Gag-Pol precursor from retroviruses which represents 1 to 10% of the translational events (30). Furthermore, these proteins, as exemplified by the L* protein from the Theiler's murine encephalitis virus, can be involved in viral persistence in vivo (40). In the case of HCV, our data are too preliminary to appreciate the role of the observed ARFP class I-restricted T-cell responses in the evolution of HCV pathogenesis. Mechanisms associated with the generation of such epitopes are unclear. A translational mechanism such as ribosomal frameshifting has been proposed to be responsible for the synthesis of the HCV alternative core protein (4, 41, 44), similar to what has been described for the L* protein from Theiler's murine encephalitis virus (40). The presence of Abs reactive to the ARFP in HCV patients strongly suggests that this frameshifted protein is expressed in vivo and that these nontraditional epitopes may not only derive from misdirected splicing events or defective ribosomal products (25). Once synthesized, the ARFP may then be processed either by conventional or cross-presentation-linked pathways, issues which remain to be studied.
Contrasting with the dynamic nature of the anti-HCV T-cell responses, the detection of anti-ARFP and anti-core protein Abs in the longitudinal follow-up was much more stable over time. Overall, anti-core protein Abs were detected in all scenarios tested, as expected and as previously reported for HCV infection (31). The detection of anti-ARFP Abs was more cryptic, although unambiguous. In our study, an overall 41.6% of tested patients displayed anti-ARFP Abs. This frequency is slightly lower than that reported by Komurian-Pradel et al. (21a), who document, by using the same ELISA, a 62% seroprevalence among 154 HCV patients tested. The observed differences may simply be due to the difference in the size and nature of the cohorts involved in the respective studies. In our study, anti-ARFP Abs were not necessarily correlated with ARFP-specific T-cell responses. This could be easily observed in our longitudinal analysis but was also seen in the 47 patients analyzed nonlongitudinally (Fig. 1). In these patients, different patterns of response were observed, with 5 patients displaying both cellular and humoral anti-ARFP responses, 10 patients displaying only humoral responses, and 2 patients with only cellular immune responses.
In conclusion, we report for the first time that T lymphocytes specific to an alternative form of core protein exist during and/or after resolution of chronic HCV infection. Altogether, our data provide additional arguments supporting the hypothesis that HCV encodes for 10 and not 9 antigens. Whether or not such responses contribute to HCV pathogenesis or control of infection warrants additional studies.
Acknowledgments
This work was supported by a grant from ANRS (Agence Nationale pour la Recherche sur le SIDA), the European Community (QLRT-PL 1999-00356 and 2001-01329), and bioMérieux SA.
We thank M. Lucas and P. Klenerman for useful comments through the course of the study.
REFERENCES
- 1.Bain, C., A. Fatmi, F. Zoulim, J. P. Zarski, C. Trépo, and G. Inchauspé. 2001. Impaired allostimulatory function of dendritic cells in chronic hepatitis C infection. Gastroenterology 120:512-524. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, E., G. Harcourt, D. Brown, M. Lucas, R. Phillips, G. Dusheiko, and P. Klenerman. 2002. The dynamics of T-lymphocyte responses during combination therapy for chronic hepatitis C virus infection. Hepatology 36:743-754. [DOI] [PubMed] [Google Scholar]
