Abstract
The membrane-proximal region of the ectodomain of the gp41 envelope glycoprotein of human immunodeficiency virus type 1 (HIV-1) is the target of three of the five broadly neutralizing anti-HIV-1 antibodies thus far isolated. We have determined crystal structures of the antigen-binding fragment for one of these antibodies, 2F5, in complex with 7-mer, 11-mer, and 17-mer peptides of the gp41 membrane-proximal region, at 2.0-, 2.1-, and 2.2-Å resolutions, respectively. The structures reveal an extended gp41 conformation, which stretches over 30 Å in length. Contacts are made with five complementarity-determining regions of the antibody as well as with nonpolymorphic regions. Only one exclusive charged face of the gp41 epitope is bound by 2F5, while the nonbound face, which is hydrophobic, may be hidden due to occlusion by other portions of the ectodomain. The structures reveal that the 2F5 antibody is uniquely built to bind to an epitope that is proximal to a membrane surface and in a manner mostly unaffected by large-scale steric hindrance. Biochemical studies with proteoliposomes confirm the importance of lipid membrane and hydrophobic context in the binding of 2F5 as well as in the binding of 4E10, another broadly neutralizing antibody that recognizes the membrane-proximal region of gp41. Based on these structural and biochemical results, immunization strategies for eliciting 2F5- and 4E10-like broadly neutralizing anti-HIV-1 antibodies are proposed.
Of primary concern in recent efforts to develop a vaccine against human immunodeficiency virus type 1 (HIV-1) has been the design of immunogens that have the capacity to elicit potently neutralizing antibodies which are effective against a wide spectrum of HIV-1 strains. Infection by HIV-1 generates many antibodies, but most of these are nonneutralizing (63) or are easily bypassed by minor viral changes (50, 62). Indeed, only five broadly neutralizing HIV-1-reactive monoclonal antibodies have thus far been isolated; two (2G12 and b12) are directed against the HIV-1 exterior gp120 envelope glycoprotein (47, 51, 60, 61), and the other three (2F5, 4E10, and Z13) are directed against the transmembrane gp41 envelope glycoprotein (40, 41, 49, 57, 68). Study of these antibodies has provided insight into which regions of HIV might be vulnerable to the immune system and, in some cases, through what mechanism. The scarcity of these broadly neutralizing antibodies, however, may be a result of the unique characteristics through which they neutralize. Of the antibodies directed against gp120, 2G12 recognizes a carbohydrate epitope on gp120 in a way which requires its domains to be swapped (7), and b12 must bypass conformational masking of the CD4-binding site (28, 54). The anti-gp41 broadly neutralizing antibodies, 2F5, 4E10, and Z13, have also proven scarce, but for reasons that are less clear. Although the three neutralize at different capacities, with 2F5 being the most potent, 4E10 being the most broad, and Z13 being the least potent or broad, the fact that all three bind to the same continuous membrane-proximal region of gp41 (68) suggests that unlike anti-gp120 broadly neutralizing antibodies, which bind to disparate regions of gp120, these antibodies may be less unique or difficult to elicit.
The membrane-proximal region of the gp41 ectodomain spans roughly 30 residues, ending at Lys683 (HXBc2 numbering) immediately upstream of the transmembrane domain, and encompasses the 2F5, 4E10, and Z13 epitopes. This region is one of the most highly conserved sequence stretches in the envelope ectodomain (25, 68). Approximately half of the residues are hydrophobic, with five being tryptophan. Mutational analysis of these tryptophans and of this region in general suggests that it plays a crucial, nonredundant role in membrane fusion (39, 53).
The 2F5 antibody was first isolated from an immortalized B cell in a screen for anti-HIV-1 antibodies, and its core epitope was determined to span residues Glu662 to Ala667 of gp41 (6, 49). Since then, protease protection, phage display, and peptide binding experiments have shown its epitope to be larger (2, 46, 59, 68), with addition of the two leucines flanking the core epitope increasing the affinity of 2F5 by 2,000-fold (59). The affinity of 2F5 for the membrane-proximal region is also enhanced when gp41 is presented in the presence of lipid (21), suggesting that the idiotope of the membrane-proximal region in vivo may depend on the presence of membrane.
The difficulty in eliciting neutralizing antibodies with the use of the membrane-proximal region of gp41 as an immunogen, in a number of different contexts (10, 17, 18, 22, 32, 33, 65), has prompted studies to elucidate its structure (2, 3, 44, 56). These include nuclear magnetic resonance (NMR) studies of the membrane-proximal region (2, 3) and crystal structures of 2F5 alone or in complex with a core 7-mer peptide (5, 44). These studies have generally shown that the core epitope contains a β-turn conformation and have subsequently led to the hypothesis that cyclically constrained immunogens might more accurately mimic the 2F5 epitope and thus might more effectively induce 2F5-like neutralizing antibodies (36, 59). However, immunogenic studies based on the 7-mer core epitope, using cyclically constrained peptides, have thus far failed to elicit broadly neutralizing antibodies (36). Some of the more recent NMR studies (2), furthermore, have shown that free peptides can adopt conformations analogous to that of the 2F5-complexed 7-mer (44) if they are extended beyond the core heptapeptide (2, 3).
To gain a fuller picture of this region of HIV-1, we have solved crystal structures of 2F5 in complex with gp41 peptides corresponding to the 2F5 epitope. Through analysis of these structures, we address not only the conformation of the 2F5-bound gp41 peptide but also two additional constraints that the virus may have evolved to protect this crucially conserved region from neutralization by the immune system: membrane proximity and steric accessibility. We show that 2F5 is uniquely built to bind to an epitope in the vicinity of a hydrophobic surface, perhaps membrane, and in a way that may bypass steric constraints on access to this region. We address through biochemical means the validity of this mechanistic hypothesis and explore the implications of our studies for the design of immunogens that have the capacity to effectively elicit 2F5- and 4E10-like broadly neutralizing anti-HIV-1 antibodies.
MATERIALS AND METHODS
2F5 IgG and Fab production.
Human monoclonal antibody 2F5 was produced by recombinant expression in Chinese hamster ovary cells (27). The 2F5 antigen-binding fragment (Fab) was prepared by reducing 2F5 immunoglobulin G (IgG) (13 mg/ml) in 100 mM dithiothreitol (1 h, 37°C), alkylating in 2 mM iodoacetamide (48 h, 4°C), and then cleaving with endoproteinase Lys-C (0.01 μg/μl; Roche Applied Sciences) in 25 mM Tris Cl and 1 mM EDTA (pH 8.5) for 4 h at 37°C. The cleavage reaction was stopped with 1 mM TLCK (Nα-p-tosyl-l-lysine chloromethyl ketone) and 0.4 mM leupeptin, and the cleavage products were passed over a protein A-Sepharose column (Sigma). Flowthrough fractions were collected and subjected to cation-exchange chromatography (Mono S HR 5/5; Pharmacia-Amersham Biosciences) with a gradient ranging from 0 to 0.5 M NaCl in 50 mM Na acetate, pH 5.0. Peak fractions were combined and further purified by size exclusion chromatography (Superdex 200 HiLoad 26/60; Pharmacia-Amersham Biosciences) in gel filtration buffer (350 mM NaCl, 2.5 mM Tris-SO4 [pH 7.1], 0.02% NaN3).
2F5 Fab-gp41 peptide complex formation.
The peptides gp41662-668 (ELDKWAS), gp41659-671 (ELLELDKWASLWN), and gp41656-674 (NEQELLELDKWASLWNWFD) and N-terminally acetylated and C-terminally amidated peptides gp41661-669 (ac-LELDKWASL-n), gp41660-670 (ac-LLELDKWASLW-n), and gp41654-670 (ac-EKNEQELLELDKWASLW-n) were obtained through the National Institute of Allergy and Infectious Diseases (NIAID) peptide facility (J. Lukszo, Princeton Biomolecules, and American Peptide). Each lyophilized peptide was resuspended in 30% acetonitrile, combined with purified 2F5 Fab to a molar ratio of 3:1, placed in 0.5× gel filtration buffer (gel filtration buffer diluted 50% with water), and concentrated with a Centricon-10 instrument (Amicon) to between 5 and 10 mg/ml.
2F5 Fab-gp41 peptide complex crystallization, data collection, structure determination, and refinement.
All crystals were grown by hanging-drop vapor diffusion. Crystallization droplets were set up with 0.5 μl of protein (in 0.5× gel filtration buffer) and 0.35 μl of reservoir solution. Crystals of 2F5 complexed with 13-mer were grown with a reservoir solution of Hampton Crystal Screen reagent 18, composed of 0.2 M Mg acetate, 0.1 M Na cacodylate (pH 6.5), and 20% polyethylene glycol 8000 (PEG 8000). Crystals of 2F5 Fab complexed with 7-mer, 11-mer, or 17-mer were grown with a reservoir solution composed primarily of Hampton Crystal Screen reagent 40. Microseeding (58) and slight alterations in the concentration of the reservoir salt enhanced crystallization and increased reproducibility. For example, with the 2F5 Fab-17-mer complex, the reservoir solution was composed of 20% PEG 4000, 20% isopropanol, and 125 mM Na citrate (pH 5.6). After crystallization droplets were set up, additional NaCl was added to the reservoir to bring the final reservoir NaCl concentration to 100 mM, and crystals were obtained by heterologous cross-seeding with crystals of the 2F5-11-mer complex.
Before data were collected at 100 K, crystals were placed for a few minutes in a stabilizing cryoprotectant solution composed of the reservoir solution supplemented with PEG 4000 and ethylene glycol to final concentrations of 30% each. Flash cooling was aided by Paratone-N (29). X-ray data were collected with 1.000-Å radiation at the Advanced Photon Source, SER-CAT beamline 22, Argonne National Laboratory. Data were processed with HKL2000 (43).
