Abstract
Human immunodeficiency virus type 1 (HIV-1) subtype C is responsible for more than 55% of HIV-1 infections worldwide. When this subtype first emerged is unknown. We have analyzed all available gag (p17 and p24) and env (C2-V3) subtype C sequences with known sampling dates, which ranged from 1983 to 2000. The majority of these sequences come from the Karonga District in Malawi and include some of the earliest known subtype C sequences. Linear regression analyses of sequence divergence estimates (with four different approaches) were plotted against sample year to estimate the year in which there was zero divergence from the reconstructed ancestral sequence. Here we suggest that the most recent common ancestor of subtype C appeared in the mid- to late 1960s. Sensitivity analyses, by which possible biases due to oversampling from one district were explored, gave very similar estimates.
The diversity of human immunodeficiency virus type 1 (HIV-1) is expanding globally. Currently, there are nine recognised subtypes of HIV-1, 14 circulating recombinant forms, and many divergent strains (28, 29). While subtype B predominates in North America and Europe (23), it is estimated that more than 55% of worldwide HIV-1 infections are caused by HIV-1 subtype C (6). This high prevalence is due to the predominance of subtype C in southern and eastern Africa (1, 4, 21, 27, 38) and India (32) and its increasing prevalence in Brazil (33) and China (30).
It is not clear whether the predominance of subtype C is a reflection of a founder effect or whether it reflects a relatively high fitness of this virus subtype (22). Subtype C viruses display certain unique genetic characteristics that, by altering biological activity, may have contributed to the success of this subtype (19, 20). Examples include the presence of three or four NF-κB enhancer copies in the long terminal repeat region (23), Tat and Rev proteins that are prematurely truncated, and a 15-bp insertion of the 5′ end of the vpu reading frame (9). Subtype C also shows a preference for CCR5 coreceptor usage, and the variable V3 loop is relatively conserved in subtype C compared to subtype B (23, 32). Recent evidence suggests the emergence of CXCR4-utilizing subtype C viruses in South Africa (39) and a rapidly growing subtype C epidemic in southern Brazil (33). Understanding the origin and subsequent diversification (the extent and rate of change) of this subtype is vitally important in efforts to design prevention and control therapies such as vaccines.
Estimating the date of the most recent common ancestor of viral strains, including dating the origin of HIV-1 and HIV-2, has proven to be possible with molecular data for well-characterized sequences with known sampling dates (12, 13, 31, 37). Thus, the HIV-1 pandemic is thought to have originated in the 1930s from a cross-species transmission of simian immunodeficiency virus (SIV) from chimpanzees (12, 31). Subsequently, this founder strain diversified into the various subtypes that we see worldwide today.
The most recent common ancestor of subtype B is thought to have appeared between 1960 and 1976 (12, 16, 31). Abebe et al. (2, 3) estimated that subtype C was introduced into Ethiopia in the early 1980s (1980 to 1984), and it was first recorded in Karonga District, northern Malawi, in 1983. The earliest subtype C DNA sequences available from other parts of Africa are from the mid- to late 1980s in South Africa (36, 42), Zambia (9, 18, 34), Somalia (9, 14, 15, 34), Tanzania (43), Gabon (5), and Rwanda (11). To date there have been no published attempts to estimate the year in which the most recent common ancestor of this subtype may have appeared. Population-based studies in Karonga District from the early 1980s to the present day (8, 17, 24, 25; Glynn et al., XIth Int. Conf. AIDS STDs Africa, 1999) have yielded HIV-1 subtype C gag and env gene sequence fragments with known sampling dates ranging from 1983 to 2000 from a large number of HIV-1-positive individuals. With these sequences along with all other available subtype C sequences with known sampling dates from the HIV-1 sequence database (http://hiv-web.lanl.gov) and a number of different phylogenetic approaches, we estimated the date of the most recent common ancestor of subtype C and of the epidemic in Karonga District, Malawi.
MATERIALS AND METHODS
Sequence information.
Non-intersubtype-recombinant subtype C sequences spanning approximately 620 bp of the gag p17-p24 region and approximately 420 bp of the env C2-V3 region with known sampling dates were available from molecular epidemiological studies of HIV-1 in the Karonga District, Malawi (17). Only one member of each linked spouse pair was included. All available sequences covering the same gene regions with known sampling dates were retrieved from the Los Alamos HIV database (http://hiv-web.lanl.gov), excluding recombinants and sequences from multiple clones. Alignments were produced that contained only subtype C sequences from the Karonga District, Malawi, and that contained all available subtype C sequences for each gene region (Table 1).