- 3.Boaz, M. J., A. Waters, S. Murad, P. J. Easterbrook, and A. Vyakarnam. 2002. Presence of HIV-1 Gag-specific IFN-gamma+IL-2+ and CD28+IL-2+ CD4 T cell responses is associated with nonprogression in HIV-1 infection. J. Immunol. 169:6376-6385. [DOI] [PubMed] [Google Scholar]
- 4.Boulant, S., M. Becchi, F. Penin, and J. Lavergne. 2003. Unusual multiple recoding events leading to alternative forms of hepatitis C virus core protein from genotype 1b. J. Biol. Chem. 278:45785-45792. [DOI] [PubMed] [Google Scholar]
- 5.Bullock, T. N., and L. C. Eisenlohr. 1996. Ribosomal scanning past the primary initiation codon as a mechanism for expression of CTL epitopes encoded in alternative reading frames. J. Exp. Med. 184:1319-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cramp, M. E., S. Rossol, S. Chokshi, P. Carucci, R. Williams, and N. V. Naoumov. 2000. Hepatitis C virus-specific T-cell reactivity during interferon and ribavirin treatment in chronic hepatitis C. Gastroenterology 118:346-355. [DOI] [PubMed] [Google Scholar]
- 6a.Currier, J. R., E. G. Kuta, E. Turk, L. B. Earhart, L. Lommis-Price, S. Janetzi, G. Ferrari, D. L. Birx, and J. H. Cox. 2002. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. J. Immunol. Methods 260:157-172. [DOI] [PubMed] [Google Scholar]
- 7.Day, C. L., G. Lauer, G. K. Robbins, B. McGovern, A. G. Wurcel, R. T. Gandhi, R. T. Chung, and B. D. Walker. 2002. Broad specificity of virus-specific CD4+ T-helper-cell responses in resolved hepatitis C virus infection. J. Virol. 76:12584-12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diepolder, H. M., R. Zachoval, R. M. Hoffmann, E. A. Wierenga, T. Santantonio, M. C. Jung, D. Eichenlaub, and G. R. Pape. 1995. Possible mechanism involving T-lymphocyte response to non-structural protein 3 in viral clearance in acute hepatitis C virus infection. Lancet 346:1006-1007. [DOI] [PubMed] [Google Scholar]
- 9.Doetze, A., J. Satoguina, G. Burchard, T. Rau, C. Loliger, B. Fleischer, and A. Hoerauf. 2000. Antigen-specific cellular hyporesponsiveness in a chronic human helminth infection is mediated by T(h)3/T(r)1-type cytokines IL-10 and transforming growth factor-beta but not by a T(h)1 to T(h)2 shift. Int. Immunol. 12:623-630. [DOI] [PubMed] [Google Scholar]
- 10.Dolganiuc, A., K. Kodys, A. Kopasz, C. Marshall, T. Do, L. Romics, Jr., P. Mandrekar, M. Zapp, and G. Szabo. 2003. Hepatitis C virus core and nonstructural protein 3 proteins induce pro- and anti-inflammatory cytokines and inhibit dendritic cell differentiation. J. Immunol. 170:5615-5624. [DOI] [PubMed] [Google Scholar]
- 11.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 12.Gale, M., Jr., B. Kwieciszewski, M. Dossett, H. Nakao, and M. G. Katze. 1999. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J. Virol. 73:6506-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gale, M. J., Jr., M. J. Korth, N. M. Tang, S. L. Tan, D. A. Hopkins, T. E. Dever, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1997. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology 230:217-227. [DOI] [PubMed] [Google Scholar]
- 14.Gerlach, J. T., H. M. Diepolder, M.-C. Jung, N. H. Gruener, W. W. Schraut, R. Zachoval, R. Hoffmann, C. A. Schirren, T. Santantonio, and G. R. Pape. 1999. Recurrence of Hepatitis C virus after loss of virus-specific CD4+ T-cell response in acute hepatitis C. Gastroenterology 117:933-941. [DOI] [PubMed] [Google Scholar]
- 15.Giannini, C., and C. Brechot. 2003. Hepatitis C virus biology. Cell Death Differ. 10(Suppl. 1):S27-38. [DOI] [PubMed] [Google Scholar]