The structure of the 2F5-gp41662-668 7-mer complex was solved by using molecular replacement with the program AmoRe as implemented in the CCP4 package (11). Molecular replacement searches with 10- to 4-Å data were carried out by using Fab molecules with a variety of different elbow angles until one (PDB accession code 1RZG [23]) gave a highly significant translation peak. CNS (4) was used for structure refinement, employing simulated torsion angle annealing, standard positional minimization, automated water picking and deletion, low-resolution solvent modeling, and individual restrained isotropic B-value refinement, all aided by interactive model rebuilding in O (24) and geometric analysis with Procheck (31) as implemented in CCP4 (11). The 2F5-gp41660-670 11-mer and 2F5-gp41654-670 17-mer complexes were virtually isomorphous to the 7-mer complex and could be solved by rigid-body refinement. Refinements for the 2F5-11-mer and 2F5-17-mer complexes were carried out as described for the 7-mer.
Sequencing of 2F5.
The published amino acid sequence of 2F5 (5, 26, 44) was confirmed by mass spectroscopy of trypsin-digested 2F5 fragments, which had been purified by reverse-phase chromatography. Peptides with masses that did not agree with the published sequence were characterized by Edman sequencing. In the light chain, one tryptic peptide had an anomalous mass of 1,798.01 Da. This peptide was isolated by high-pressure liquid chromatography and sequenced as SGTASVVCLLNNFYPR, producing a calculated mass of 1,798.038 Da. It differed from the published sequence, SGTASWCLLNNFYPR, by the substitution of two Val residues for one Trp residue. In the heavy chain, one tryptic peptide had a mass difference, being 30 Da heavier than the published sequence. Edman sequencing produced a sequence, GPVNAMDVWGQITVTISSTSTK, which contained an Ala-to-Thr change, accounting for the observed mass difference.
Construction of Env glycoprotein expression plasmids.
A codon-optimized construct of the JRFL gp160 envelope glycoprotein was obtained from the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, National Institutes of Health [NIH]). To generate JRFL gp145 cleavage-minus, C9-tagged protein, the plasmid was first modified to encode cleavage mutants by replacing two Arg residues at the gp120-gp41 cleavage junction with Ser residues (REKR to SEKS at positions 508 and 511). Next, the cleavage-minus mutant, along with the entire transmembrane domain, the first five residues of the cytoplasmic tail, and a C9 tag (TETSQVAPA), were cloned into the CMV/R vector (66). QuikChange (Stratagene) was used to create expression plasmids encoding variants of the 2F5-4E10 membrane-proximal region: 2F5-4E10 (656NEQELLELDKWASLWNWFDITNWLWYIK683-TM-C9 tag), HA-2F5-4E10 (HA-656NEQELLELDKWASLWNWFDITNWLWYIK683-TM-C9 tag.), 2F5-HA-4E10 (656NEQELLELDKWASL-HA-WNWFDITNWLWYIK683-TM-C9 tag), and 2F5-4E10-HA (656NEQELLELDKWASLWNWFDITNWLW-HA-YIK683-TM-C9 tag). HA refers to the hemagglutinin sequence, YPYDVPDYA, which is recognized by the anti-HA antibody sc-7392 (Santa Cruz). The sequences of all constructs were confirmed by DNA sequencing.
Production of HIV-1 EnvPLs.
Expression plasmids were transfected into 293 cells by the calcium phosphate method (Invitrogen). Forty-eight hours after transfection, cells were harvested at 4°C with phosphate-buffered saline (PBS) containing 5 mM EDTA, washed once with PBS without EDTA, and then used for production of envelope glycoprotein proteoliposomes (EnvPLs), as previously described (1, 21). CHAPSO was used for lysis, and 10% glycerol was used in the dialysis buffer. The lipids used were 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, and1,2-dioleoyl-sn-glycero-3-phosphate (Avanti Polar) at a molar ratio of 6:3:1. The presence of the reconstituted lipid membrane was confirmed by staining with R-phycoerythrin-streptavidin (Caltag) and fluorescence-activated flow cytometry. To produce EnvPLs without lipid membrane, EnvPLs were washed extensively with the detergent CHAPSO and then with PBS.
Flow cytometry analysis of antibody binding.
Anti-HA antibody sc-7392 (Santa Cruz), antibody b12 (D. Burton), and the broadly neutralizing gp41 antibodies 2F5 and 4E10 (H. Katinger) were conjugated with phycoerythrin (M. Roederer, http://www.drmr.com/abcon/) (52). Proteoliposomes (PLs) (106 beads) were separately stained with each of the four phycoerythrin-conjugated antibodies at concentrations ranging from 2 to 200 μg/ml, in a final volume of 100 μl of flow cytometry buffer (PBS containing 3% fetal bovine serum and 0.02% NaN3). All PLs were incubated for 20 min at 4°C, washed twice with flow cytometry buffer, and then analyzed with a FACScan flow cytometer with CellQuest software (Becton Dickinson).
Virus neutralization assays.
Virus neutralization data were derived by using CD4 T cells as target cells and flow cytometry to enumerate the number of infected cells after a single round of virus infection (34). Briefly, 40 μl of virus stock was incubated with 10 μl of antibody. After incubation for 30 min at 37°C, 20 μl of mitogen-stimulated CD4 T cells (1.5 × 105 cells) was added to each well. The multiplicity of infection was approximately 0.1. Cells were maintained in interleukin-2 culture medium containing 1 μM indinavir and were fed on day 1 with 150 μl of interleukin-2 culture medium. On day 2 after infection, cells were stained for intracellular p24 antigen by using the Beckman Coulter KC57 anti-p24 antibody, followed by quantification of HIV-1-infected cells by flow cytometry. The percentage of neutralization was defined as the reduction in the number of p24-positive cells in antibody-containing wells compared with the number in wells incubated with mock antibody. Virus isolate JRFL was obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Structural analysis and figure preparation.
Structural comparisons were made with the CCP4 program lsqkab (11). Interactive surfaces were analyzed with MS (14), HBPLUS (35), and Grasp (42). Figures were made with Grasp (42), Xtalview (37), and Raster3D (38).
Data deposition.
Coordinates for 2F5 complexed to 7-mer, 11-mer, and 17-mer gp41 peptides have been deposited with the Protein Data Bank under PDB accession codes 1TJG, 1TJH, and 1TJI, respectively.
RESULTS AND DISCUSSION
Crystallization strategy.
To obtain a structure of 2F5 with its complete gp41 epitope, we adopted an iterative crystallization procedure that allowed us to extend stepwise from the core epitope to encompass the entire 2F5 epitope. We began by trying to reproduce the published 2F5 crystals (5), both alone or in complex with a 7-mer peptide. Perhaps because our 2F5 contained slightly different C termini (produced by endoproteinase Lys-C digestion of alkylated and reduced IgG as opposed to natural digestion), we were unable to reproduce the previous crystals. We used the Hampton Crystal Screen to find alternative crystallization conditions. We were able to grow needles with Hampton Crystal Screen reagent 13 (PEG 400) and to grow diamond plates with reagent 40 (PEG 4000 and isopropanol). The needles did not diffract, whereas the diamond plates diffracted to Bragg spacings of better than 2 Å and turned out to have a lattice similar to that described by Bryson et al. (5) for their 2F5-7-mer complex. Structure solution and refinement yielded an Rcrystal of 19.87% and an Rfree of 22.58% (Table 1). Similar screening was carried out with peptides extending from the core 7-mer an additional three residues on either flank (gp41659-671; 13-mer) and an additional six residues on either flank (gp41656-674; 19-mer). No crystals were obtained with the 2F5-19-mer complex. Screening with the 2F5-13-mer complex produced crystals with Hampton Crystal Screen reagent 18 (PEG 8000). These crystals were small needles with 622 Laue symmetry and diffracted to only 4 Å.
TABLE 1.
Crystallographic data and refinement statistics
| Parameter (unit) | Valuea for:
|
||
|---|---|---|---|
| 2F5 Fab + 17-mer-gp41654-670 | 2F5 Fab + 11-mer-gp41660-670 | 2F5 Fab + 7-mer-gp41662-668 | |
| Crystallographic data | |||
| Space group | P212121 | P212121 | P212121 |
| Unit cell dimensions (Å) | |||
| a | 58.66 | 58.29 | 57.86 |
| b | 63.68 | 63.52 | 64.02 |
| c | 178.38 | 178.94 | 179.41 |
| Resolution (Å) | 50-2.20 (2.28-2.20) | 50-2.10 (2.18-2.10) | 20-2.00 (2.07-2.00) |
| Reflections (no.) | 204,793 | 254,046 | 175,644 |
| Unique reflections (no.) | 32,958 (2,538) | 38,349 (2,808) | 43,466 (4,039) |
| Completeness (%) | 96.0 (75.5) | 96.4 (72.4) | 97.1 (92.1) |
| I/σ | 14.5 (1.95) | 19.4 (1.57) | 12.5 (1.47) |
| Rsym (%)b | 12.6 (49.4) | 8.4 (45.1) | 8.9 (53.0) |
| Refinement statistics (F > 0) | |||
| Nonhydrogen atoms (no.) | |||
| Total | 4,156 | 3,991 | 3,915 |
| Peptide | 125 | 100 | 60 |
| Solvent | 610 | 470 | 434 |
| Resolution (Å) | 20-2.20 | 20-2.10 | 20-2.00 |
| Rcrystal (%)c | 18.12 | 20.05 | 19.87 |
| Rfree (%)cd | 22.22 | 23.33 | 22.58 |
| RMSD bond length (Å) | 0.0058 | 0.0057 | 0.0058 |
| RMSD bond angles (°) | 1.320 | 1.374 | 1.333 |
| Avg B factor (Å2) | |||
| Protein | 31.8 | 39.0 | 29.7 |
| Solvent | 63.6 | 54.8 | 46.1 |
Data in parentheses are for the highest-resolution shell.