TABLE 1.
Data sets used in the studya
| Data set | Description | Length (bp) | Size (no. of taxa) | Years represented |
|---|---|---|---|---|
| Entire gag | All subtype C gag sequences with known sampling date and with desired fragment | 660 | 376 | 1983-1984, 1986-2000 |
| Karonga gag | All Karonga region subtype C gag sequences (including only one from each linked spouse pair) | 618 | 208 | 1983-1984, 1987-1989, 1997-2000 |
| Representative entire gag | Selected from entire gag to give a balanced representation of countries for each sampling year (as far as possible) | 639 | 59 | 1983-1984, 1986-2000 |
| Malawi clade gag | Clade observed in Karonga subtype C phylogeny by McCormack et al. (17) | 618 | 73 | 1983-1984, 1987-1989, 1997-2000 |
| Entire env | All subtype C env sequences with known sampling date and with desired fragment | 435 | 299 | 1984, 1986-2000 |
| Karonga env | All Karonga region subtype C env sequences (including only one from each linked spouse pair) | 417 | 125 | 1984, 1987-1989, 1997-2000 |
| Representative entire env | Selected from entire env set to give a balanced representation of countries for each sampling year (as far as possible) | 435 | 59 | 1984, 1986-2000 |
gag sequences used are fragments of the p17-p24 region, and env fragment are from the C2-V3 region.
Ancestral sequence and phylogeny reconstruction.
The most recent common ancestor of the subtype C clade and of the subtype C epidemic in the Karonga District, Malawi, were reconstructed for both gene regions. Reference sequences from each HIV-1 group M subtype were obtained from the Los Alamos HIV database and used to root the subtype C phylogeny in eight separate ancestral sequence reconstructions with MrBayes 2 (10). A consensus sequence of the eight resulting ancestral sequences was taken as the most recent common ancestor and used as the outgroup in subsequent phylogeny reconstructions. Modeltest (26) was used to select the optimal model of substitution for each data set, and model parameters were further optimized in PAUP* (35). Phylogeny reconstruction was carried out with both distance and Bayesian methods. With distance methods, a heuristic search strategy was used, with starting trees obtained by random stepwise addition of taxa (10 replicates) and branch swapping, as implemented in PAUP* (35). Bayesian phylogeny reconstruction was performed with MrBayes (10) for 4 million generations, sampling every 100 generations, with the final tree being constructed from generations 2.5 to 4 million to ensure settling of the likelihood values. All tree files and alignments are available from the authors on request.
Dating methods.
Four independent estimates of the genetic divergence of individual sequences from the subtype C ancestral sequence were calculated from each data set: synonymous distances (dS), calculated with the codeml program from the PAML package (41); genetic distances (with the optimal substitution model) calculated with PAUP* (35); branch lengths from individual sequences to the ancestral node on phylogenies reconstructed from distance data (optimized with maximum likelihood and the appropriate substitution model in PAUP* (35) and extracted from the tree file with p4 (3; P. Foster, www.nhm.ac.uk/zoology/external/p4.htm); and branch lengths from individual sequences to the ancestral node of the Bayesian phylogeny.
Correlation between the above divergence values and the sampling year were examined with linear regression analysis. The year at which the regression line crossed the x axis was taken as an indication of the year of origin, i.e., when sequence divergence from the ancestral sequence was zero. Both individual sequence values and mean values per year were plotted against sampling year for each data set. For each analysis, the significance of the correlation was calculated with one-tailed t tests, and 95% confidence intervals for the year in which the most recent common ancestor occurred were calculated (confidence interval for the x intercept) with the Minitab statistical software.
Method validation.
The most recent common ancestor of a large clade within the gag subtype C phylogeny, restricted primarily to sequences originating in Malawi and possibly introduced by a single individual shortly before 1983 (17), was reconstructed, and its date of origin was estimated in one approach to test the ability of our methods to produce a sensible date for an ancestral sequence. To ensure that tree space was well searched, 500 heuristic searches, from a random starting point with total branch reconstruction branch swapping, were carried out on both Karonga env and gag sequence alignments. Branch lengths for each tree were plotted against sampling date in linear regression analyses as described above. Further, 500 bootstrap replicates were performed on the same datasets in PAUP (35), and maximum-likelihood optimized branch lengths of each replicate were similarly plotted.