- 16.Godkin, A. J., H. C. Thomas, and P. J. Openshaw. 2002. Evolution of epitope-specific memory CD4(+) T cells after clearance of hepatitis C virus. J. Immunol. 169:2210-2214. [DOI] [PubMed] [Google Scholar]
- 17.Hahn, C. S., Y. G. Cho, B. S. Kang, I. M. Lester, and Y. S. Hahn. 2000. The HCV core protein acts as a positive regulator of fas-mediated apoptosis in a human lymphoblastoid T cell line. Virology 276:127-137. [DOI] [PubMed] [Google Scholar]
- 18.Hahn, Y. S., V. L. Braciale, and T. J. Braciale. 1991. Presentation of viral antigen to class I major histocompatibility complex-restricted cytotoxic T lymphocyte. Recognition of an immunodominant influenza hemagglutinin site by cytotoxic T lymphocyte is independent of the position of the site in the hemagglutinin translation product. J. Exp. Med. 174:733-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamal, S. M., J. Fehr, B. Roesler, T. Peters, and J. W. Rasenack. 2002. Peginterferon alone or with ribavirin enhances HCV-specific CD4 T-helper 1 responses in patients with chronic hepatitis C. Gastroenterology 123:1070-1083. [DOI] [PubMed] [Google Scholar]
- 20.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komurian-Pradel, F., G. Paranhos-Baccala, M. Sodoyer, P. Chevallier, B. Mandrand, V. Lotteau, and P. Andre. 2001. Quantitation of HCV RNA using real-time PCR and fluorimetry. J. Virol. Methods 95:111-119. [DOI] [PubMed] [Google Scholar]
- 21a.Komurian-Pradel, F. Hepatology, in press.
- 22.Li, J. S., S. P. Tong, L. Vitvitski, D. Lepot, and C. Trepo. 1991. Two French genotypes of hepatitis C virus: homology of the predominant genotype with the prototype American strain. Gene 105:167-172. [DOI] [PubMed] [Google Scholar]
- 23.MacDonald, A. J., M. Duffy, M. T. Brady, S. McKirnan, W. Hall, J. Hegarty, M. Curry, and K. H. G. Mills. 2002. CD4 T helper type 1 and regulatory T cells induced against the same epitopes on the core protein in Hepatitis C virus-infected persons. J. Infect. Dis. 185:720-727. [DOI] [PubMed] [Google Scholar]
- 24.Marusawa, H., M. Hijikata, T. Chiba, and K. Shimotohno. 1999. Hepatitis C virus core protein inhibits Fas- and tumor necrosis factor alpha-mediated apoptosis via NF-κB activation. J. Virol. 73:4713-4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayrand, S. M., and W. R. Green. 1998. Non-traditionally derived CTL epitopes: exceptions that prove the rules? Immunol. Today 19:551-556. [DOI] [PubMed] [Google Scholar]
- 26.Mayrand, S. M., D. A. Schwarz, and W. R. Green. 1998. An alternative translational reading frame encodes an immunodominant retroviral CTL determinant expressed by an immunodeficiency-causing retrovirus. J. Immunol. 160:39-50. [PubMed] [Google Scholar]
- 27.McGuirk, P., C. McCann, and K. H. Mills. 2002. Pathogen-specific T regulatory 1 cells induced in the respiratory tract by a bacterial molecule that stimulates interleukin 10 production by dendritic cells: a novel strategy for evasion of protective T helper type 1 responses by Bordetella pertussis. J. Exp. Med. 195:221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McNeil, A. C., W. L. Shupert, C. A. Iyasere, C. W. Hallahan, J. A. Mican, R. T. Davey, Jr., and M. Connors. 2001. High-level HIV-1 viremia suppresses viral antigen-specific CD4(+) T cell proliferation. Proc. Natl. Acad. Sci. USA 98:13878-13883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Missale, G., R. Bertoni, V. Lamonaca, A. Valli, M. Massari, C. Mori, M. G. Rumi, M. Houghton, F. Fiaccadori, and C. Ferrari. 1996. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J. Clin. Investig. 98:706-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montagnier, L. 1999. Human immunodeficiency viruses (Retroviridae), p. 763-774. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology, 2nd ed., vol. 2. Academic Press, New York, N.Y. [Google Scholar]