Rsym = Σ|Iobs − Iavg|/ΣIavg.
R = Σhkl||Fobs| − |Fcalc||/Σhkl|Fobs|.
Test set comprised 10% of reflections.
To overcome the sensitivity of complex crystallization to extensions on the core peptide, we proceeded in smaller steps. We extended the 7-mer peptide by one residue on either flank (gp41661-669; 9-mer) or by two residues (gp41660-670; 11-mer) and tested the 2F5 complexes for crystallization. Diamond plates of the 2F5-11-mer complex could be grown from Hampton Crystal Screen reagent 40 and seeding with the 2F5-7-mer crystals. These 2F5-11-mer crystals diffracted to 2.1 Å, and structure solution and refinement yielded an Rcrystal of 20.05% and an Rfree of 23.33% (Table 1). Analysis of the 2F5-11-mer structure showed lattice interactions at the 11-mer C terminus but room to accommodate a longer N terminus. We exploited this information with a 17-mer, gp41654-670, which added six residues to the N terminus but left the C terminus intact. This 17-mer encompasses the entire 2F5 epitope, as defined by phage display and protease protection (46, 68), with additional N-terminal residues.
Crystallization of the 2F5-17-mer complex produced diamond shaped crystals with Hampton Crystal Screen reagent 40 after cross-seeding with the 2F5-11-mer crystals. These crystals diffracted to 2.2 Å and were virtually isomorphous with both the 2F5-7-mer and 2F5-11-mer crystals. Structure solution and refinement of the 2F5-17-mer yielded an Rcrystal of 18.12% and an Rfree of 22.22% (Table 1), with root-mean square deviations (RMSDs) from ideality on bonds of 0.0058 Å and 87.6% of the residues in the most favored Ramachandran angles. The two Ramachandran outliers, AlaL51 and ThrL30, are typically observed as such in antibodies and occur in well-ordered turns. These outliers were also observed in both the 2F5-7-mer and 2F5-11-mer complexes. Because the 17-mer peptide, gp41654-670 (ac-EKNEQELLELDKWASLW-n), encompassed both the shorter 7-mer and 11-mer peptides, we placed it at the focus of our structural analysis.
Overall structure of the 2F5-gp41 complex.
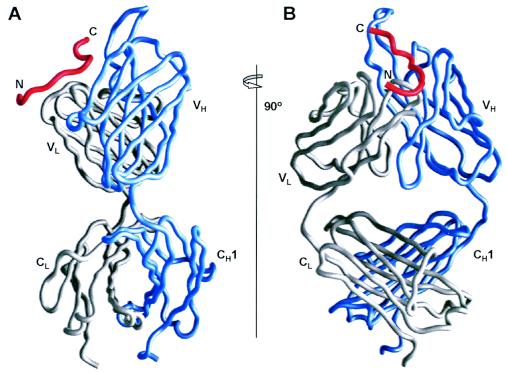
The overall structure of the 2F5 Fab complexed to a 17-mer gp41 epitope is shown in Fig. 1. Main-chain electron density was observed for all 451 residues of the Fab. Density for the heavy chain extended to LysH218 (Kabat IgG numbering) in the hinge region, the predicted cleavage site of the endoproteinase Lys-C used in the Fab preparation. Density for the light chain extended all the way to CysL214, the final light-chain residue. Because reduction and alkylation were performed on the interchain disulfides during preparation of the Fab, CysL214 of the light chain and also CysH216 of the heavy chain were modified by acetamide groups, and these chemical modifications could be observed in the electron density. All six complementarity-determining region (CDR) loops were well ordered, with an average B factor of 26.4 Å2, slightly lower than the average B factor of 31.3 Å2 for the entire Fab. Residue GlyH100c of the 2F5 CDR H3 loop makes a hydrophobic lattice contact with ArgL18 of a symmetry mate, while peptide contacts at the base of the CDR H3 appear to be predominantly responsible for ordering the loop, which is disordered when not bound to the epitope (5, 44). The relative orientation (elbow angle) between the variable and constant domains was 150.3°.
FIG. 1.
Overall structure of the 2F5-Fab-gp41654-670 17-mer complex. (A) Cα traces of the 2F5 Fab heavy chain (blue) and light chain (gray) and of the gp41 peptide (red) from residue Glu657 to Trp670; (B) 90° view of the same traces.
Because we pursued a strategy to visualize the entire 2F5 epitope, the peptide was extended until the N terminus was no longer constrained by interaction with 2F5. Thus, in contrast to the entirely ordered Fab, the first three residues of the gp41 peptide, Glu654, Lys655, and Asn656, could not be discerned, while the next two residues, Glu657 and Gln658, were marginally ordered. Starting at Glu659, the electron density improved, and from Leu660 all the way to the C terminus of the peptide at Trp670, the structure was clearly defined (Fig. 2C). This improvement in order was reflected in the B factors, with Glu657 and Gln658 exhibiting an average B factor of 120.20 Å2, while residues Glu659 to Trp670 had an average B factor of 35.39 Å2, similar to the average B value of the Fab.
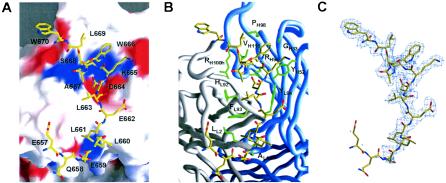
FIG. 2.
Contact interface between gp41 and 2F5. The orientation shown is similar to that in Fig. 1B, with the CDR H3 protruding off the top of the figure. (A) Electrostatic potential. An atomic bond representation of the peptide (yellow, carbon; red, oxygen; blue, nitrogen) is shown, with gp41 residues labeled. The molecular surface of 2F5 is colored by electrostatic potential: red for electronegative, blue for electropositive, and white for apolar (42). Extensive hydrogen bonds and salt bridges are observed between the peptide and antibody, including contacts with the CDRs of both 2F5 chains, as well as contacts with nonpolymorphic regions of the 2F5 light-chain N terminus. (B) Hydrophobic interactions. A representation of the peptide similar to that for panel A is shown, but in this case it is shown against the Cα worm of the 2F5 heavy (blue) and light (gray) chains, with side chains labeled and shown in green for 2F5 residues that form hydrophobic contacts with gp41. (C) Electron density of the gp41 peptide. Shown (blue) is the electron density (2Fo − Fc) around the gp41 peptide contoured at 1σ. The electron density around residues Glu657 and Asn658 cannot be seen at this contour level, consistent with the tenuous nature of their contacts with 2F5. Beginning with residue Glu659, the density improves and is maintained through Trp670.
2F5-bound conformation of gp41.
The gp41654-670 peptide perches in relatively extended conformation at the CDR junction between the heavy and light chains (Fig. 1 and 2). The peptide spans roughly 25 Å from the Cα of residue Glu659 to the Cα of Trp670, while the maximal side chain distance extends over 30 Å. Progressing from the N terminus, the peptide makes tenuous contact with the CDR L1, bends roughly 90° while passing over and contacting the N terminus of the light chain, and then proceeds in a roughly linear manner 15 Å, past the CDR L3 and H2, with the final seven residues contacting the 22-amino-acid-long CDR H3.
The overall extended conformation of the peptide is reflected in its generally β-type Ramachandran angles. Because the peptide N terminus is poorly ordered (Fig. 2C), it may adopt an alternative conformation in the context of the rest of the envelope ectodomain. Nonetheless, we note that the initial bend occurs at a modified gamma-like turn. The carbonyl oxygen of Gln658 is outside hydrogen bonding distance (4.9 Å) from the backbone amide of Leu661; still, this bend in the peptide allows hydrophobic interactions to occur between the side chains of Glu659 and Leu661. Within the 14 ordered residues of the peptide, other than the modified gamma-like turn, only two other turns are observed, both of which are type I β-turns: one between residues Asp664 and Ala667 and the other between residues Trp666 and Leu669. These overlapping turns reverse directional changes, roughly canceling each other so that the overall path of the peptide remains essentially straight (Fig. 1 and 2). Only three intrapeptide hydrogen bonds occur, one between the backbone amide of Trp666 and the side chain carboxylic acid of Asp664, one between the backbone amide of Ala667 and the carbonyl oxygen of Asp664, and one between the backbone amide of Leu669 and the carbonyl oxygen of Trp666. These intrapeptide hydrogen bonds constrain the conformations of only six residues (residues 664 to 669), suggesting that when excised from the rest of the HIV-1 ectodomain, the conformation of a free gp41 peptide would not be strongly constrained into the conformation recognized by 2F5.
Contact interface between gp41 and 2F5.
The overall surface area of 2F5 that is buried by gp41 in the structure is 634.7 Å2, an increase of more than 50% over the area buried by the 7-mer core epitope (418.8 Å2 is buried in the 2F5-7-mer structure). Conversely, the surface area on gp41 that is buried in the interaction with 2F5 is 563.4 Å2 (versus 377.7 Å2 in the 2F5-7-mer structure). This falls into the range that is typical for protein-antibody interactions (15). When analyzed according to the chemical nature of the interaction, 43.6% of the interactive surface on the peptide involves nonhydrophobic residues: 28.8% acidic, 11.3% basic, and 3.5% polar. Similarly, many of the residues on the antibody that are buried by gp41 are polar or charged, with 30.7% of the interactive surface contributed by polar residues, 27.6% by basic residues, and 4.8% by acidic residues, with the remaining 36.8% contributed by hydrophobic residues (Table 2). When the surfaces of the antibody and the gp41 peptide are colored by electrostatic potential (Fig. 2A and 3E), their comparison reveals a general complementarity of charge throughout the contact interface.
TABLE 2.