All sequences available for analysis dating from 1983 and 1984 had been collected in Karonga District, and to investigate their possible effect on the estimated date of the most recent common ancestor, alignments were produced that had all sequences dating from 1983 and 1984 removed and that had every possible combination of these sequences included for both gene regions. For each alignment, maximum-likelihood optimized branch lengths of heuristic trees were plotted against sampling date. Finally, to attempt to account for possible bias due to the disproportionate number of sequences from Karonga District present in the entire subtype C alignments (55.4 and 42.6% of sequences in the entire gag and env data sets, respectively), representative data sets containing a more balanced geographic representation of sequences were assembled and analyzed for each gene region.
RESULTS
Origin of the subtype C epidemic.
When all available gag p17-p24 subtype C sequences were included for analysis with each sequence contributing equally, linear regression analysis indicated a date between 1966 and 1969 for the timing of the origin of subtype C; the 95% confidence intervals ranged from 1959 to 1973 (entire gag, Table 2). There was a significant correlation between genetic divergence and time (P < 0.005), consistent with progressive evolution over time, i.e., clocklike behavior. Linear regression analysis of genetic distances and tree branch length data from the env alignments indicated a date between 1962 and 1965 (1955 to 1972) (entire env, Table 2) for the time at zero divergence, while the dS values suggested 1972 (95% confidence interval, 1966 to 1976). All correlations were significant (P < 0.005). Estimates produced by allowing each year to contribute equally to the analysis (plotting through the mean value for each year) were less consistent (data not shown).
TABLE 2.
Estimated date of origin of subtype C and of the subtype C epidemic in the Karonga District, Malawi, from gag and env gene sequences
| Gene | Methoda | Date of originb (95% confidence interval)
|
||
|---|---|---|---|---|
| Entire data set | Karonga data set | Representative entire data set | ||
| gag | dS | 1967 (1960-1970) | 1960 (1948-1966) | 1968 (1958-1975) |
| Genetic distance | 1968 (1963-1972) | 1968 (1962-1973) | 1970 (1961-1976) | |
| ML Opt. BLens | 1966 (1959-1970) | 1966 (1960-1971) | 1971 (1961-1976) | |
| MrBayes BLens | 1969 (1965-1973) | 1969 (1964-1973) | 1974 (1967-1978) | |
| env | dS | 1972 (1966-1976) | 1975 (1969-1979) | 1971 (1946-1978) |
| Genetic distance | 1962 (1956-1968) | 1966 (1961-1972) | 1950 (1875-1960) | |
| ML Opt. BLens | 1964 (1955-1971) | 1967 (1961-1972) | 1953 (NA) (P < 0.025) | |
| MrBayes BLens | 1965 (1958-1972) | 1973 (1968-1977) | 1947 (NA) (P < 0.05) | |
dS, synonymous distances; ML Opt Blens, maximum-likelihood optimized heuristic distance tree branch lengths; MrBayes BLens, branch lengths obtained from phylogeny inferred with MrBayes. NA: confidence intervals incalculable
Data significant at P < 0.005 except as noted. Four estimates of genetic divergence of individual sequences to the subtype C ancestral sequence were used: (i) synonymous distances, dS (codeml program, PAML package); (ii) genetic distances with the optimal substitution model (PAUP*); (iii) branch lengths from individual sequences to the ancestral node on phylogenies reconstructed from distance data (optimized by maximum likelihood and the appropriate substitution model in PAUP* and extracted from the tree file with p4); and (iv) branch lengths from individual sequences to the ancestral node of the Bayesian phylogeny reconstructed with MrBayes.
Origin of the subtype C epidemic in the Karonga District, Malawi.
Linear regression analysis of all gag sequences from Karonga indicated a date between 1966 and 1969 for the ancestor of the Karonga subtype C epidemic when genetic distances and branch length data were used (95% confidence interval, 1960 to 1973, Karonga gag; Table 2). However, linear regression analysis of dS values calculated from the Karonga gag sequences indicated 1960 to 1961 as a likely date for the ancestor of the Karonga epidemic (P < 0.005; confidence interval, 1948 to 1966). Analysis of the Karonga env data set indicated that the origin of the epidemic was dated at 1966 to 1973 with genetic distance and branch length data and 1975 for dS data (Karonga env, Table 2).