- 31.Nasoff, M. S., S. L. Zebedee, G. Inchauspe, and A. M. Prince. 1991. Identification of an immunodominant epitope within the capsid protein of hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:5462-5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer, B. E., E. Boritz, N. Blyveis, and C. C. Wilson. 2002. Discordance between frequency of human immunodeficiency virus type 1 (HIV-1)-specific gamma interferon-producing CD4+ T cells and HIV-1-specific lymphoproliferation in HIV-1-infected subjects with active viral replication. J. Virol. 76:5925-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prezzi, C., M. A. Casciaro, V. Francavilla, E. Schiaffella, L. Finocchi, L. V. Chircu, G. Bruno, A. Sette, S. Abrignani, and V. Barnaba. 2001. Virus-specific CD8+ T cells with type 1 or type 2 cytokine profile are related to different disease activity in chronic hepatitis C virus infection. Eur. J. Immunol. 31:894-906. [DOI] [PubMed] [Google Scholar]
- 34.Sarobe, P., J. J. Lasarte, N. Casares, A. Lopez-Diaz de Cerio, E. Baixeras, P. Labarga, N. Garcia, F. Borras-Cuesta, and J. Prieto. 2002. Abnormal priming of CD4+ T cells by dendritic cells expressing hepatitis C virus core and E1 proteins. J. Virol. 76:5062-5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmittel, A., U. Keilholz, S. Bauer, U. Kuhne, S. Stevanovic, E. Thiel, and C. Scheibenbogen. 2001. Application of the IFN-gamma ELISPOT assay to quantify T cell responses against proteins. J. Immunol. Methods 247:17-24. [DOI] [PubMed] [Google Scholar]
- 36.Seeff, L. B. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35-S46. [DOI] [PubMed] [Google Scholar]
- 37.Soguero, C., M. Joo, K. A. Chianese-Bullock, D. T. Nguyen, K. Tung, and Y. S. Hahn. 2002. Hepatitis C virus core protein leads to immune suppression and liver damage in a transgenic murine model. J. Virol. 76:9345-9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. C. Lai. 1999. Inhibition of the interferon-inducible kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 39.Ulsenheimer, A., J. T. Gerlach, N. H. Gruener, M. C. Jung, C. A. Schirren, W. Schraut, R. Zachoval, G. R. Pape, and H. M. Diepolder. 2003. Detection of functionally altered hepatitis C virus-specific CD4 T cells in acute and chronic hepatitis C. Hepatology 37:1189-1198. [DOI] [PubMed] [Google Scholar]
- 40.van Eyll, O., and T. Michiels. 2002. Non-AUG-initiated internal translation of the L* protein of Theiler's virus and importance of this protein for viral persistence. J. Virol. 76:10665-10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varaklioti, A., N. Vassilaki, U. Georgopoulou, and P. Mavromara. 2002. Alternate translation occurs within the core coding region of the hepatitis C viral genome. J. Biol. Chem. 277:17713-17721. [DOI] [PubMed] [Google Scholar]
- 42.Walewski, J. L., T. R. Keller, D. D. Stump, and A. D. Branch. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7:710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wedemeyer, H., X. S. He, M. Nascimbeni, A. R. Davis, H. B. Greenberg, J. H. Hoofnagle, T. J. Liang, H. Alter, and B. Rehermann. 2002. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J. Immunol. 169:3447-3458. [DOI] [PubMed] [Google Scholar]
- 44.Xu, Z., J. Choi, T. S. Yen, W. Lu, A. Strohecker, S. Govindarajan, D. Chien, M. J. Selby, and J. Ou. 2001. Synthesis of a novel hepatitis C virus protein by ribosomal frameshift. EMBO J. 20:3840-3848. [DOI] [PMC free article] [PubMed] [Google Scholar]