Buried surface areas on gp41 and 2F5, by residue
| gp41 residue | Buried surface area (Å2) | 2F5 residue | Buried surface area (Å2) | |
|---|---|---|---|---|
| Glu657 | 16.77 | |||
| Gln658 | 19.80 | |||
| Glu659 | 24.26 | |||
| Leu660 | 0 | |||
| Leu661 | 68.18 | |||
| Glu662 | 56.52 | |||
| Leu663 | 35.65 | |||
| Asp664 | 64.80 | |||
| Lys665 | 63.63 | |||
| Trp666 | 99.84 | |||
| Ala667 | 27.98 | |||
| Ser668 | 0 | |||
| Leu669 | 51.60 | |||
| Trp670 | 34.40 | |||
| Total | 563.42 |
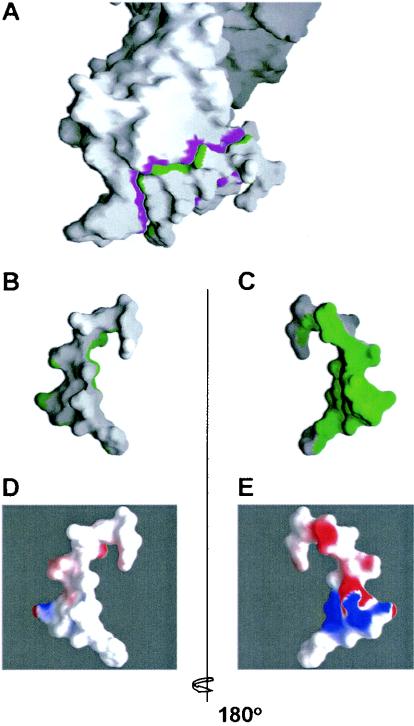
FIG. 3.
2F5 binding surface on gp41. (A) Molecular surface representations of 2F5 complexed to gp41. The orientation shown is similar to that in Fig. 4. Colored in magenta is the surface on 2F5 that is buried by the interaction with gp41, and colored in green is the surface on gp41 that is buried by the interaction with 2F5. (B and C) Close-ups of the peptide. The gp41 peptide molecular surface from panel A is oriented so that Trp670 is at the bottom of the panel and the N terminus of the peptide is at the top; 180° views are shown. Roughly 40% of the surface of the gp41 peptide is buried by 2F5 (green), while the remaining surface remains hidden (white). (D and E) When the molecular surface of the peptide is colored by electrostatic potential, it becomes apparent that the surface that is bound by 2F5 is charged (E), while the surface that is hidden from 2F5 is hydrophobic (D). The electrostatic potential shown is colored at the same potential contour as in Fig. 2A, with red for electronegative, blue for electropositive, and white for apolar (42).
All of the residues of the gp41 peptide between Gln657 and Trp670, with the exception of Leu660 and Ser668, interact directly with the antibody, either through hydrogen bonds and salt bridges or through hydrophobic interactions (Tables 2 and 3). These interactions confirm phage display, protease protection, and peptide affinity assays (2, 46, 59, 68) which show that the 2F5 epitope is larger than the originally defined core heptapeptide. At the peptide N terminus, the contacts for the relatively disordered Glu657 and Gln658 are tenuous, but beginning at Glu659, the contact surface becomes well defined, in concordance with peptide affinity assays which show that the presence of Glu659 enhances the affinity of 2F5 by sixfold (2). Unexpectedly, the contact interface between 2F5 and gp41 includes residues not only within the variable CDRs of 2F5 but also within nonpolymorphic regions, namely, residues of the N terminus of the light chain. A salt bridge is observed between the side chain carboxylic acid of Glu659 and the positively charged light-chain amino terminus, while hydrophobic contacts are observed between the side chain of Leu661 and AlaL1 and LeuL2. These interactions confirm results of binding studies which show that alteration of the 2F5 light-chain N terminus can ablate 2F5's interaction with gp41 (M. Zwick, personal communication), as well as peptide affinity assays which show that extension of the core peptide to Glu659 enhances 2F5 affinity (2). Leu661, which is also critical for optimal 2F5 binding (59), not only forms interactions with the nonpolymorphic N terminus of the 2F5 light chain but also forms a hydrophobic contact with the side chain of CDR L3 PheL93.
TABLE 3.
Hydrogen bonds and salt bridges between gp41 and 2F5
| gp41 residue | 2F5 residue | Distance (Å) |
|---|---|---|
| Gln658O | GlnL27Nɛ2 | 3.18 |
| Glu659Oɛ2 | AlaL1N | 2.88 |
| Glu662O | TyrL94N | 2.78 |
| Glu662Oɛ2 | ArgH58NH2 | 2.76 |
| Glu662Oɛ1 | ArgH58Nɛ | 2.75 |
| Asp664Oδ1 | HisL96Nɛ2 | 2.88 |
| Asp664Oδ1 | ArgH95NH1 | 2.83 |
| Asp664Oδ2 | ArgH95NH2 | 2.85 |
| Asp664N | HisL92O | 2.83 |
| Lys665N | TyrL94OH | 3.32 |
| Lys665Nζ | AspH54Oδ1 | 2.75 |
| Lys665Nζ | AspH56Oδ1 | 2.88 |
| Trp670O | ArgH100hN | 2.89 |
Proceeding further from the peptide N terminus, the gp41 residues Asp664, Lys665, and Trp666, which lie at the core of the 2F5 epitope and have been shown to be essential for 2F5 binding (13, 41, 48), account for over 40% of the surface area that is buried by 2F5 and for almost half of the hydrogen bonds between gp41 and 2F5 (Tables 2 and 3). The side chain of Asp664 makes hydrogen bonds with the guanidinium atoms of CDR H3 ArgH95 and the imidazole side chain of CDR L3 HisL96, while the backbone amide of Asp664 hydrogen bonds with the carbonyl oxygen of CDR L3 HisL92. These polar interactions are enhanced by hydrophobic interactions between the Cβ of Asp664 and the main chain of CDR L3 HisL92, as well as between the main-chain carbons of Asp664 and the aromatic ring of CDR L3 TyrL94 (Fig. 2B). Lys665 makes hydrogen bonds through its backbone amide and the ring hydroxyl of TyrL94, as well as between its side chain amine and the carboxylic acids of CDR H2 AspH54 and AspH56. The aliphatic base of Lys665 is stabilized by packing against the aromatic ring of CDR H2 TyrH52 (Fig. 2B). Residue Trp666, out of all of the gp41 residues, makes the most extensive interactions with 2F5, in terms of buried surface area (Table 2). Its ring carbons make hydrophobic contacts with CDR H1 GlyH33, as well as CDR H3 ArgH95, ProH98, and ValH100k. Taken together, these interactions confirm not only why mutation of these three core gp41 residues ablates 2F5 binding but also why mutations in 2F5 residues (HisL92, TyrL94, and PheH32) affect binding (67).
Proceeding further along gp41 toward the 2F5 epitope C terminus, the remaining contacts are made predominantly with the CDR H3, although only with nonapical residues. The side chain of gp41 Leu669 interacts with CDR H3 ProH98, confirming its importance for 2F5 binding (59), while the carbonyl oxygen of Trp670 hydrogen bonds with the backbone amide of ArgH100h. It is rather surprising that out of the 22 amino acids of the 2F5 CDR H3, only 10 show any interaction with gp41 (Table 2), leaving open the possibility that the 2F5 CDR H3 may have functions outside the realm of direct gp41 binding (see below).
Exclusive 2F5-bound face of gp41.
In general, antibodies that bind to peptides envelop them, whereas antibodies that bind to protein surfaces have much flatter surfaces of interaction (12). In the case of 2F5, out of a total accessible surface area of 1,377.2 Å2 on the peptide (638.7 Å2 for the core epitope from position 661 to 670), a surface of only 563.4 Å2 (377.7 Å2 for the core) is actually buried by the interaction with 2F5 (Fig. 3A to C). Thus, only 41% (59% for the core) of the gp41 epitope is buried by 2F5. Instead of enveloping the antigen, 2F5 only binds the gp41 peptide on one exclusive face (Fig. 3A to C). This mode of interaction is suggestive of an antibody-protein interface in which a non-2F5-bound hidden face is occluded by other portions of the envelope ectodomain.
In order to further address the characteristics of the gp41 peptide surface that is bound by 2F5, as well as the one that is perhaps occluded from 2F5 binding, we analyzed the electrostatic potentials of these surfaces. As shown in Fig. 3D and E, only the 2F5-bound face of gp41 is charged (Fig. 3E), while the face that is hidden or occluded from 2F5 binding is much more hydrophobic (Fig. 3D). The hydrophobicity of this surface provides further support for the possibility that this face of gp41 may be involved in protein contacts which may occlude it from recognition by 2F5. Because antibodies against the hidden hydrophobic face may be preferentially selected due to favorable binding energies at hydrophobic surfaces, this may explain in part why immunizations with nonoccluded peptides have thus far failed to elicit 2F5-like antibodies.
Characterization of the 2F5 CDR H3 loop.
In a survey of human antibodies, the mean length of the CDR H3 loops of antibodies directed against viral antigens was 16.5 residues, which is longer than those of antibodies directed against any other class of antigen (12). The selection bias for these long loops is not entirely clear, but broadly neutralizing anti-HIV-1 antibodies follow this trend as well, with the CDR H3 loop of 2F5 extending 22 amino acids in length. From analysis of the 2F5-17-mer structure it is clear that the interactions between the CDR H3 loop of 2F5 and gp41 occur predominantly at the base of the CDR H3, while the rest of the residues of the CDR H3, namely, at the apex, remain largely unbound to gp41.