Method validation.
The regression analysis carried out on sequences belonging to a Malawi clade evident within the subtype C phylogeny indicated 1979 to 1981 as the date of the most recent common ancestor of this clade. Maximum-likelihood optimized branch lengths of 500 heuristic trees from both Karonga gag and env alignments plotted against the sampling year of each sequence estimated the likely range for the date of the most recent common ancestor of the Karonga epidemic (Fig. 1) as either 1964 to 1972 (gag) or 1956 to 1970 (env), with the 500 bootstrap replicate estimates being 1964 to 1974 and 1954 to 1974 for gag and env, respectively.
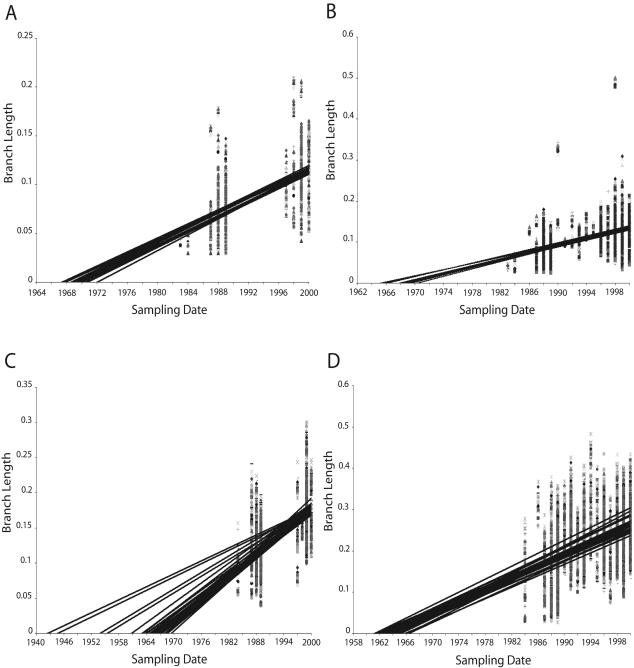
FIG. 1.
Sensitivity plots for Karonga gag (A), entire gag (B), Karonga env (C), and entire env (D). Alignments that had all sequences dating from 1983 and 1984 removed and that had every possible combination of these sequences included were produced. For each alignment, a distance-based heuristic strategy was used (starting trees obtained by random stepwise addition of taxa, 10 replicates) and branch swapping as implemented in PAUP* (35). Branch lengths from individual sequences to the reconstructed ancestor were optimized with PAUP* (35), extracted from the tree file with p4 (8), and plotted against the sampling date.
The influence of the early subtype C sequences collected in Karonga District in 1983 and 1984 on the estimated dates of the appearance of the most recent common ancestor of the subtype C epidemic itself and of the subtype C epidemic in Karonga is shown in Fig. 1. Regression analysis of branch lengths from heuristic trees drawn from gag sequences suggest a date between 1967 and 1972 for the Karonga epidemic (Fig. 1A) and 1965 to 1970 for the entire subtype C epidemic (Fig. 1B), while similar analysis on the env sequences suggested 1942 to 1970 for the Karonga epidemic (Fig. 1C) and 1961 to 1967 for the entire subtype C epidemic (Fig. 1D). Estimated dates of the subtype C most recent common ancestor calculated from the representative alignments ranged from 1968 to 1974 (95% confidence interval, 1958 to 1978) for gag and 1947 to 1971 (95% confidence interval, 1875 to 1978) for env (see Table 2).
DISCUSSION
In one of the most intensive studies of its kind, we have estimated the date of origin of the most prevalent subtype of HIV-1, subtype C. Our work suggests that the most recent common ancestor of subtype C appeared in the late 1960s and that the most recent common ancestor of the subtype C epidemic in Karonga District, Malawi, also dates back to this period. This is consistent with the origin of HIV-1 group M (1930s) (12, 31) and the first evidence of subtype C (1983) in Malawi (17) as well as with estimates of the origins of subtype B (some time between 1960 and 1976) (12, 16, 31). A number of sensitivity analyses were used to account for known biases in the data, and these supported the main result.