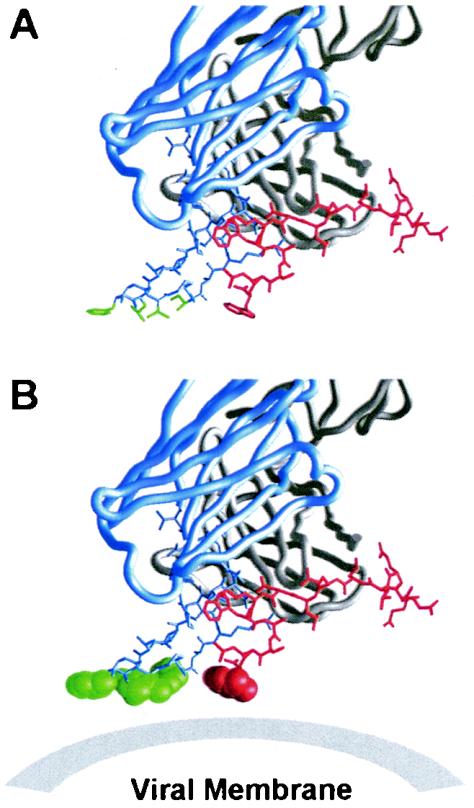
Analysis of the 2F5 CDR H3 loop shows that the apex of the loop (which does not contact gp41) contains a stretch of hydrophobic amino acids, namely, Leu100a, Phe100b, Val100d, and Ile100f (Fig. 4A). Interestingly, the side chains of these residues all face in the same general direction and define a protruding flat hydrophobic surface (Fig. 4B). In some ways, the 2F5 CDR H3 resembles the structure of a foot, with the sole of the foot defining a hydrophobic plane.
FIG. 4.
CDR H3 loop of 2F5. 2F5 is shown with the heavy chain in blue and the light chain in gray. Atomic bond representations are shown for the 2F5 CDR H3 loop (blue rods), and the gp41 peptide (red rods). (A) Side chains of hydrophobic residues at the apex of the CDR H3 loop (LeuH100a, PheH100b, ValH100d, and IleH100f) are colored in green. These residues define a hydrophobic surface which if extended as a plane intersects the indole ring of Trp670. (B) The coplanarity of these hydrophobic side chains is evident when the atoms of the side chains are shown in a space-filling representation. The hydrophobic plane defined by these apical CDR H3 residues may be an adaptation that allows 2F5 to bind at or in close proximity to the viral membrane.
It has been suggested that D3-3 is the D(H) genomic precursor of this region of 2F5 (26), in that 13 out of 15 2F5 CDR H3 nucleotides match with D3-3. If D3-3 is the genomic precursor of this region, then three of the hydrophobic “sole” residues, i.e., Phe100b, Val100d, and Ile100f, would be directly encoded by this D segment. The interspersing proline Pro100e would change during affinity maturation from a D3-3-encoded valine. While valine is compatible with the sole structure, proline with its confining Ramachandran angles serves to restrict conformational flexibility of the loop apex.
Although the hydrophobic apex of the 2F5 CDR H3 does not contact the gp41 peptide directly, the close proximity of the 2F5 epitope to the viral membrane suggests that this hydrophobic surface might interact directly with the viral membrane (Fig. 4B), or at least allow 2F5 to accommodate an epitope that is in close proximity to the membrane, perhaps by binding to membrane and allowing for enhanced scanning for the gp41 epitope. Perhaps relevant to this, we observed the terminal residue of the 2F5 epitope on gp41, Trp670, to be positioned with its hydrophobic indole side chain aligned perfectly with the plane defined by the protruding 2F5 CHR H3 (Fig. 4B).
Conformation of the 2F5 epitope within the context of the entire envelope ectodomain.
The gp41 ectodomain undergoes large conformational changes related to its function as a class I fusion protein (9). Although we define here the structure of the 2F5 epitope, it is important to further define the context of this structure, both overall, with respect to the fusogenic state of gp41, and locally, with respect to neighboring regions.
In terms of overall conformation, 2F5 is believed to bind optimally to a prefusion or intermediate conformation of gp41 (16, 19, 20, 55), although it may also recognize the postfusion six-helix bundle conformation of gp41 (20, 68). The structure of the prefusion viral spike has not been determined, but several structures of the postfusion six-helix bundle have been determined (8, 64). Compared to the six-helix conformation (Fig. 5), which extends to Lys665 of gp41 (64), the RMSD of the overlapping main-chain atoms of residues Glu657 to Lys665 is 3.9 Å2, with numerous steric clashes (Fig. 5). If both the postfusion bundle and the 2F5-bound extended conformation of gp41 were rigidly fixed, these would be incompatible. However, there is considerable flexibility at the overlapping termini of both. The C-terminal region of the six-helix bundle is flexible and susceptible to proteolytic digestion all the way to residue Leu661 (8), whereas our analysis shows that the 2F5-bound extended conformation of gp41 is not constrained by 2F5 until Leu659. Superposition of a truncated HR2 helix (to Leu661) with a truncated 2F5 epitope (from Glu659) leads to an RMSD of 0.3 Å2, which would be compatible with 2F5 binding, although perhaps at lower affinity.
FIG. 5.
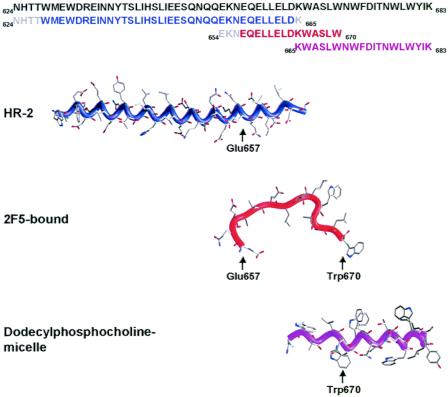
Comparison of the 2F5-bound conformation of gp41 with other gp41 conformations. In blue is the structure of the HR2 helix observed in the structure of the postfusion six-helix bundle (64), in red is the structure of 2F5-bound gp41 presented here, and in magenta is the NMR structure of the membrane-proximal region of gp41 in the context of dodecylphosphocholine micelles (56). The structures are aligned not according to biological context, but relative to the Cα positions of residues Glu657 and Trp670. The helices of both the six-helix bundle and the downstream micellar structure would have to partially unravel for gp41 to adopt the 2F5-bound conformation. The sequences of the three structures are aligned at the top, with structurally ordered residues colored according to their respective structures and the sequences of disordered residues colored gray.
In terms of the downstream region, NMR analysis of residues Lys665 to Lys683, in dodecylphosphocholine micelles (56), shows an entirely helical conformation (Fig. 5). This would be incompatible with the 2F5-bound structure, with an RMSD for the overlapping six residues (residues 665 to 670) of 1.2 Å2. However, when helices form, unless they encounter a helix-breaking residue or alternative structure, the stability gained by the intrachain hydrogen bonds of the helix often allows them to propagate, extending the helical conformation. Our analysis of the 2F5-bound conformation of gp41 suggests that in the absence of the rest of the gp41 ectodomain, it is held in this conformation primarily by interactions with 2F5. Thus, in the absence of 2F5, it is not unexpected that a helix, formed downstream of the 2F5 epitope, might propagate further upstream. Nonetheless, in light of the many direct contacts of 2F5 in the region of residues 661 to 670 (Fig. 2), and because 2F5 can bind to the membrane-proximal region of gp41 in the presence of lipid, when 2F5 is bound, we would expect the region of residues 661 to 670 to adopt the extended conformation that we observe in the 2F5-gp41 crystal structure.
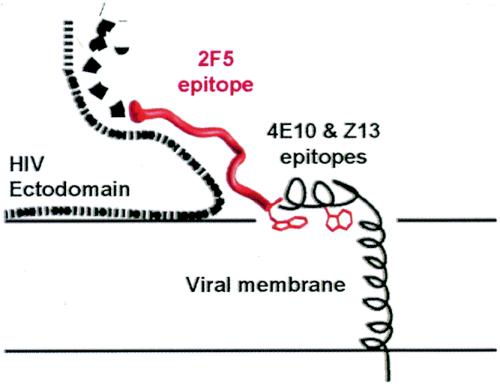
Taken together, the data suggest that the membrane-proximal region at the start of the 2F5 epitope is relatively flexible, perhaps assuming different conformations depending on the fusogenic state of gp41. From the center to the end of the 2F5 epitope (residues 661 to 670), an extended mostly β conformation, containing two overlapping type 1 β-turns, stretches for roughly 25 Å. In the prefusogenic state, this extended structure is presumably stabilized by interactions through its hidden face with the rest of the HIV ectodomain. Then, from residues 670 to 683, a hydrophobic helix, perhaps lying parallel with the viral membrane (56), would complete the ectodomain (Fig. 6).
FIG. 6.
Transmembrane-proximal region of gp41. This schematic shows the 2F5 epitope (red) with its hidden face occluded by the HIV ectodomain. The conformation of the N-terminal adjoining region is not known. The C-terminal adjoining 4E10 and Z13 epitopes are shown as a helix lying parallel to the viral membrane, with Trp670 and Trp678 (red) embedded in the lipid bilayer. Although the 4E10-Z13 epitope is shown for visual clarity as extending away from the rest of the HIV ectodomain, the angle it makes with the 2F5 epitope is unknown, and it is likely to be partially occluded in the viral spike.
Binding of 2F5 to gp41 is enhanced by lipid membrane and hydrophobic context.
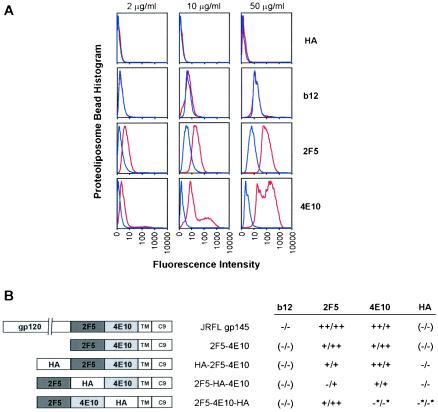
The structural model (Fig. 6) predicts that proper formation of the 2F5 and 4E10 epitopes should be strongly dependent on the presence of membrane. In order to confirm biochemically that an intimate association exists between the membrane-proximal region of gp41 and the viral membrane, we performed binding studies using PLs in which various envelope constructs were presented in either the presence or absence of lipid membrane. As shown in Fig. 7A, when JRFL gp145 was presented on PLs in the presence of lipid, the affinity of both 2F5 and 4E10 increased. In contrast, the presence of lipid had no effect on the affinity of the anti-gp120 antibody b12.
FIG. 7.
Biochemical analysis of 2F5 and 4E10 binding: effect of lipid membrane and hydrophobic context. PLs composed of paramagnetic beads conjugated to the 1D4 antibody were used to capture different envelope constructs through C-terminal C9 tags, which are recognized by 1D4. Each of the envelope constructs retained the native JRFL transmembrane domain (TM), so that envelope-TM-captured PLs incubated with lipids simulated the native TM in a lipid bilayer. Alternatively PLs could be washed extensively with detergent to remove bound lipid and expose the naked TM. In this manner, PLs with and without lipid membrane could be prepared. (A) Flow cytometry analysis of b12, 2F5, and 4E10 binding to PLs with and without lipid membrane. Fluorescently labeled antibodies at 2 μg/ml (left panels), 10 μg/ml (middle panels), and 50 μg/ml (right panels) were incubated with JRFL gp145-captured PLs. Histograms (normalized for 100,000 events) of the flow cytometry-sorted PL fluorescence intensity are shown for PLs with membrane (red) and without membrane (blue). (B) Effect of context on the binding of 2F5 and 4E10. Five different PLs were analyzed, each with the 2F5 and 4E10 epitopes placed in a different context. A schematic of each context is shown, with gp120 referring to the cleavage-minus N-terminal gp120 attached to gp41; 2F5, 4E10, and HA referring to the respective epitopes of 2F5, 4E10, and the antihemagglutinin antibody sc-7392; and TM and C9 referring to the JRFL transmembrane region and 1D4-recognized tag, respectively. Each PL was tested for binding to fluorescently labeled antibodies b12, 2F5, 4E10, and HA, either in the presence of membrane or after extensive detergent washing to remove membrane. The relative fluorescence with and without membrane was tested over an antibody concentration range of 2 to 200 μg/ml. The results of two independent experiments are shown (presented as result for experiment 1/result for experiment 2). ++, 10- to 50-fold-greater fluorescence in the presence of membrane; +, 2- to 10-fold-greater fluorescence; −, 0.5- to 2.0-fold-greater fluorescence. Parentheses indicate that the particular antibody epitope was not present on the construct, and asterisks indicate that the overall level of antibody binding was low, both with and without membrane. As can be seen, not only the presence of lipid membrane but also the surrounding sequence influences optimal 2F5 and 4E10 epitope recognition.
To confirm that antibody binding to membrane-proximal epitopes is not merely inhibited by aggregation or large-scale structural rearrangement of the transmembrane region in the absence of lipid, we interspersed the HA antibody epitope into three different locations of the membrane-proximal region: at the N terminus of the 2F5 epitope, between the 2F5 and 4E10 epitopes, and downstream of the 4E10 epitope (Fig. 7B). With the exception of 2F5-4E10-HA, which displayed lower anti-HA antibody binding regardless of the presence of lipid, the binding of the HA antibody to the constructs was not affected by the presence of lipid (Fig. 7B). Binding of 2F5, however, to HA-2F5-4E10 and 2F5-4E10-HA increased in the presence of lipid, as did binding of 4E10 to HA-2F5-4E10 and 2F5-HA-4E10 (Fig. 7B). These results suggest that the membrane-proximal region of these constructs is accessible even in the absence of membrane and that the lower binding affinity of 2F5 and 4E10 to gp41 after the loss of lipid is not an artifact but rather is an actual effect of the loss of a hydrophobic binding environment.
In addition to the presence of membrane, these studies also suggest that the hydrophobic continuity of the membrane-proximal region is an important factor in optimal 2F5 binding. In construct 2F5-HA-4E10 (Fig. 7B), where an HA tag disrupts the continuity of the membrane-proximal region by being placed between the 2F5 and 4E10 epitopes, the effect of enhanced membrane context 2F5 affinity is decreased. The membrane context effect is recovered, however, when the HA tag is placed downstream of the 4E10 epitope and continuity is restored (Fig. 7B). These results may be due to the fact that when Trp670, which is predicted from our structural analysis to lie along the plane of the viral membrane (Fig. 4), is displaced from the core 2F5 epitope by an HA tag, the 2F5 epitope, and therefore 2F5 itself, is not sufficiently close to the membrane for the hydrophobic CDR H3 of 2F5 to fully enhance binding.
Taken together, these biochemical analyses confirm the structural model that the membrane-proximal region of gp41 is intimately involved with the viral membrane and that the integrity of the 2F5 and 4E10 epitopes is highly membrane- and sequence-context dependent. Because the viral membrane is derived from host lipids, it is seen as self by the immune system. Such an intimate membrane association with the viral envelope may help to obscure humoral immune recognition; the membrane-proximal region would “sit” on the membrane, restricting steric accessibility to otherwise conserved potential neutralization epitopes. In this context we note that an examination of viral envelope class I fusion proteins shows that many of the lentiviruses, which form persistent infections, have similar long hydrophobic membrane-proximal regions after their HR2 helices. In contrast, for nonpersistent infections such as influenza and Ebola virus infections, the membrane-proximal regions are much less hydrophobic, while those of morbillivirus (measles) and rubulavirus (mumps) are far shorter.
Large-scale steric accessibility.
Because 2F5 and 4E10 do not block gp120-receptor interactions, these antibodies may bind after virus-cell surface attachment. But in order to do so, they might face steric barriers related to this crowded interface, as has been observed with CD4i antibodies, where steric constraints preclude IgG binding (30). Thus, we sought to address whether the membrane-proximal region of gp41 is subject to any large-scale steric hinderances.
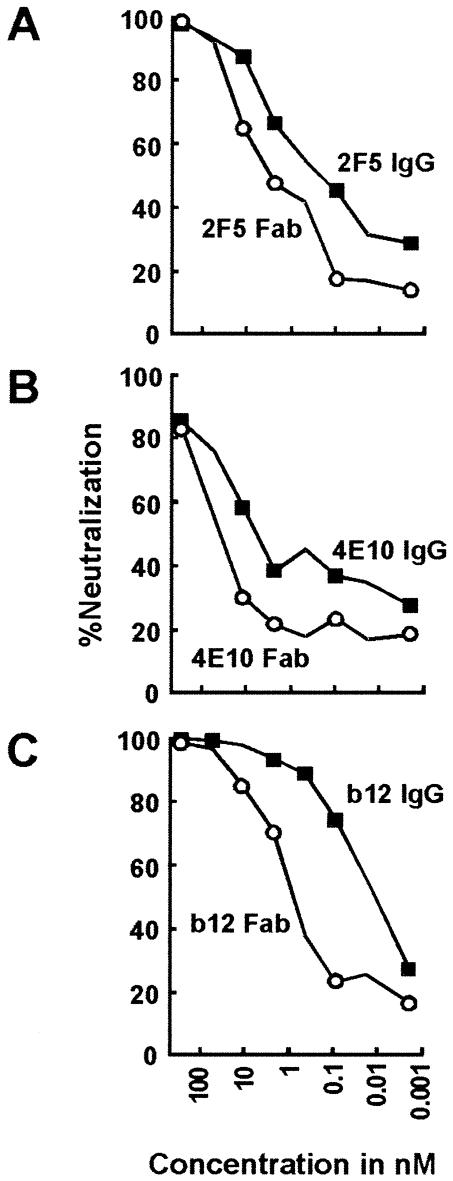
For this purpose, we compared the neutralization capacities of the Fab versus the IgG for the respective antibodies. Since a Fab is approximately one-third the size of an IgG, if large-scale steric clashes that hinder binding occur, then one would expect the Fab to neutralize better than the IgG, as is the case for the CD4i antibodies that bind at the sterically restricted virus-cell interface (30). In general, however, for antibodies that are not sterically restricted, Fabs neutralize less well than the bulkier bivalent IgGs. As shown in Fig. 8, the 2F5 and 4E10 Fabs (Fig. 8A and B), as well as the gp120-reactive b12 Fab (Fig. 8C), were less effective than their respective IgGs in neutralizing the primary HIV-1 strain JRFL, suggesting that 2F5 and 4E10 are not affected by large-scale steric hindrance of IgG access to the membrane-proximal region.
FIG. 8.
Neutralization of HIV-1 isolate JRFL by IgG versus Fab. In order to assess the existence of steric hindrance of access of IgG molecules to membrane-proximal region epitopes, neutralization capacities of IgGs (squares) versus Fabs (circles) were compared for 2F5 (A), 4E10 (B), and b12 (C) as a control. The concentrations of the IgGs and Fabs ranged from 0.002 to 300 nM. Neutralization data were obtained with CD4 T cells as target cells and flow cytometry of anti-p24 antibody-stained cells to assess the number of infected cells after a single round of virus infection. The percentage of neutralization was calculated as the reduction in the number of infected cells compared to the number of infected cells in wells incubated with mock antibody. Similar results were obtained for HIV-1 strain SF162.
Vaccine implications.
The findings of this study suggest that an effective immunization strategy to elicit 2F5- or 4E10-like broadly neutralizing antibodies would likely have to account for viral mechanisms of immune evasion that constrain the membrane-proximal region, namely, conformation, surface occlusion, and membrane proximity, although perhaps not large-scale steric accessibility. The precise conformation that 2F5 recognizes may be difficult to stabilize. Both the upstream six-helix bundle and downstream membrane-bound helix enforce different conformations on the 2F5 epitope. The stabilization of extended structures is also not trivial. Tight turns can be stabilized with designed disulfide or lactam bridges (Fig. 9A), and such approaches are already under way (36, 59); even so, such turns account for less than half of the 2F5 epitope. One critical question will be the degree of flexibility of the 2F5 epitope in the context of the full envelope ectodomain. Does the entire 2F5-bound conformation observed in this structure have to be stabilized, or is only a critical substructure essential for broad immune recognition? Such questions should be answerable by immunizations with structurally constrained antigens. Alternatively, structures of the 2F5 epitope in a more complete ectodomain context, or even in complex with another neutralizing antibody that recognizes the 2F5 epitope, may also provide answers.