We applied our methods to the estimation of the most recent common ancestor of a well-described (17) clade within the subtype C phylogeny with gag sequences. Regression analysis of four divergence estimates versus time suggested that the ancestor of this clade appeared between 1979 and 1981. This date is consistent with our previous conclusion, from molecular and epidemiological evidence, that a single individual may have been responsible for the introduction of this cluster shortly before 1983 (17). The good agreement between the methods and the plausible date produced give us a certain degree of confidence in the ability of these methods to estimate the most recent common ancestor of the larger data sets. For the most part, the correlation between genetic divergence and time was significant, consistent with clocklike behavior. Furthermore, estimates of the date of the most recent common ancestor resulting from plotting 500 heuristic trees were consistent with dates produced with the four different methods of estimating divergence (1964 to 1972 for gag, 1956 to 1970 for env), as were estimates from plotting 500 bootstrap trees (1964 to 1974 for gag, 1954 to 1974 for env).
Individual divergent sequences had a large effect when each year contributed equally to the analysis (rather than each individual sequence contributing equally), and we therefore believe that ancestral dates estimated from regression analyses of mean values per sampling year are less accurate than those where all sequences contribute equally. Our ability to estimate the date of the most recent subtype C common ancestor did not rely on the presence of the small number of sequences dating from the early 1980s. However, the estimated date of the most recent common ancestor of the Karonga epidemic with env sequences was severely affected when some of the early sequences were removed (Fig. 1C). Dates estimated from the gag representative alignment were pushed slightly forward towards the present, while estimates with env sequences were pushed further back into the past. Only 59 sequences were used in each alignment, and the smaller numbers widened the confidence intervals and reduced the significance of each correlation. The large numbers of available sequences from the Karonga data set, which are spread over a number of sampling years, contributed hugely to our ability to estimate the year of the most recent subtype C common ancestor. However, the rate of dS substitutions over time appeared to be slower overall for the Karonga gag data set than for other subtype C sequences.
In general, estimates of the date of origin of the most recent subtype C common ancestor were more consistent between methods used for the gag than for the env alignments. The dates estimated with env ranged from 1947 to 1975, whereas the dates ranged from 1966 to 1974 for gag. Estimates derived from dS data (expected to appear clock-like) ranged from 1967 to 1969 for gag and 1970 to 1975 for env. The later dates inferred with env can be explained by the higher rate of substitution in env (tests for substitution saturation with DAMBE [40] indicated no substitution saturation within the env data). The shorter fragment size used for env (420 bp) compared to gag (620 bp) may also have contributed to this.
Attempts to estimate the date of origin of the most recent common ancestor of the different subtypes have been hampered by a lack of early sequences, lack of enough sequences with known sampling dates, and lack of sequences representing a range of sampling years. Population-based studies in Karonga District, Malawi, have provided a large number of subtype C sequences ranging in date between 1983 and 2000 which have largely overcome these problems for estimating the date of origin of this subtype. While full gene or genome sequences were not available, and while we accept that the large number of Karonga sequences may have biased the reconstruction of the subtype C ancestral sequence as well as the date of the most recent subtype C common ancestor, our data indicate that the most recent common ancestor of subtype C probably originated in the 1960s. The gag data (which we believe is the more reliable) suggest a date between 1966 and 1969 (95% confidence interval, 1959 to 1973). The Karonga epidemic is very diverse and may be a good model for the subtype C epidemic.
Acknowledgments
Many thanks to Peter Foster for assistance in the use of p4 and to Marco Salemi for critical reading of the manuscript. Thanks also to the two independent reviewers for helpful comments and suggestions.
Until 1996, the Karonga Prevention Study was funded primarily by LEPRA (The British Leprosy Relief Association) and ILEP (The International Federation of Anti-Leprosy Organizations) with contributions from the WHO/UNDP/World Bank Special Programme for Research and Training in Tropical Diseases. Since 1996, the Wellcome Trust has been the principal funder. J.R.G. is funded by the United Kingdom Department of Health (Public Health Career Scientist award).
Permission for the study was received from the National Health Sciences Research Committee, Malawi, and the Ethics Committee of the London School of Hygiene & Tropical Medicine, United Kingdom.