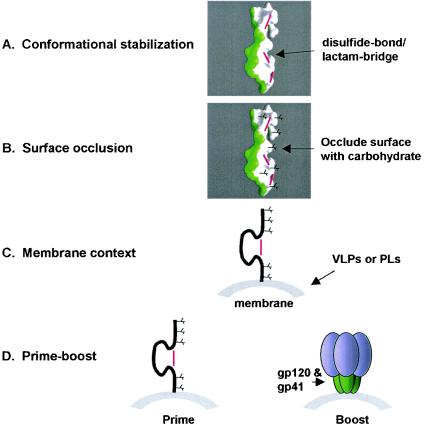
FIG. 9.
Vaccine immunization strategy. Shown is a four-part strategy to elicit 2F5-like antibodies. (A) Conformational stabilization of the 2F5-bound extended conformation of gp41. The molecular surface of a potential immunogen is shown in a color scheme similar to that in Fig. 3B and C, with the surface bound by 2F5 colored in green and the surface hidden from 2F5 colored in white. Disulfide bonds or lactam bridges that stabilize conformation are shown in magenta. (B) Occlusion of the hidden face of gp41. Carbohydrate (black) is shown occluding the hidden hydrophobic surface of gp41 from humoral immune recognition. Due to the size of the epitope, N-linked glycans may be too large to use here, but smaller O-linked glycans may allow more precise masking. (C) Membrane context. To elicit antibodies that are able to accommodate an epitope that is proximal to membrane, one could immunize with a conformationally stabilized, surface-occluded immunogen in the context of membrane, either on virus-like particles (VLPs) or on PLs. (D) Prime-boost. Various prime-boost strategies could be employed to select only those antibodies that are able to overcome accessibility barriers to the membrane-proximal region. Shown here is one example, with the prime consisting of a conformationally stabilized, surface-occluded immunogen presented in the context of membrane. A boost with the complete Env ectodomain should select antibodies that can bind to the native viral spike.
To account for local surface occlusion, immunogens that induce antibodies that only bind to the 2F5-bound surface would need to be designed. This might be accomplished in a manner similar to that tried for anti-gp120 immunogens, for example, by masking the unbound hidden surface of gp41 with carbohydrate modifications (Fig. 9B) (45). O-linked glycolsylation might be preferable in this case due to the smaller size of these glycans, which would interfere less with the 2F5-bound face of the peptide. Alternatively, one could anchor the epitope to a larger molecule or surface in a manner that would leave only the 2F5-bound surface exposed. For example, one could first attach reactive groups on to the hidden face of the gp41 epitope, then bind the 2F5 complex to a nonimmunogenic graphite or plastic surface that reacts with these groups, and then release 2F5. The latter approach not only would eliminate local surface occlusion but also would allow the reactive groups to weld the 2F5-enforced conformation into place.
In terms of membrane proximity, one could present a conformationally stabilized, surface-occluded immunogen in the context of membrane, either on virus-like particles or on PLs (21) (Fig. 9C). The enhanced 2F5 binding that we observed in the context of a PL membrane (Fig. 7) suggests that even in a highly artificial context, the presence of membrane recapitulates essential components of 2F5 and 4E10 recognition.
Elicitation of 2F5-like antibodies with any of these immunogens could be enhanced with prime-boost strategies (Fig. 9D). For example, priming with a conformationally stabilized, surface-occluded, membrane-anchored immunogen may elicit high titers of antibodies, only a small portion of which recognize virus. A boost, on the other hand, composed of the complete wild-type envelope ectodomain and presented in a membrane-anchored context, should select antibodies capable of binding wild-type virus. Such prime-boost strategies could be repeated (for example, with diverse strains of HIV or with additional peptides) to enhance antibody specificity and titer.
These immunization strategies (Fig. 9) should account for the constraints on the conserved membrane-proximal epitope suggested by our mechanistic analysis of the 2F5-gp41 crystal structure. Whether the analysis presented here defines a sufficient road map for elicitation of 2F5- or 4E10-like antibodies will need to be determined experimentally. Our studies on 2F5 and those of others on b12 and 2G12 (7, 54, 68) present a paradigm for using structural information from broadly neutralizing antibodies to understand and overcome HIV-1 mechanisms of immune evasion.
ResidueBuried surfaceAlaL130.10 LeuL214.96 SerL263.59 GlnL2745.18 LeuL9111.77 HisL9241.46 PheL9353.37 TyrL9470.08 ProL951.97 HisL969.68 PheH3211.20 GlyH3327.12 TyrH5240.92 AspH5416.52 AspH5613.98 ArgH5820.19 ArgH9528.46 ArgH966.51 GlyH972.32 ProH9832.99 ProH100e16.69 IleH100f13.42 AlaH100g20.79 ArgH100h69.44 ValH100k26.49 AsnH100l4.03 Total634.74
Acknowledgments
We thank H. Alston for assistance with manuscript preparation; D. Burton for b12 antibody; R. Cardoso and I. Wilson for sharing unpublished observations; D. Davies and J. Scott for insightful comments; M. Gawinowicz for the sequencing of 2F5; B. Hartman, T. Miller, and K. Stroud for graphical assistance; C. Huang and T. Zhou for help with the structural analysis; S. Majeed for technical assistance; G. Nabel for suggestions and the CMV/R vector; M. Roederer for fluorescent antibody-labeling reagents and for assistance with flow cytometry; M. Zwick for discussions, comments on the manuscript, and allowing us to cite unpublished observations; the NIH AIDS Research and Reference Reagent Program for JRFL virus and codon-optimized DNA; and members of SER-CAT for help with data collection.
Use of SER-CAT at the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. W-31-109-Eng-38.
REFERENCES
- 1.Babcock, G. J., T. Mirzabekov, W. Wojtowicz, and J. Sodroski. 2001. Ligand binding characteristics of CXCR4 incorporated into paramagnetic proteoliposomes. J. Biol. Chem. 276:38433-38440. [DOI] [PubMed] [Google Scholar]
- 2.Barbato, G., E. Bianchi, P. Ingallinella, W. H. Hurni, M. D. Miller, G. Ciliberto, R. Cortese, R. Bazzo, J. W. Shiver, and A. Pessi. 2003. Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion. J. Mol. Biol. 330:1101-1115. [DOI] [PubMed] [Google Scholar]
- 3.Biron, Z., S. Khare, A. O. Samson, Y. Hayek, F. Naider, and J. Anglister. 2002. A monomeric 3(10)-helix is formed in water by a 13-residue peptide representing the neutralizing determinant of HIV-1 on gp41. Biochemistry 41:12687-12696. [DOI] [PubMed] [Google Scholar]
- 4.Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54:905-921. [DOI] [PubMed] [Google Scholar]
- 5.Bryson, S., A. Cunningham, J. Ho, R. C. Hynes, D. E. Isenman, B. H. Barber, R. Kunert, H. Katinger, M. Klein, and E. F. Pai. 2001. Cross-neutralizing human monoclonal anti-HIV-1 antibody 2F5: preparation and crystallographic analysis of the free and epitope-complexed forms of its Fab′ fragment. Protein Peptide Lett. 8:413-418. [Google Scholar]
- 6.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, A. Jungbauer, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses 10:359-369. [DOI] [PubMed] [Google Scholar]
- 7.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 8.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 9.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 10.Coeffier, E., J. M. Clement, V. Cussac, N. Khodaei-Boorane, M. Jehanno, M. Rojas, A. Dridi, M. Latour, R. El Habib, F. Barre-Sinoussi, M. Hofnung, and C. Leclerc. 2000. Antigenicity and immunogenicity of the HIV-1 gp41 epitope ELDKWA inserted into permissive sites of the MalE protein. Vaccine 19:684-693. [DOI] [PubMed] [Google Scholar]
- 11.Collaborative Computational Project. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50:760-763. [DOI] [PubMed] [Google Scholar]
- 12.Collis, A. V., A. P. Brouwer, and A. C. Martin. 2003. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J. Mol. Biol. 325:337-354. [DOI] [PubMed] [Google Scholar]
- 13.Conley, A. J., J. A. Kessler II, L. J. Boots, J. S. Tung, B. A. Arnold, P. M. Keller, A. R. Shaw, and E. A. Emini. 1994. Neutralization of divergent human immunodeficiency virus type 1 variants and primary isolates by IAM-41-2F5, an anti-gp41 human monoclonal antibody. Proc. Natl. Acad. Sci. USA 91:3348-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connolly, M. L. 1983. Analytical molecular-surface calculation. J. Appl. Crystallogr. 16:548-558. [Google Scholar]
- 15.Davies, D. R., and G. H. Cohen. 1996. Interactions of protein antigens with antibodies. Proc. Natl. Acad. Sci. USA 93:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Rosny, E., R. Vassell, S. Jiang, R. Kunert, and C. D. Weiss. 2004. Binding of the 2F5 monoclonal antibody to native and fusion-intermediate forms of human immunodeficiency virus type 1 gp41: implications for fusion-inducing conformational changes. J. Virol. 78:2627-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckhart, L., W. Raffelsberger, B. Ferko, A. Klima, M. Purtscher, H. Katinger, and F. Ruker. 1996. Immunogenic presentation of a conserved gp41 epitope of human immunodeficiency virus type 1 on recombinant surface antigen of hepatitis B virus. J. Gen. Virol. 77:2001-2008. [DOI] [PubMed] [Google Scholar]