REFERENCES
- 1.Abebe, A. C. L. Kuiken, J. Goudsmit, M. Valk, T. Messels, et al. 1997. HIV type 1 subtype C in Addis Ababa, Ethiopia. AIDS Res. Hum. Retroviruses 13:1071-1075. [DOI] [PubMed] [Google Scholar]
- 2.Abebe A., V. V. Lukashov, G. Pollakis, A Kliphuis, A. L. Fontanet, J. Goudsemit, and T. F. de Wit. 2001. Timing of the HIV-1 subtype C epidemic in Ethiopia based on early virus strains and subsequent virus distribution. AIDS 15:1555-1561. [DOI] [PubMed] [Google Scholar]
- 3.Abebe, A., V. V. Lukashovm T. F. Ribske De Wit, B. Fisscha, B. Tegboro, et al. 2001. Timing of the introduction into Ethiopia of subcluster C′ of HIV type 1 subtype C. AIDS Res. Hum. Retroviruses 17:657-661. [DOI] [PubMed] [Google Scholar]
- 4.Abebe, A., G. Pollakis, A. L. Fonanet, B. Fisscha, B. Tegbaro, A. Kliphuts, et al. 2000. Identification of a genetic subcluster of HIV subtype 1 C (C′) widespread in Ethiopia. AIDS Res. Hum. Retroviruses 16:1909-1914. [DOI] [PubMed] [Google Scholar]
- 5.Delaporte, E., W. Janssesns, M. Peeters, A. Buve, G. Dibanga, J. L. Perret, V. Ditsambou, et al. 1996. Epidemiological and molecular characterization of HIV infection in Gabon, 1986-1994. AIDS 10:903-1010. [DOI] [PubMed] [Google Scholar]
- 6.Esparza, J., and N. Bhamarapravati. 2000. Accelerating the development and future availability pf HIV-1 vaccines: why, when, where and how? Lancet 355:2061-2066. [DOI] [PubMed] [Google Scholar]
- 7.Gao, F., D. L. Robertson, D. C. Carruthers, S. G. Morrison, B. Jian, V. Chen, et al. 1998. A ciomprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glynn, J. R., J. P. Clewly, B. Ngwira, S. Malema, D. K. Warndorff, A. C. Crampin, and P. E. M. Fine. 2001. The development of the HIV epidemic in Karonga District, Malawi. AIDS 15:2025-2029. [DOI] [PubMed] [Google Scholar]
- 9.Huang, D. D., T. A. Glesler, and J. W. Bremer. 2003. Sequence characterization of the protease and partial reverse transcriptase proteins of the NED panel, and international HIV type 1 subtype reference and standards panel. AIDS Res. Hum. Retroviruses 19:321-328. [DOI] [PubMed] [Google Scholar]
- 10.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Baynesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 11.Kampinga, G. A., A. Simonon, P. Van de Perre, E. Karita, P. Msellati, and J. Goudsmit. 1997. Primary infections with HIV-1 of women and their offspring in Rwanda: findings of heterogeneity at seroconversion, coinfection, and recombinants of HIV-1 subtypes A and C. Virology 227:63-76. [DOI] [PubMed] [Google Scholar]
- 12.Korber, B., M. Muldoon, J. Theiler, F. Gao, R. Gupta, A. Lapedes, B. H. Hahn, S. Wolinsky, and T. Bhattacharya. 2000. Timing the ancestor of the HIV-1 pandemic strains. Science 288:1789-1796. [DOI] [PubMed] [Google Scholar]
- 13.Lemey, P., O. G. Pybus, B. Wang, N. K. Saksena, M. Salemi, and A. M. Vandamme. 2003. Tracing the origin and history of the HIV-2 epidemic. Proc. Natl. Acad. Sci. USA 100:6588-6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louwagie, J., W. Janssens, J. Mascola, L. Heyndrickx, P. Hegerich, G. van der Groen, F. E. McCutchan, and D. S. Burke. 1995. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J. Virol. 69:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Louwagie, J., F. E. McCutchan, M. Peeters, T. P. Brennan, E. Sanders-Buell, G. A. Eddy, G. van der Groen, K. Fransen, G. M. Gershy-Damet, R. Deleys, et al. 1993. Phylogenetic analysis of gag genes from 70 international HIV-1 isolates provides evidence for multiple genotypes. AIDS 7:769-780. [DOI] [PubMed] [Google Scholar]