- 18.Ernst, W., R. Grabherr, D. Wegner, N. Borth, A. Grassauer, and H. Katinger. 1998. Baculovirus surface display: construction and screening of a eukaryotic epitope library. Nucleic Acids Res. 26:1718-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finnegan, C. M., W. Berg, G. K. Lewis, and A. L. DeVico. 2002. Antigenic properties of the human immunodeficiency virus transmembrane glycoprotein during cell-cell fusion. J. Virol. 76:12123-12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furuta, R. A., C. T. Wild, Y. Weng, and C. D. Weiss. 1998. Capture of an early fusion-active conformation of HIV-1 gp41. Nat. Struct. Biol. 5:276-279. [DOI] [PubMed] [Google Scholar]
- 21.Grundner, C., T. Mirzabekov, J. Sodroski, and R. Wyatt. 2002. Solid-phase proteoliposomes containing human immunodeficiency virus envelope glycoproteins. J. Virol. 76:3511-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho, J., K. S. MacDonald, and B. H. Barber. 2002. Construction of recombinant targeting immunogens incorporating an HIV-1 neutralizing epitope into sites of differing conformational constraint. Vaccine 20:1169-1180. [DOI] [PubMed] [Google Scholar]
- 23.Huang, C. C., M. Venturi, S. Majeed, M. J. Moore, S. Phogat, M.-Y. Zhang, D. S. Dimitrov, W. A. Hendrickson, J. Robinson, J. Sodroski, R. Wyatt, H. Choe, M. Farzan, and P. D. Kwong. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV-1 coreceptor binding site on gp120. Proc. Natl. Acad. Sci. USA 101:2706-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, T. A., J. Y. Zou, S. W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47:110-119. [DOI] [PubMed] [Google Scholar]
- 25.Kuiken, C. L., B. Foley, B. Hahn, B. Korber, P. A. Marx, F. McCutchan, J. W. Mellors, and S. Wolinksy. 2001. HIV sequence compendium. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 26.Kunert, R., F. Ruker, and H. Katinger. 1998. Molecular characterization of five neutralizing anti-HIV type 1 antibodies: identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res. Hum. Retroviruses 14:1115-1128. [DOI] [PubMed] [Google Scholar]
- 27.Kunert, R., W. Steinfellner, M. Purtscher, A. Assadian, and H. Katinger. 2000. Stable recombinant expression of the anti HIV-1 monoclonal antibody 2F5 after IgG3/IgG1 subclass switch in CHO cells. Biotechnol. Bioeng. 67:97-103. [DOI] [PubMed] [Google Scholar]
- 28.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 29.Kwong, P. D., and L. Yee. 1999. Use of cryoprotectants in combination with immiscible oils for flash cooling macromolecular crystals. J. Appl. Crystallogr. 32:102-105. [Google Scholar]
- 30.Labrijn, A. F., P. Poignard, A. Raja, M. B. Zwick, K. Delgado, M. Franti, J. Binley, V. Vivona, C. Grundner, C. C. Huang, M. Venturi, C. J. Petropoulos, T. Wrin, D. S. Dimitrov, J. Robinson, P. D. Kwong, R. T. Wyatt, J. Sodroski, and D. R. Burton. 2003. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. J. Virol. 77:10557-10565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski, R. A., M. W. MacArthur, D. S. Moss, and J. M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283-291. [Google Scholar]
- 32.Liang, X., S. Munshi, J. Shendure, G. Mark III, M. E. Davies, D. C. Freed, D. C. Montefiori, and J. W. Shiver. 1999. Epitope insertion into variable loops of HIV-1 gp120 as a potential means to improve immunogenicity of viral envelope protein. Vaccine 17:2862-2872. [DOI] [PubMed] [Google Scholar]
- 33.Liao, M., Y. Lu, Y. Xiao, M. P. Dierich, and Y. Chen. 2000. Induction of high level of specific antibody response to the neutralizing epitope ELDKWA on HIV-1 gp41 by peptide-vaccine. Peptides 21:463-468. [DOI] [PubMed] [Google Scholar]
- 34.Mascola, J. R., M. K. Louder, C. Winter, R. Prabhakara, S. C. De Rosa, D. C. Douek, B. J. Hill, D. Gabuzda, and M. Roederer. 2002. Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. J. Virol. 76:4810-4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald, I. K., and J. M. Thornton. 1994. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238:777-793. [DOI] [PubMed] [Google Scholar]
- 36.McGaughey, G. B., M. Citron, R. C. Danzeisen, R. M. Freidinger, V. M. Garsky, W. M. Hurni, J. G. Joyce, X. Liang, M. Miller, J. Shiver, and M. J. Bogusky. 2003. HIV-1 vaccine development: constrained peptide immunogens show improved binding to the anti-HIV-1 gp41 MAb. Biochemistry 42:3214-3223. [DOI] [PubMed] [Google Scholar]
- 37.McRee, D. E. 1999. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 125:156-165. [DOI] [PubMed] [Google Scholar]
- 38.Merritt, E. A., and D. J. Bacon. 1997. Macromolecular crystallography. Methods Enzymol. 277 :505-524. [DOI] [PubMed]
- 39.Munoz-Barroso, I., K. Salzwedel, E. Hunter, and R. Blumenthal. 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 73:6089-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muster, T., R. Guinea, A. Trkola, M. Purtscher, A. Klima, F. Steindl, P. Palese, and H. Katinger. 1994. Cross-neutralizing activity against divergent human immunodeficiency virus type 1 isolates induced by the gp41 sequence ELDKWAS. J. Virol. 68:4031-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicholls, A., K. A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 43.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 44.Pai, E. F., M. H. Klein, P. Chong, and A. Pedyczak. April 2000. Fab′-epitope complex from the HIV-1 cross-neutralizing monoclonal antibody 2F5. World Intellectual Property Organization patent WO-00/61618.
- 45.Pantophlet, R., I. A. Wilson, and D. R. Burton. 2003. Hyperglycosylated mutants of human immunodeficiency virus (HIV) type 1 monomeric gp120 as novel antigens for HIV vaccine design. J. Virol. 77:5889-5901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker, C. E., L. J. Deterding, C. Hager-Braun, J. M. Binley, N. Schulke, H. Katinger, J. P. Moore, and K. B. Tomer. 2001. Fine definition of the epitope on the gp41 glycoprotein of human immunodeficiency virus type 1 for the neutralizing monoclonal antibody 2F5. J. Virol. 75:10906-10911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parren, P. W., H. J. Ditzel, R. J. Gulizia, J. M. Binley, C. F. Barbas III, D. R. Burton, and D. E. Mosier. 1995. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS 9:F1-F6. [DOI] [PubMed] [Google Scholar]
- 48.Purtscher, M., A. Trkola, A. Grassauer, P. M. Schulz, A. Klima, S. Dopper, G. Gruber, A. Buchacher, T. Muster, and H. Katinger. 1996. Restricted antigenic variability of the epitope recognized by the neutralizing gp41 antibody 2F5. AIDS 10:587-593. [DOI] [PubMed] [Google Scholar]
- 49.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, A. Jungbauer, et al. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 50.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. USA 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas, 3rd, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roederer, M., S. De Rosa, R. Gerstein, M. Anderson, M. Bigos, R. Stovel, T. Nozaki, D. Parks, and L. Herzenberg. 1997. 8 color, 10-parameter flow cytometry to elucidate complex leukocyte heterogeneity. Cytometry 29:328-339. [DOI] [PubMed] [Google Scholar]
- 53.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IGG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 55.Sattentau, Q. J., S. Zolla-Pazner, and P. Poignard. 1995. Epitope exposure on functional, oligomeric HIV-1 gp41 molecules. Virology 206:713-717. [DOI] [PubMed] [Google Scholar]
- 56.Schibli, D. J., R. C. Montelaro, and H. J. Vogel. 2001. The membrane- proximal tryptophan-rich region of the HIV glycoprotein, gp41, forms a well-defined helix in dodecylphosphocholine micelles. Biochemistry 40:9570-9578. [DOI] [PubMed] [Google Scholar]
- 57.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retroviruses 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 58.Stura, E., and I. Wilson. 1991. Applications of the streak seeding technique in protein crystallization. J. Crystal Growth 110:270-282. [Google Scholar]
- 59.Tian, Y., C. V. Ramesh, X. Ma, S. Naqvi, T. Patel, T. Cenizal, M. Tiscione, K. Diaz, T. Crea, E. Arnold, G. F. Arnold, and J. W. Taylor. 2002. Structure-affinity relationships in the gp41 ELDKWA epitope for the HIV-1 neutralizing monoclonal antibody 2F5: effects of side-chain and backbone modifications and conformational constraints. J. Peptide Res. 59:264-276. [DOI] [PubMed] [Google Scholar]
- 60.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas III, D. R. Burton, D. D. Ho, et al. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 63.Weiss, R. A., P. R. Clapham, R. Cheingsong-Popov, A. G. Dalgleish, C. A. Carne, I. V. Weller, and R. S. Tedder. 1985. Neutralization of human T-lymphotropic virus type III by sera of AIDS and AIDS-risk patients. Nature 316:69-72. [DOI] [PubMed] [Google Scholar]
- 64.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 65.Xiao, Y., Y. Zhao, Y. Lu, and Y. H. Chen. 2000. Epitope-vaccine induces high levels of ELDKWA-epitope-specific neutralizing antibody. Immunol. Investig. 29:41-50. [DOI] [PubMed] [Google Scholar]
- 66.Yang, Z. Y., Y. Huang, L. Ganesh, K. Leung, W. P. Kong, O. Schwartz, K. Subbarao, and G. J. Nabel. 2004. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 78:5642-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zwick, M. B., H. K. Komori, R. L. Stanfield, S. Church, M. Wang, P. W. Parren, R. Kunert, H. Katinger, I. A. Wilson, and D. R. Burton. 2004. The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5. J. Virol. 78:3155-3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]