- 16.Lukashov, V. V., and J. Goudsmit. 2002. Recent evolutionary history of human immunodeficiency virus type 1 subtype B: reconstruction of epidemic onset based on sequence distances to the common ancestor. J. Mol. Evol. 54:680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCormack, G. P., J. R. Glynn, A. C. Crampin, F. Sibande, D. Mulawa, L. Bliss, P. Broadbent, K. Abarca, J. M. Ponnighaus, P. E. Fine, and J. P. Clewley. 2002. Early evolution of the human immunodeficiency virus type 1 subtype C epidemic in rural Malawi. J. Virol. 76:12890-12899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCutchan, F. E., B. L. Ungar, P. Hegerich, C. R. Roberts, A. K. Fowler, S. K. Hira, P. L. Perine, and D. S. Burke. 1992. Genetic analysis of HIV-1 isolates from Zambia and an expanded phylogenetic tree for HIV-1. J. Acquir. Immune Defic. Syndr. 5:441-449. [PubMed] [Google Scholar]
- 19.Ndung'u, T., B. Renjifo, and M. Essex. 2001. Construction and analysis of an infectious human Immunodeficiency virus type 1 subtype C molecular clone. J. Virol. 75:4964-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ndung'u, T., B. Renjifo, V. A. Novitsky, M. F. McLane, S. Gaolekwe, and M. Essex. 2000. Molecular cloning and biological characterization of full-length HIV-1 subtype C from Botswana. Virology 278:390-399. [DOI] [PubMed] [Google Scholar]
- 21.Novitsky, V. A., M. A. Montano, M. F. McLane, B. Renjifo, F. Vannberg, B. T. Foley, T. P. Ndung'u, M. Rahman, M. J. Makhema, R. Marlink, and M. Essex. 1999. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J. Virol. 73:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oelrichs, R. B., I. L. Shrestha, D. A. Anderson, and N. J. Deacon. 2000. The explosive human immunodeficiency virus type 1 epidemic among injecting drug users of Kathmandu, Nepal, is caused by a subtype C virus of restricted genetic diversity. J. Virol. 74:1149-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peeters, M., and P. M. Sharp. 2000. Genetic diversity of HIV-1: the moving target. AIDS 14(Suppl. 3):S129-S140. [PubMed] [Google Scholar]
- 24.Ponnighaus, J. M., P. E. Fine, L. Bliss, P. J. Gruer, B. Kapira-Mwamondwe, E. Msosa, R. J. Rees, D. Clayton, M. C. Pike, J. A. Sterne, et al. 1993. The Karonga Prevention Trial: a leprosy and tuberculosis vaccine trial in northern Malawi. I. Methods of the vaccination phase. Lepr. Rev. 64:338-356. [DOI] [PubMed] [Google Scholar]
- 25.Ponninghaus, J. M., P. E. Fine, L. Bliss, I. J. Sliney, D. J. Bradley, and R. J. Rees. 1987. The Lepra Evaluation Project (LEP), an epidemiological study of leprosy in Northern Malawi. I. Methods. Lepr. Rev. 58:359-375. [DOI] [PubMed] [Google Scholar]
- 26.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 27.Renjifo, B., B. Chaplin, D. Mwakagile, P. Shah, F. Vannberg, G. Msamanga, D. Hunter, W. Fawzi, and M. Essex. 1998. Epidemic expansion of HIV type 1 subtype C and recombinant genotypes in Tanzania. AIDS Res. Hum. Retroviruses 14:635-638. [DOI] [PubMed] [Google Scholar]
- 28.Robbins, K. E., P. Lemey, O. G. Pybus, H. W. Jaffe, A. S. Youngpairoj, T. M. Brown, M. Salemi, A. M. Vandamme, and M. L. Kalish. 2003. U.S. human immunodeficiency virus type 1 epidemic: date of origin, population history, and characterization of early strains. J. Virol. 77:6359-6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson, D. L., J. P. Anderson, J. A. Bradac, J. K. Carr, B. Foley, R. K. Funkhouser, F. Gao, B. H. Hahn, M. L. Kalish, C. Kuiken, G. H. Learn, T. Leitner, F. McCutchan, S. Osmanov, M. Peeters, D. Pieniazek, M. Salminen, P. M. Sharp, S. Wolinsky, and B. Korber. 2000. HIV-1 nomenclature proposal. Science 288:55-56. [DOI] [PubMed] [Google Scholar]
- 30.Rodenburg, C. M., Y. Li, S. A. Trask, Y. Chen, J. Decker, D. L. Robertson, M. L. Kalish, G. M. Shaw, S. Allen, B. H. Hahn, and F. Gao. 2001. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res. Hum. Retroviruses 17:161-168. [DOI] [PubMed] [Google Scholar]
- 31.Salemi, M., K. Strimmer, W. W. Hall, M. Duffy, E. Delaporte, S. Mboup, M. Peeters, and A. M. Vandamme. 2001. Dating the common ancestor of SIVcpz and HIV-1 group M and the origin of HIV-1 subtypes using a new method to uncover clock-like molecular evolution. FASEB J. 15:276-278. [DOI] [PubMed] [Google Scholar]
- 32.Shankarappa, R., R. Chatterjee, G. H. Learn, D. Neogi, M. Ding, P. Roy, A. Ghosh, L. Kingsley, L. Harrison, J. I. Mullins, and P. Gupta. 2001. Human immunodeficiency virus type 1 env sequences from Calcutta in eastern India: identification of features that distinguish subtype C sequences in India from other subtype C sequences. J. Virol. 75:10479-10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soares, M. A., T. De Oliveira, R. M. Brindeiro, R. S. Diaz, E. C. Sabino, L. Brigido, I. L. Pires, M. G. Morgado, M. C. Dantas, D. Barreira, P. R. Teixeira, S. Cassol, and A. Tanuri. 2003. A specific subtype C of human immunodeficiency virus type 1 circulates in Brazil. AIDS 17:11-21. [DOI] [PubMed] [Google Scholar]
- 34.Swanson, P., S. G. Devare, and J. Hackett, Jr. 2003. Molecular characterization of 39 HIV isolates representing group M (subtypes A-G) and group O: sequence analysis of gag p24, pol integrase, and env gp41. AIDS Res. Hum. Retroviruses 19:625-629. [DOI] [PubMed] [Google Scholar]
- 35.Swofford, D. L. 1998. PAUP*: phylogenetic analysis using parsimny (*and other methods), 4th ed. Sinauer Associates, Sunderland, Mass.
- 36.Treurnicht, F. K., T. L. Smith, S. Engelbrecht, M. Claassen, B. A. Robson, M. Zeier, and E. J. van Rensburg. 2002. Genotypic and phenotypic analysis of the env gene from South African HIV-1 subtype B and C isolates. J. Med. Virol. 68:141-146. [DOI] [PubMed] [Google Scholar]
- 37.Van Dooren, S., M. Salemi, and A. M. Vandamme. 2001. Dating the origin of the African human T-cell lymphotropic virus type-i (HTLV-I) subtypes. Mol. Biol. Evol. 18:661-671. [DOI] [PubMed] [Google Scholar]
- 38.Van Harmelen, J. H., E. Van der Ryst, A. S. Loubser, D. York, S. Madurai, S. Lyons, R. Wood, and C. Williamson. 1999. A predominantly HIV type 1 subtype C-restricted epidemic in South African urban populations. AIDS Res. Hum. Retroviruses 15:395-398. [DOI] [PubMed] [Google Scholar]
- 39.van Rensburg, E. J., T. L. Smith, M. Zeier, B. Robson, C. Sampson, F. Treurnicht, and S. Engelbrecht. 2002. Change in co-receptor usage of current South African HIV-1 subtype C primary isolates. AIDS 16:2479-2480. [DOI] [PubMed] [Google Scholar]
- 40.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]
- 41.Yang, Z. 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13:555-556. [DOI] [PubMed] [Google Scholar]
- 42.Zacharova, V., M. L. Becker, V. Zachar, P. Ebbesen, and A. S. Goustin. 1997. DNA sequence analysis of the long terminal repeat of the C subtype of human immunodeficiency virus type 1 from Southern Africa reveals a dichotomy between B subtype and African subtypes on the basis of upstream NF-IL-6 motif. AIDS Res. Hum. Retroviruses 13:719-724. [DOI] [PubMed] [Google Scholar]
- 43.Zwart, G., T. F. Wolfs, R. Bookelman, S. Hartman, M. Bakker, C. A. Boucher, C. Kuiken, and J. Goudsmit. 1993. Greater diversity of the HIV-1 V3 neutralization domain in Tanzania compared with the Netherlands: serological and genetic analysis. AIDS 7:467-474. [DOI] [PubMed] [Google Scholar]